Enhanced Expression of Human Endogenous Retroviruses, TRIM28 and SETDB1 in Autism Spectrum Disorder
Abstract
:1. Introduction
2. Results
2.1. Study Populations
2.2. Influence of Age on Expression of HERVs, TRIM28, and SETDB1
2.3. Expression Levels of the Housekeeping Gene
2.4. Expression Levels of the Pol Genes of HERV-H, HERV-K, and HERV-W
2.5. Expression Levels of Env Genes of SYN1, SYN2, and MSRV
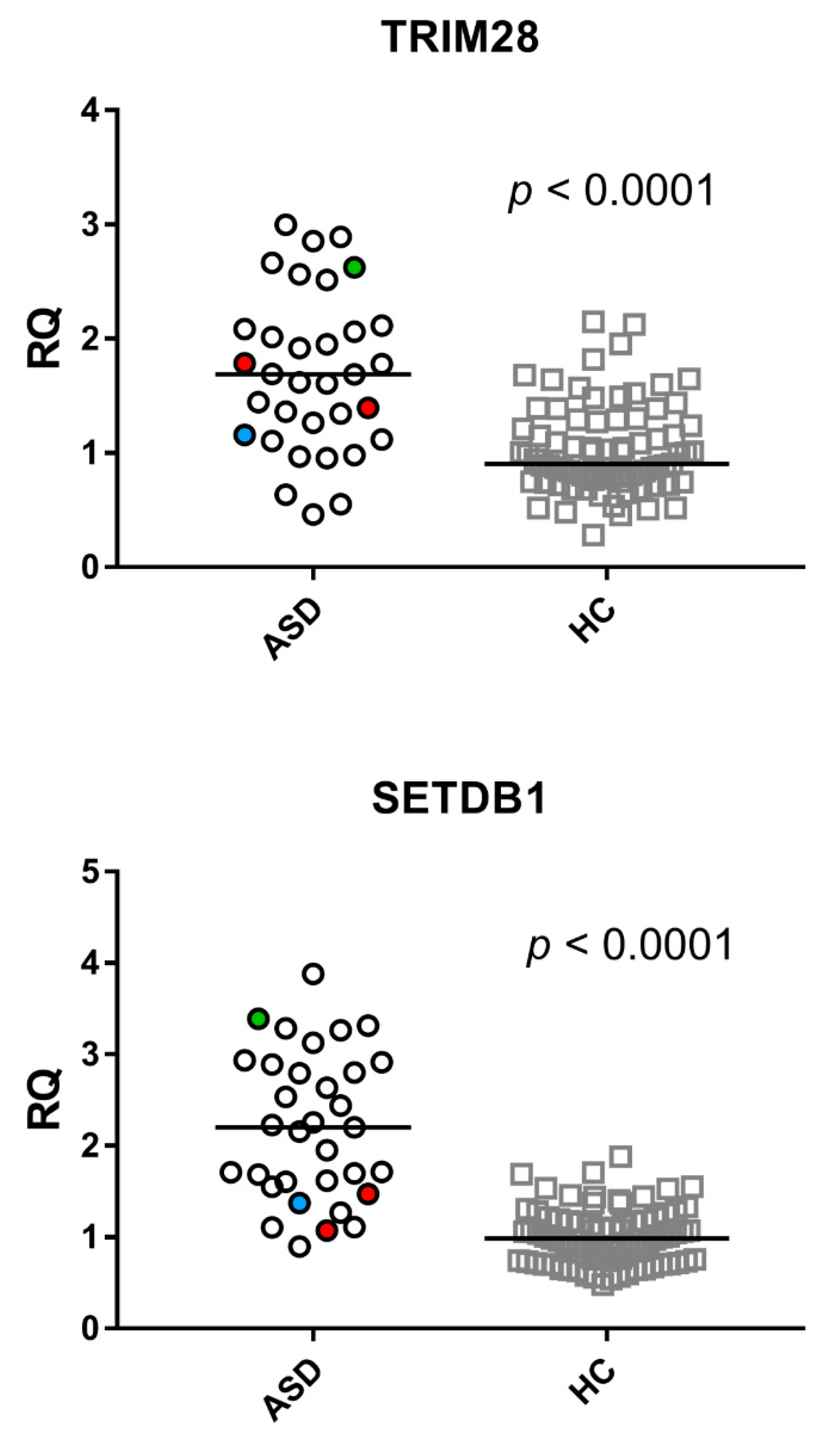
2.6. Expressions of TRIM28 and SETDB1
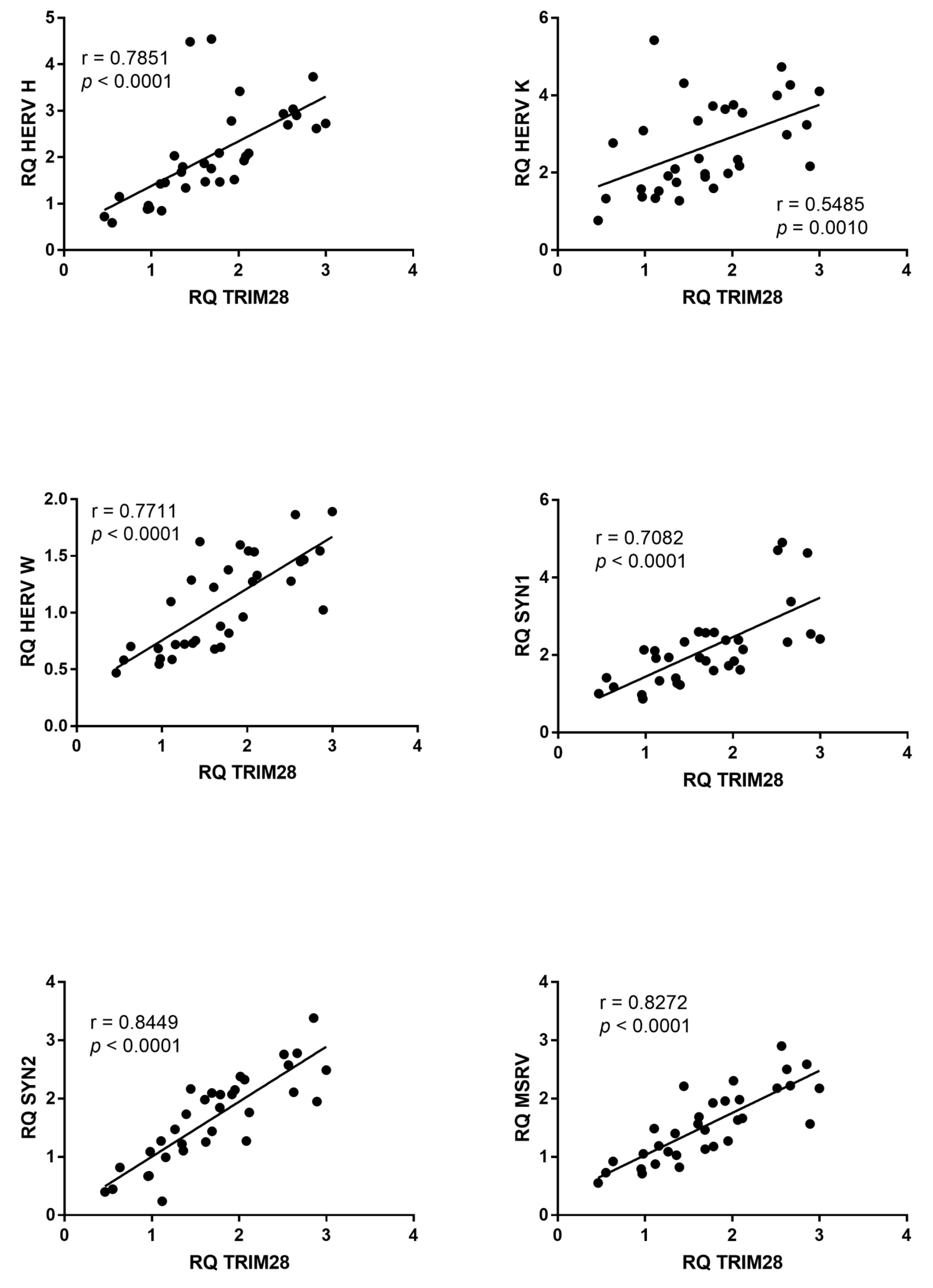
2.7. Correlations between Expressions of HERVs and TRIM28 or SETDB1
3. Discussion
4. Materials and Methods
4.1. Study Populations
4.2. Total RNA Extraction
4.3. Reverse Transcription
4.4. Transcription Levels of Pol Genes of HERV-H, -K, and -W, of Env Genes of SYN1, SYN2, and MSRV As Well As of TRIM28 and SETDB1 by Real-Time PCR Assay
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ASD | autism spectrum disorder |
| DC | dendritic cell |
| HERVs | human endogenous retroviruses |
| KRAB-ZFPs | Krüppel-associated box domain zinc finger proteins |
| NF-kB | nuclear factor kB |
| MSRV | multiple sclerosis retrovirus |
| PBMCs | peripheral blood mononuclear cells |
| PRR | pattern recognition receptor |
| SETDB1 | SET domain bifurcated histone lysine methyltrasferase 1 |
| SYN1 | syncytin 1 |
| SYN2 | syncytin 2 |
| TRIM28 | tripartite motif containing 28 |
| TLR | toll-like receptor |
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Arlington, VA, USA, 2013. [Google Scholar]
- Leblond, C.S.; Le, T.L.; Malesys, S.; Cliquet, F.; Tabet, A.C.; Delorme, R.; Rolland, T.; Bourgeron, T. Operative list of genes associated with autism and neurodevelopmental disorders based on database review. Mol. Cell. Neurosci. 2021, 113, 103623. [Google Scholar] [CrossRef]
- Tick, B.; Bolton, P.; Happé, F.; Rutter, M.; Rijsdijk, F. Heritability of autism spectrum disorders: A meta-analysis of twin studies. J. Child Psychol. Psychiatry 2016, 57, 585–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bölte, S.; Girdler, S.; Marschik, P.B. The contribution of environmental exposure to the etiology of autism spectrum disorder. Cell. Mol. Life Sci. 2019, 76, 1275–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thapar, A.; Rutter, M. Genetic advances in autism. J. Autism Dev. Disord. 2021, 51, 4321–4332. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Smith, S.E.; Malkova, N.; Tse, D.; Su, Y.; Patterson, P.H. Activation of the maternal immune system alters cerebellar development in the offspring. Brain Behav. Immun. 2009, 23, 116–123. [Google Scholar] [CrossRef] [Green Version]
- Malkova, N.V.; Yu, C.Z.; Hsiao, E.Y.; Moore, M.J.; Patterson, P.H. Maternal immune activation yields offspring displaying mouse versions of the three core symptoms of autism. Brain Behav. Immun. 2012, 26, 607–616. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, E.Y.; McBride, S.W.; Chow, J.; Mazmanian, S.K.; Patterson, P.H. Modeling an autism risk factor in mice leads to permanent immune dysregulation. Proc. Natl. Acad. Sci. USA 2012, 109, 12776–12781. [Google Scholar] [CrossRef] [Green Version]
- Robinson-Agramonte, M.d.L.A.; Noris García, E.; Fraga Guerra, J.; Vega Hurtado, Y.; Antonucci, N.; Semprún-Hernández, N.; Schultz, S.; Siniscalco, D. Immune dysregulation in autism spectrum disorder: What do we know about It? Intern. J. Mol. Sci. 2022, 23, 3033. [Google Scholar] [CrossRef]
- Goines, P.E.; Croen, L.A.; Braunschweig, D.; Yoshida, C.K.; Grether, J.; Hansen, R.; Kharrazi, M.; Ashwood, P.; Van de Water, J. Increased midgestational IFN-γ, IL-4 and IL-5 in women bearing a child with autism: A case-control study. Mol. Autism 2011, 2, 13. [Google Scholar] [CrossRef] [Green Version]
- Abdallah, M.W.; Larsen, N.; Grove, J.; Nørgaard-Pedersen, B.; Thorsen, P.; Mortensen, E.L.; Hougaard, D.M. Amniotic fluid inflammatory cytokines: Potential markers of immunologic dysfunction in autism spectrum disorders. World J. Biol. Psychiatry 2013, 14, 528–538. [Google Scholar] [CrossRef]
- Zawadzka, A.; Cieślik, M.; Adamczyk, A. The role of maternal immune activation in the pathogenesis of autism: A review of the evidence, proposed mechanisms and implications for treatment. Int. J. Mol. Sci. 2021, 22, 11516. [Google Scholar] [CrossRef]
- Lu, J.; Wang, Z.; Liang, Y.; Yao, P. Rethinking autism: The impact of maternal risk factors on autism development. Am. J. Transl. Res. 2022, 14, 1136–1145. [Google Scholar]
