Impact of Doxorubicin on Cell-Substrate Topology
Abstract
:1. Introduction
2. Theory
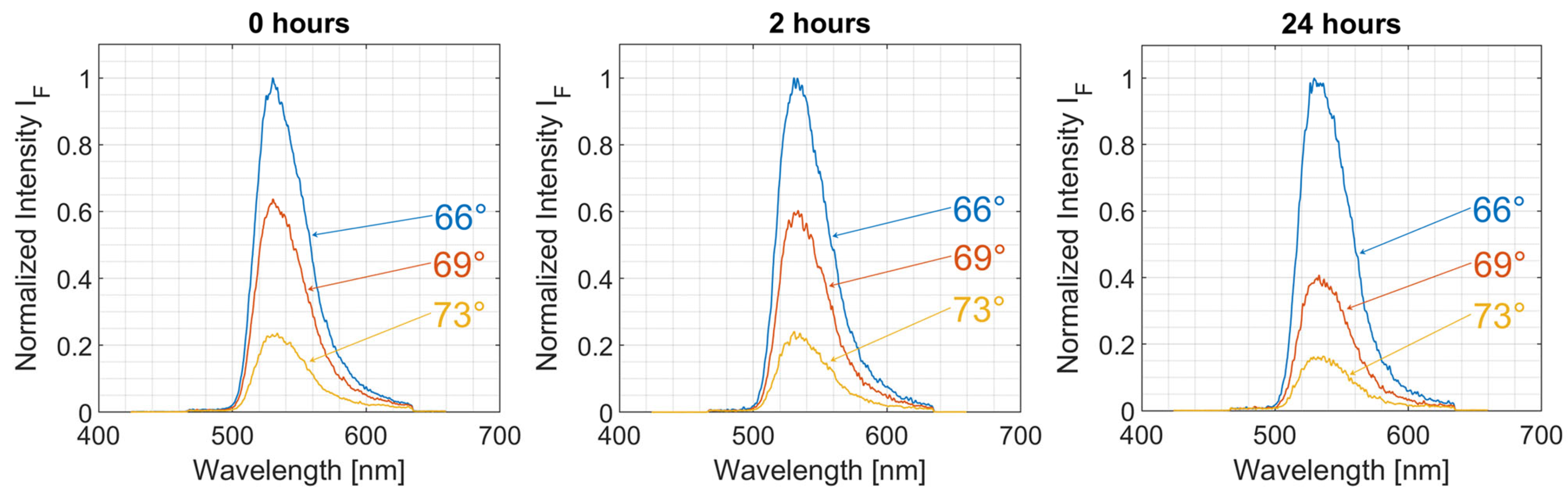