- Masi, A.; Glozier, N.; Dale, R.; Guastella, A.J. The immune system, cytokines, and biomarkers in autism spectrum disorder. Neurosci. Bull. 2017, 33, 194–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balestrieri, E.; Cipriani, C.; Matteucci, C.; Benvenuto, A.; Coniglio, A.; Argaw-Denboba, A.; Toschi, N.; Bucci, I.; Miele, M.T.; Grelli, S.; et al. Children with autism spectrum disorder and their mothers share abnormal expression of selected endogenous retroviruses families and cytokines. Front. Immunol. 2019, 10, 2244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, W.E. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 2019, 17, 355–370. [Google Scholar] [CrossRef] [PubMed]
- Blond, J.L.; Lavillette, D.; Cheynet, V.; Bouton, O.; Oriol, G.; Chapel-Fernandes, S.; Mandrand, B.; Mallet, F.; Cosset, F.L. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 2000, 74, 3321–3329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaise, S.; de Parseval, N.; Benit, L.; Heidmann, T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 13013–13018. [Google Scholar] [CrossRef] [Green Version]
- Mangeney, M.; Renard, M.; Schlecht-Louf, G.; Bouallaga, I.; Heidmann, O.; Letzelter, C.; Richaud, A.; Ducos, B.; Heidmann, T. Placental syncytins: Genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 20534–20539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holder, B.S.; Tower, C.L.; Forbes, K.; Mulla, M.J.; Aplin, J.D.; Abrahams, V.M. Immune cell activation by trophoblast-derived microvesicles is mediated by syncytin 1. Immunology 2012, 136, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Lokossou, A.G.; Toudic, C.; Nguyen, P.T.; Elisseeff, X.; Vargas, A.; Rassart, É.; Lafond, J.; Leduc, L.; Bourgault, S.; Gilbert, C.; et al. Endogenous retrovirus-encoded Syncytin-2 contributes to exosome-mediated immunosuppression of T cells. Biol. Reprod. 2020, 102, 185–198. [Google Scholar] [CrossRef]
- Garcia-Montojo, M.; Rodriguez-Martin, E.; Ramos-Mozo, P.; Ortega-Madueño, I.; Dominguez-Mozo, M.I.; Arias-Leal, A.; García-Martínez, M.Á.; Casanova, I.; Galan, V.; Arroyo, R.; et al. Syncytin-1/HERV-W envelope is an early activation marker of leukocytes and is upregulated in multiple sclerosis patients. Eur. J. Immunol. 2020, 50, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Isbel, L.; Whitelaw, E. Endogenous retroviruses in mammals: An emerging picture of how ERVs modify expression of adjacent genes. Bioessays 2012, 34, 734–738. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 2016, 351, 1083–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Wang, L.; Liu, H.; Chen, J.; Liu, D. Transcriptome analyses implicate endogenous retroviruses involved in the host antiviral immune system through the interferon pathway. Virol. Sin. 2021, 36, 1315–1326. [Google Scholar] [CrossRef] [PubMed]
- Rolland, A.; Jouvin-Marche, E.; Viret, C.; Faure, M.; Perron, H.; Marche, P.N. The envelope protein of a human endogenous retrovirus-W family activates innate immunity through CD14/TLR4 and promotes Th1-like responses. J. Immunol. 2006, 176, 7636–7644. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Ahmad, S.; Hur, S. Endogenous retroelements and the host innate immune sensors. Adv. Immunol. 2016, 132, 47–69. [Google Scholar] [CrossRef] [Green Version]
- Mameli, G.; Astone, V.; Arru, G.; Marconi, S.; Lovato, L.; Serra, C.; Sotgiu, S.; Bonetti, B.; Dolei, A. Brains and peripheral blood mononuclear cells of multiple sclerosis (MS) patients hyperexpress MS-associated retrovirus/HERV-W endogenous retrovirus, but not Human herpesvirus 6. J. Gen. Virol. 2007, 88, 264–274. [Google Scholar] [CrossRef]
- Perron, H.; Dougier-Reynaud, H.L.; Lomparski, C.; Popa, I.; Firouzi, R.; Bertrand, J.B.; Marusic, S.; Portoukalian, J.; Jouvin-Marche, E.; Villiers, C.L.; et al. Human endogenous retrovirus protein activates innate immunity and promotes experimental allergic encephalomyelitis in mice. PLoS ONE 2013, 8, e80128. [Google Scholar] [CrossRef]
- Madeira, A.; Burgelin, I.; Perron, H.; Curtin, F.; Lang, A.B.; Faucard, R. MSRV envelope protein is a potent, endogenous and pathogenic agonist of human toll-like receptor 4: Relevance of GNbAC1 in multiple sclerosis treatment. J. Neuroimmunol. 2016, 291, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Hummel, J.; Kämmerer, U.; Müller, N.; Avota, E.; Schneider-Schaulies, S. Human endogenous retrovirus envelope proteins target dendritic cells to suppress T-cell activation. Eur. J. Immunol. 2015, 45, 1748–1759. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Z.; Wang, P.; Li, S.; Zeng, J.; Tu, X.; Yan, Q.; Xiao, Z.; Pan, M.; Zhu, F. Syncytin-1, an endogenous retroviral protein, triggers the activation of CRP via TLR3 signal cascade in glial cells. Brain Behav. Immun. 2018, 67, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Yu, P. The potential role of retroviruses in autoimmunity. Immunol. Rev. 2016, 269, 85–99. [Google Scholar] [CrossRef]
- Grandi, N.; Tramontano, E. Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front. Immunol. 2018, 9, 2039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tovo, P.A.; Rabbone, I.; Tinti, D.; Galliano, I.; Trada, M.; Daprà, V.; Cerutti, F.; Bergallo, M. Enhanced expression of human endogenous retroviruses in new-onset type 1 diabetes: Potential pathogenetic and therapeutic implications. Autoimmunity 2020, 53, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Tovo, P.A.; Opramolla, A.; Pizzol, A.; Calosso, G.; Daprà, V.; Galliano, I.; Calvi, C.; Pinon, M.; Cisarò, F.; Rigazio, C.; et al. Overexpression of endogenous retroviruses in children with celiac disease. Eur. J. Pediatr. 2021, 180, 2429–2434. [Google Scholar] [CrossRef]
- Huang, W.J.; Liu, Z.C.; Wei, W.; Wang, G.H.; Wu, J.G.; Zhu, F. Human endogenous retroviral pol RNA and protein detected and identified in the blood of individuals with schizophrenia. Schizophr. Res. 2006, 83, 193–199. [Google Scholar] [CrossRef]
- Küry, P.; Nath, A.; Créange, A.; Dolei, A.; Marche, P.; Gold, J.; Giovannoni, G.; Hartung, H.P.; Perron, H. Human endogenous retroviruses in neurological diseases. Trends Mol. Med. 2018, 24, 379–394. [Google Scholar] [CrossRef] [Green Version]
- Johansson, E.M.; Bouchet, D.; Tamouza, R.; Ellul, P.; Morr, A.S.; Avignone, E.; Germi, R.; Leboyer, M.; Perron, H.; Groc, L. Human endogenous retroviral protein triggers deficit in glutamate synapse maturation and behaviors associated with psychosis. Sci. Adv. 2020, 6, eabc0708. [Google Scholar] [CrossRef]
- Sobocińska, J.; Molenda, S.; Machnik, M.; Oleksiewicz, U. KRAB-ZFP transcriptional regulators acting as oncogenes and tumor suppressors: An Overview. Int. J. Mol. Sci. 2021, 23, 2212. [Google Scholar] [CrossRef]
- Friedman, J.R.; Fredericks, W.J.; Jensen, D.E.; Speicher, D.W.; Huang, X.P.; Neilson, E.G.; Rauscher, F.J., 3rd. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996, 10, 2067–2078. [Google Scholar] [CrossRef] [Green Version]
- Schultz, D.C.; Ayyanathan, K.; Negorev, D.; Maul, G.G.; Rauscher, F.J., 3rd. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002, 16, 919–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodge, J.E.; Kang, Y.-K.; Beppu, H.; Lei, H.; Li, E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. Biol. 2004, 24, 2478–2486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turelli, P.; Castro-Diaz, N.; Marzetta, F.; Kapopoulou, A.; Raclot, C.; Duc, J.; Tieng, V.; Quenneville, S.; Trono, D. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res. 2014, 24, 1260–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Liu, Y.; Lu, H.; Sun, S.C.; Jin, W.; Wang, X.; Dong, C. Epigenetic activation during T helper 17 cell differentiation is mediated by Tripartite motif containing 28. Nat. Commun. 2018, 12, 1424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gehrmann, U.; Carpier, J.M.; Burgdorf, N.; Hoyler, T.; Suarez, G.; Joannas, L.; Heurtebise-Chrétien, S.; Durand, S.; Panes, R.; Bellemare-Pelletier, A.; et al. Critical role for TRIM28 and HP1β/γ in the epigenetic control of T cell metabolic reprograming and effector differentiation. Proc. Natl. Acad. Sci. USA 2019, 116, 25839–25849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czerwinska, P.; Jaworska, A.M.; Wlodarczyk, N.A.; Mackiewicz, A.A. Melanoma stem cell-like phenotype and significant suppression of immune response within a tumor are regulated by TRIM28 protein. Cancers 2020, 12, 2998. [Google Scholar] [CrossRef]
- Fasching, L.; Kapopoulou, A.; Sachdeva, R.; Petri, R.; Jönsson, M.E.; Männe, C.; Turelli, P.; Jern, P.; Cammas, F.; Trono, D.; et al. TRIM28 represses transcription of endogenous retroviruses in neural progenitor cells. Cell. Rep. 2015, 10, 20–28. [Google Scholar] [CrossRef]
- Kawabe, H.; Stegmüller, J. The role of E3 ubiquitin ligases in synapse function in the healthy and diseased brain. Mol. Cell. Neurosci. 2021, 112, 103602. [Google Scholar] [CrossRef]
- Ibi, D.; González-Maeso, J. Epigenetic signaling in schizophrenia. Cell. Signal. 2015, 27, 2131–2136. [Google Scholar] [CrossRef] [Green Version]
- Markouli, M.; Strepkos, D.; Chlamydas, S.; Piperi, C. Histone lysine methyltransferase SETDB1 as a novel target for central nervous system diseases. Prog. Neurobiol. 2021, 200, 101968. [Google Scholar] [CrossRef]
- Iwase, S.; Martin, D.M. Chromatin in nervous system development and disease. Mol. Cell. Neurosci. 2018, 87, 1–3. [Google Scholar] [CrossRef]
- Yoon, S.H.; Choi, J.; Lee, W.J.; Do, J.T. Genetic and epigenetic etiology underlying autism spectrum disorder. J. Clin. Med. 2020, 9, 966. [Google Scholar] [CrossRef] [Green Version]
- Ladd-Acosta, C.; Fallin, M.D.; Kaufmann, W.E.; Feinberg, A.P. Common DNA methylation alterations in multiple brain regions in autism. Mol. Psychiatry 2014, 19, 862–871. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S.V.; Ellis, S.E.; Bakulski, K.M.; Sheppard, B.; Croen, L.A.; Hertz-Picciotto, I.; Newschaffer, C.J.; Feinberg, A.P.; Arking, D.E.; Ladd-Acosta, C.; et al. Cross-tissue integration of genetic and epigenetic data offers insight into autism spectrum disorder. Nat. Commun. 2017, 8, 1011. [Google Scholar] [CrossRef] [Green Version]
- Balestrieri, E.; Arpino, C.; Matteucci, C.; Sorrentino, R.; Pica, F.; Alessandrelli, R.; Coniglio, A.; Curatolo, P.; Rezza, G.; Macciardi, F.; et al. HERVs expression in autism spectrum disorders. PLoS ONE 2012, 7, e48831. [Google Scholar] [CrossRef] [Green Version]
- Rowe, H.M.; Kapopoulou, A.; Corsinotti, A.; Fasching, L.; Macfarlan, T.S.; Tarabay, Y.; Viville, S.; Jakobsson, J.; Pfaff, S.L.; Trono, D. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013, 23, 452–461. [Google Scholar] [CrossRef] [Green Version]
- Manghera, M.; Ferguson-Parry, J.; Lin, R.; Douville, R.N. NF-kB and IRF1 induce endogenous retrovirus K expression via interferon-stimulated response elements in its 5′ long terminal repeat. J. Virol. 2016, 90, 9338–9349. [Google Scholar] [CrossRef] [Green Version]
- Patterson, P.H. Immune involvement in schizophrenia and autism: Etiology, pathology and animal models. Behav. Brain Res. 2009, 204, 313–321. [Google Scholar] [CrossRef]
- Estes, M.L.; McAllister, A.K. Maternal immune activation: Implications for neuropsychiatric disorders. Science 2016, 353, 772–777. [Google Scholar] [CrossRef] [Green Version]
- Shuid, A.N.; Jayusman, P.A.; Shuid, N.; Ismail, J.; Kamal Nor, N.; Mohamed, I.N. Association between viral infections and risk of autistic disorder: An overview. Int. J. Environ. Res. Public Health 2021, 18, 2817. [Google Scholar] [CrossRef]
- Pendyala, G.; Chou, S.; Jung, Y.; Coiro, P.; Spartz, E.; Padmashri, R.; Li, M.; Dunaevsky, A. Maternal immune activation causes behavioral impairments and altered cerebellar cytokine and synaptic protein expression. Neuropsychopharmacology 2017, 42, 1435–1446. [Google Scholar] [CrossRef] [Green Version]
- Assinger, A.; Yaiw, K.C.; Göttesdorfer, I.; Leib-Mösch, C.; Söderberg-Nauclér, C. Human cytomegalovirus (HCMV) induces human endogenous retrovirus (HERV) transcription. Retrovirology 2013, 10, 132. [Google Scholar] [CrossRef] [Green Version]
- Ruprecht, K.; Obojes, K.; Wengel, V.; Gronen, F.; Kim, K.S.; Perron, H.; Schneider-Schaulies, J.; Rieckmann, P. Regulation of human endogenous retrovirus W protein expression by herpes simplex virus type 1: Implications for multiple sclerosis. J. Neurovirol. 2006, 12, 65–71. [Google Scholar] [CrossRef]
- Mameli, G.; Poddighe, L.; Mei, A.; Uleri, E.; Sotgiu, S.; Serra, C.; Manetti, R.; Dolei, A. Expression and activation by Epstein Barr virus of human endogenous retroviruses-W in blood cells and astrocytes: Inference for multiple sclerosis. PLoS ONE 2012, 7, e44991. [Google Scholar] [CrossRef]
- Van der Kuyl, A.C. HIV infection and HERV expression: A review. Retrovirology 2012, 9, 6. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, N.; Domingues, P.; Golebiowski, F.; Patzina, C.; Tatham, M.H.; Hay, R.T.; Hale, B.G. An influenza virus-triggered SUMO switch orchestrates co-opted endogenous retroviruses to stimulate host antiviral immunity. Proc. Natl. Acad. Sci. USA 2019, 116, 17399–17408. [Google Scholar] [CrossRef] [Green Version]
- Tovo, P.A.; Garazzino, S.; Daprà, V.; Alliaudi, C.; Silvestro, E.; Calvi, C.; Montanari, P.; Galliano, I.; Bergallo, M. Chronic HCV infection is associated with overexpression of human endogenous retroviruses that persists after drug-induced viral clearance. Int. J. Mol. Sci. 2020, 21, 3980. [Google Scholar] [CrossRef]
- Tovo, P.A.; Garazzino, S.; Daprà, V.; Pruccoli, G.; Calvi, C.; Mignone, F.; Alliaudi, C.; Denina, M.; Scolfaro, C.; Zoppo, M.; et al. COVID-19 in children: Expressions of Type I/II/III interferons, TRIM28, SETDB1, and endogenous retroviruses in mild and severe cases. Int. J. Mol. Sci. 2021, 22, 7481. [Google Scholar] [CrossRef]
- Bergallo, M.; Canosa, S.; Galliano, I.; Daprà, V.; Montanari, P.; Sestero, M.; Gennarelli, G.; Benedetto, C.; Revelli, A.; Tovo, P.A. Impaired transcription of human endogenous retroviruses in the sperm with exception of syncytin 1: Short communication. Mol. Biol. Rep. 2021, 48, 5803–5808. [Google Scholar] [CrossRef]
- Bjerregaard, B.; Lemmen, J.G.; Petersen, M.R.; Østrup, E.; Iversen, L.H.; Almstrup, K.; Larsson, L.I.; Ziebe, S. Syncytin-1 and its receptor is present in human gametes. J. Assist. Reprod. Genet. 2014, 31, 533–539. [Google Scholar] [CrossRef] [Green Version]
- Grow, E.J.; Flynn, R.A.; Chavez, S.L.; Bayless, N.L.; Wossidlo, M.; Wesche, D.J.; Martin, L.; Ware, C.B.; Blish, C.A.; Chang, H.Y.; et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 2015, 522, 221–225. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Zhu, H.; Song, J.; Jiang, Y.; Ouyang, H.; Huang, R.; Zhang, G.; Fan, X.; Tao, R.; Jiang, J.; et al. Upregulation of leukocytic syncytin-1 in acute myeloid leukemia patients. Med. Sci. Monit. 2016, 22, 2392–2403. [Google Scholar] [CrossRef] [Green Version]
- Tolosa, J.M.; Parsons, K.S.; Hansbro, P.M.; Smith, R.; Wark, P.A. The placental protein syncytin-1 impairs antiviral responses and exaggerates inflammatory responses to influenza. PLoS ONE 2015, 10, e0118629. [Google Scholar] [CrossRef] [Green Version]
- Perron, H.; Lang, A. The human endogenous retrovirus link between genes and environment in multiple sclerosis and in multifactorial diseases associating neuroinflammation. Clin. Rev. Allergy Immunol. 2010, 39, 51–61. [Google Scholar] [CrossRef]
- Bhat, R.K.; Ellestad, K.K.; Wheatley, B.M.; Warren, R.; Holt, R.A.; Power, C. Age- and disease-dependent HERV-W envelope allelic variation in brain: Association with neuroimmune gene expression. PLoS ONE 2011, 6, e19176. [Google Scholar] [CrossRef]
- Wang, X.; Huang, J.; Zhu, F. Human endogenous retroviral envelope protein syncytin-1 and inflammatory abnormalities in neuropsychological diseases. Front. Psychiatry 2018, 9, 422. [Google Scholar] [CrossRef]
- Charvet, B.; Reynaud, J.M.; Gourru-Lesimple, G.; Perron, H.; Marche, P.N.; Horvat, B. Induction of proinflammatory multiple sclerosis-associated retrovirus envelope protein by human Herpesvirus-6A and CD46 receptor engagement. Front. Immunol. 2018, 9, 2803. [Google Scholar] [CrossRef]
- Hartung, H.P.; Derfuss, T.; Cree, B.A.; Sormani, M.P.; Selmaj, K.; Stutters, J.; Prados, F.; MacManus, D.; Schneble, H.M.; Lambert, E.; et al. Efficacy and safety of temelimab in multiple sclerosis: Results of a randomized phase 2b and extension study. Mult. Scler. 2021, 9, 22–440. [Google Scholar] [CrossRef]
- Curtin, F.; Champion, B.; Davoren, P.; Duke, S.; Ekinci, E.I.; Gilfillan, C.; Morbey, C.; Nathow, T.; O’Moore-Sullivan, T.; O’Neal, D.; et al. A safety and pharmacodynamics study of temelimab, an antipathogenic human endogenous retrovirus type W envelope monoclonal antibody, in patients with type 1 diabetes. Diabetes Obes. Metab. 2020, 22, 1111–1121. [Google Scholar] [CrossRef]
- Wu, S.; Ding, Y.; Wu, F.; Li, R.; Xie, G.; Hou, J.; Mao, P. Family history of autoimmune diseases is associated with an increased risk of autism in children: A systematic review and meta-analysis. Neurosci. Biobehav. Rev. 2015, 55, 322–332. [Google Scholar] [CrossRef]
- Chen, S.W.; Zhong, X.S.; Jiang, L.N.; Zheng, X.Y.; Xiong, Y.Q.; Ma, S.J.; Qiu, M.; Huo, S.T.; Ge, J.; Chen, Q. Maternal autoimmune diseases and the risk of autism spectrum disorders in offspring: A systematic review and meta-analysis. Behav. Brain Res. 2016, 296, 61–69. [Google Scholar] [CrossRef]
- Edmiston, E.; Ashwood, P.; Van de Water, J. Autoimmunity, autoantibodies, and autism spectrum disorder. Biol. Psychiatry 2017, 81, 383–390. [Google Scholar] [CrossRef] [Green Version]
- Mazón-Cabrera, R.; Vandormael, P.; Somers, V. Antigenic targets of patient and maternal autoantibodies in autism spectrum disorder. Front. Immunol. 2019, 10, 1474. [Google Scholar] [CrossRef] [Green Version]
- Heidmann, O.; Béguin, A.; Paternina, J.; Berthier, R.; Deloger, M.; Bawa, O.; Heidmann, T. HEMO, an ancestral endogenous retroviral envelope protein shed in the blood of pregnant women and expressed in pluripotent stem cells and tumors. Proc. Natl. Acad. Sci. USA 2017, 114, E6642–E6651. [Google Scholar] [CrossRef] [Green Version]
- Cipriani, C.; Ricceri, L.; Matteucci, C.; De Felice, A.; Tartaglione, A.M.; Argaw-Denboba, A. High expression of endogenous retroviruses from intrauterine life to adulthood in two mouse models of autism spectrum disorders. Sci. Rep. 2018, 8, 629. [Google Scholar] [CrossRef] [Green Version]
- Gropman, A.L.; Batshaw, M.L. Epigenetics, copy number variation, and other molecular mechanisms underlying neurodevelopmental disabilities: New insights and diagnostic approaches. J. Dev. Behav. Pediatr. 2010, 3, 582–591. [Google Scholar] [CrossRef] [Green Version]
- LaSalle, J.M. Epigenomic strategies at the interface of genetic and environmental risk factors for autism. J. Hum. Genet. 2013, 58, 396–401. [Google Scholar] [CrossRef] [Green Version]
- Mouat, J.S.; LaSalle, J.M. The promise of DNA methylation in understanding multigenerational factors in autism spectrum disorders. Front. Genet. 2022, 13, 831221. [Google Scholar] [CrossRef]
- Tangsuwansri, C.; Saeliw, T.; Thongkorn, S.; Chonchaiya, W.; Suphapeetiporn, K.; Mutirangura, A.; Tencomnao, T.; Hu, V.W.; Sarachana, T. Investigation of epigenetic regulatory networks associated with autism spectrum disorder (ASD) by integrated global LINE-1 methylation and gene expression profiling analyses. PLoS ONE 2018, 13, e0201071. [Google Scholar] [CrossRef] [Green Version]
- García-Ortiz, M.V.; Torre-Aguilar, M.J.; Morales-Ruiz, T.; Gómez-Fernández, A.; Flores-Rojas, K.; Gil-Campos, M.; Martin-Borreguero, P.; Ariza Rafael, R.; Roldán-Arjona, T.; Perez-Navero, J.L. Analysis of global and local DNA methylation patterns in blood samples of patients with autism spectrum disorder. Front. Pediatr. 2021, 9, 1066. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Peng, H.; Yurchenko, V.; Yap, K.L.; Negorev, D.G.; Schultz, D.C.; Psulkowski, E.; Fredericks, W.J.; White, D.E.; Maul, G.G.; et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell. 2007, 28, 823–837. [Google Scholar] [CrossRef] [Green Version]
- Liang, Q.; Deng, H.; Li, X.; Wu, X.; Tang, Q.; Chang, T.H.; Peng, H.; Rauscher, F.J., 3rd; Ozato, K.; Zhu, F. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J. Immunol. 2011, 187, 4754–4763. [Google Scholar] [CrossRef] [Green Version]
- Kamitani, S.; Ohbayashi, N.; Ikeda, O.; Togi, S.; Muromoto, R.; Sekine, Y.; Ohta, K.; Ishiyama, H.; Matsuda, T. KAP1 regulates type I interferon/STAT1-mediated IRF-1 gene expression. Biochem. Biophys. Res. Commun. 2008, 370, 366–370. [Google Scholar] [CrossRef] [Green Version]
- Cuellar, T.L.; Herzner, A.M.; Zhang, X.; Goyal, Y.; Watanabe, C.; Friedman, B.A.; Janakiraman, V.; Durinck, S.; Stinson, J.; Arnott, D.; et al. Silencing of retrotransposons by SETDB1 inhibits the interferon response in acute myeloid leukemia. J. Cell. Biol. 2017, 216, 3535–3549. [Google Scholar] [CrossRef] [Green Version]
- Krischuns, T.; Günl, F.; Henschel, L.; Binder, M.; Willemsen, J.; Schloer, S.; Rescher, U.; Gerlt, V.; Zimmer, G.; Nordhof, C.; et al. Phosphorylation of TRIM28 enhances the expression of IFN-β and proinflammatory cytokines during HPAIV infection of human lung epithelial cells. Front. Immunol. 2018, 9, 2229. [Google Scholar] [CrossRef] [Green Version]
- Santoni de Sio, F.R.; Barde, I.; Offner, S.; Kapopoulou, A.; Corsinotti, A.; Bojkowska, K.; Genolet, R.; Thomas, J.H.; Luescher, I.F.; Pinschewer, D.; et al. KAP1 regulates gene networks controlling T-cell development and responsiveness. FASEB J. 2012, 26, 4561–4575. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.F.; Yu, J.; Chang, M.; Zhang, M.; Zhou, D.; Cammas, F.; Sun, S.C. TRIM28 mediates chromatin modifications at the TCRα enhancer and regulates the development of T and natural killer T cells. Proc. Natl. Acad. Sci. USA 2012, 109, 20083–20088. [Google Scholar] [CrossRef] [Green Version]
- Chikuma, S.; Yamanaka, S.; Nakagawa, S.; Ueda, M.T.; Hayabuchi, H.; Tokifuji, Y.; Kanayama, M.; Okamura, T.; Arase, H.; Yoshimura, A. TRIM28 expression on dendritic cells prevents excessive T cell priming by silencing endogenous retrovirus. J. Immunol. 2021, 206, 1528–1539. [Google Scholar] [CrossRef]
- Grassi, D.A.; Jönsson, M.E.; Brattås, P.L.; Jakobsson, J. TRIM28 and the control of transposable elements in the brain. Brain Res. 2019, 1705, 43–47. [Google Scholar] [CrossRef]
- Minkovsky, A.; Sahakyan, A.; Rankin-Gee, E.; Bonora, G.; Patel, S.; Plath, K. The Mbd1-Atf7ip-Setdb1 pathway contributes to the maintenance of X chromosome inactivation. Epigenetics Chromatin 2014, 7, 12. [Google Scholar] [CrossRef] [Green Version]
- Taherian, M.; Maghsoudi, H.; Bidaki, K.; Taherian, R. The relationship between skewed X-chromosome Inactivation and neurological disorders development: A Review. Int. Clin. Neurosci. J. 2016, 23, 81–91. [Google Scholar] [CrossRef]
- Cukier, H.N.; Lee, J.M.; Ma, D.; Young, J.I.; Mayo, V.; Butler, B.L.; Ramsook, S.S.; Rantus, J.A.; Abrams, A.J.; Whitehead, P.L.; et al. The expanding role of MBD genes in autism: Identification of a MECP2 duplication and novel alterations in MBD5, MBD6, and SETDB1. Autism Res. 2015, 5, 385–397. [Google Scholar] [CrossRef] [Green Version]
- Spyropoulou, A.; Gargalionis, A.; Dalagiorgou, G.; Adamopoulos, C.; Papavassiliou, K.A.; Lea, R.W.; Piperi, C.; Papavassiliou, A.G. Role of histone lysine methyltransferases SUV39H1 and SETDB1 in gliomagenesis: Modulation of cell proliferation, migration, and colony formation. Neuromol. Med. 2014, 16, 70–82. [Google Scholar] [CrossRef]
- Xu, Q.; Goldstein, J.; Wang, P.; Gadi, I.K.; Labreche, H.; Rehder, C.; Wang, W.P.; McConkie, A.; Xu, X.; Jiang, Y.H. Chromosomal microarray analysis in clinical evaluation of neurodevelopmental disorders-reporting a novel deletion of SETDB1 and illustration of counseling challenge. Pediatric Res. 2016, 80, 371–381. [Google Scholar] [CrossRef]
- Matsui, T.; Leung, D.; Miyashita, H.; Maksakova, I.A.; Miyachi, H.; Kimura, H.; Tachibana, M.; Lorincz, M.C.; Shinkai, Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 2010, 464, 927–931. [Google Scholar] [CrossRef] [Green Version]
- Wiznerowicz, M.; Jakobsson, J.; Szulc, J.; Liao, S.; Quazzola, A.; Beermann, F.; Aebischer, P.; Trono, D. The Kruppel-associated box repressor domain can trigger de novo promoter methylation during mouse early embryogenesis. J. Biol. Chem. 2007, 282, 34535–34541. [Google Scholar] [CrossRef] [Green Version]
- Bojkowska, K.; Aloisio, F.; Cassano, M.; Kapopoulou, A.; Santoni de Sio, F.; Zangger, N.; Offner, S.; Cartoni, C.; Thomas, C.; Quenneville, S.; et al. Liver-specific ablation of Krüppel-associated box-associated protein 1 in mice leads to male-predominant hepatosteatosis and development of liver adenoma. Hepatology 2012, 56, 1279–1290. [Google Scholar] [CrossRef] [Green Version]
- Diem, O.; Schäffner, M.; Seifarth, W.; Leib-Mösch, C. Influence of antipsychotic drugs on human endogenous retrovirus (HERV) transcription in brain cells. PLoS ONE 2012, 7, e30054. [Google Scholar] [CrossRef] [Green Version]
- Dong, E.; Nelson, M.; Grayson, D.R.; Costa, E.; Guidotti, A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc. Natl. Acad. Sci. USA 2008, 105, 13614–13619. [Google Scholar] [CrossRef] [Green Version]
- Christensen, D.L.; Baio, J.; Van Naarden, B.K.; Bilder, D.; Charles, J.; Costantino, J.N. Centers for Disease Control and Prevention. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States. MMWR Surveill. Summ. 2016, 65, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Zablotsky, B.; Black, L.I.; Maenner, M.J.; Schieve, L.A.; Danielson, M.L.; Bitsko, R.H.; Blumberg, S.J.; Kogan, D.; Boyle, C.A. Prevalence and trends of developmental disabilities among children in the United States: 2009–2017. Pediatrics 2019, 144, e20190811. [Google Scholar] [CrossRef] [PubMed]
- EmbertiGialloreti, L.; Mazzone, L.; Benvenuto, A.; Fasano, A.; Alcon, A.G.; Kraneveld, A.; Moavero, R.; Raz, R.; Riccio, M.P.; Siracusano, M.; et al. Risk and protective environmental factors associated with autism spectrum disorder: Evidence-based principles and recommendations. J. Clin. Med. 2019, 8, 217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frye, R.E.; Cakir, J.; Rose, S.; Palmer, R.F.; Austin, C.; Curtin, P. Physiological mediators of prenatal environmental influences in autism spectrum disorder. Bioessays 2021, 43, e2000307. [Google Scholar] [CrossRef]
- Hertz-Picciotto, I.; Korrick, S.A.; Ladd-Acosta, C.; Karagas, M.R.; Lyall, K.; Schmidt, R.J.; Dunlop, A.L.; Croen, L.A.; Dabelea, D.; Daniels, J.L.; et al. Program collaborators for environmental influences on child health outcomes (ECHO). Maternal tobacco smoking and offspring autism spectrum disorder or traits in ECHO cohorts. Autism Res. 2022, 15, 551–569. [Google Scholar] [CrossRef]
- Azébi, S.; Batsché, E.; Michel, F.; Kornobis, E.; Muchardt, C. Expression of endogenous retroviruses reflects increased usage of atypical enhancers in T cells. EMBO J. 2019, 38, e101107. [Google Scholar] [CrossRef]
- Bergallo, M.; Galliano, I.; Daprà, V.; Pirra, A.; Montanari, P.; Pavan, M.; Calvi, C.; Bertino, E.; Coscia, A.; Tovo, P.A. Transcriptional activity of human endogenous retroviruses in response to prenatal exposure of maternal cigarette smoking. Am. J. Perinatol. 2019, 36, 1060–1065. [Google Scholar] [CrossRef] [PubMed]
- Laderoute, M.P.; Giulivi, A.; Larocque, L.; Bellfoy, D.; Hou, Y.; Wu, H.X.; Fowke, K.; Wu, J.; Diaz-Mitoma, F. The replicative activity of human endogenous retrovirus K102 (HERV-K102) with HIV viremia. AIDS 2007, 21, 2417–2424. [Google Scholar] [CrossRef]
- Bowen, L.N.; Tyagi, R.; Li, W.; Alfahad, T.; Smith, B.; Wright, M.; Singer, E.J.; Nath, A. HIV-associated motor neuron disease: HERV-K activation and response to antiretroviral therapy. Neurology 2016, 87, 1756–1762. [Google Scholar] [CrossRef] [Green Version]
- Tyagi, R.; Li, W.; Parades, D.; Bianchet, M.A.; Nath, A. Inhibition of human endogenous retrovirus-K by antiretroviral drugs. Retrovirology 2017, 14, 21. [Google Scholar] [CrossRef] [Green Version]
- Morandi, E.; Tanasescu, R.; Tarlinton, R.E.; Constantin-Teodosiu, D.; Gran, B. Do antiretroviral drugs protect from multiple sclerosis by inhibiting expression of MS-associated retrovirus? Front. Immunol. 2019, 9, 3092. [Google Scholar] [CrossRef]
- Piccinini, M.; Rinaudo, M.T.; Chiapello, N.; Ricotti, E.; Baldovino, S.; Mostert, M.; Tovo, P.A. The human 26S proteasome is a target of antiretroviral agents. AIDS 2002, 16, 693–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piccinini, M.; Rinaudo, M.T.; Anselmino, A.; Buccinnà, B.; Ramondetti, C.; Dematteis, A.; Ricotti, E.; Palmisano, L.; Mostert, M.; Tovo, P.A. The HIV protease inhibitors nelfinavir and saquinavir, but not a variety of HIV reverse transcriptase inhibitors, adversely affect human proteasome function. Antivir. Ther. 2005, 10, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Rutter, M.; DiLavore, P.C.; Risi, S. Autism Diagnostic Observation Schedule, 2nd ed.; (ADOS-2) manual (Part II): Toddler module; Western Psychological Services: Torrance, CA, USA, 1999. [Google Scholar]
- Schanab, O.; Humer, J.; Gleiss, A.; Mikula, M.; Sturlan, S.; Grunt, S.; Okamoto, I.; Muster, T.; Pehamberger, H.; Waltenberger, A. Expression of human endogenous retrovirus K is stimulated by ultraviolet radiation in melanoma. Pigment Cell. Melanoma Res. 2011, 24, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Mameli, G.; Poddighe, L.; Astone, V.; Delogu, G.; Arru, G.; Sotgiu, S.; Serra, C.; Dolei, A. Novel reliable real-time PCR for differential detection of MSRVenv and syncytin-1 in RNA and DNA from patients with multiple sclerosis. J. Virol. Methods 2009, 161, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Soygur, B.; Sati, L. The role of syncytins in human reproduction and reproductive organ cancers. Reproduction 2016, 152, R167–R178. [Google Scholar] [CrossRef] [Green Version]



| Total sample, n | 33 |
| Males, n (%) | 28 (85) |
| Age, yr, median (IQR) a | 3.8 (3.0–6.0) |
| Autism severity (ADOS-CSS) b, median (IQR) | 7.5 (5.5–8.5) c |
| Intellectual disability, d n (%) | 15 (45) |
| Seizures, n (%) | 3 (9) |
| In treatment with valproic acid (%) | 2 (6) |
| In treatment with other psychotropic medication e (%) | 2 (6) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tovo, P.-A.; Davico, C.; Marcotulli, D.; Vitiello, B.; Daprà, V.; Calvi, C.; Montanari, P.; Carpino, A.; Galliano, I.; Bergallo, M. Enhanced Expression of Human Endogenous Retroviruses, TRIM28 and SETDB1 in Autism Spectrum Disorder. Int. J. Mol. Sci. 2022, 23, 5964. https://doi.org/10.3390/ijms23115964
Tovo P-A, Davico C, Marcotulli D, Vitiello B, Daprà V, Calvi C, Montanari P, Carpino A, Galliano I, Bergallo M. Enhanced Expression of Human Endogenous Retroviruses, TRIM28 and SETDB1 in Autism Spectrum Disorder. International Journal of Molecular Sciences. 2022; 23(11):5964. https://doi.org/10.3390/ijms23115964
Chicago/Turabian StyleTovo, Pier-Angelo, Chiara Davico, Daniele Marcotulli, Benedetto Vitiello, Valentina Daprà, Cristina Calvi, Paola Montanari, Andrea Carpino, Ilaria Galliano, and Massimiliano Bergallo. 2022. "Enhanced Expression of Human Endogenous Retroviruses, TRIM28 and SETDB1 in Autism Spectrum Disorder" International Journal of Molecular Sciences 23, no. 11: 5964. https://doi.org/10.3390/ijms23115964






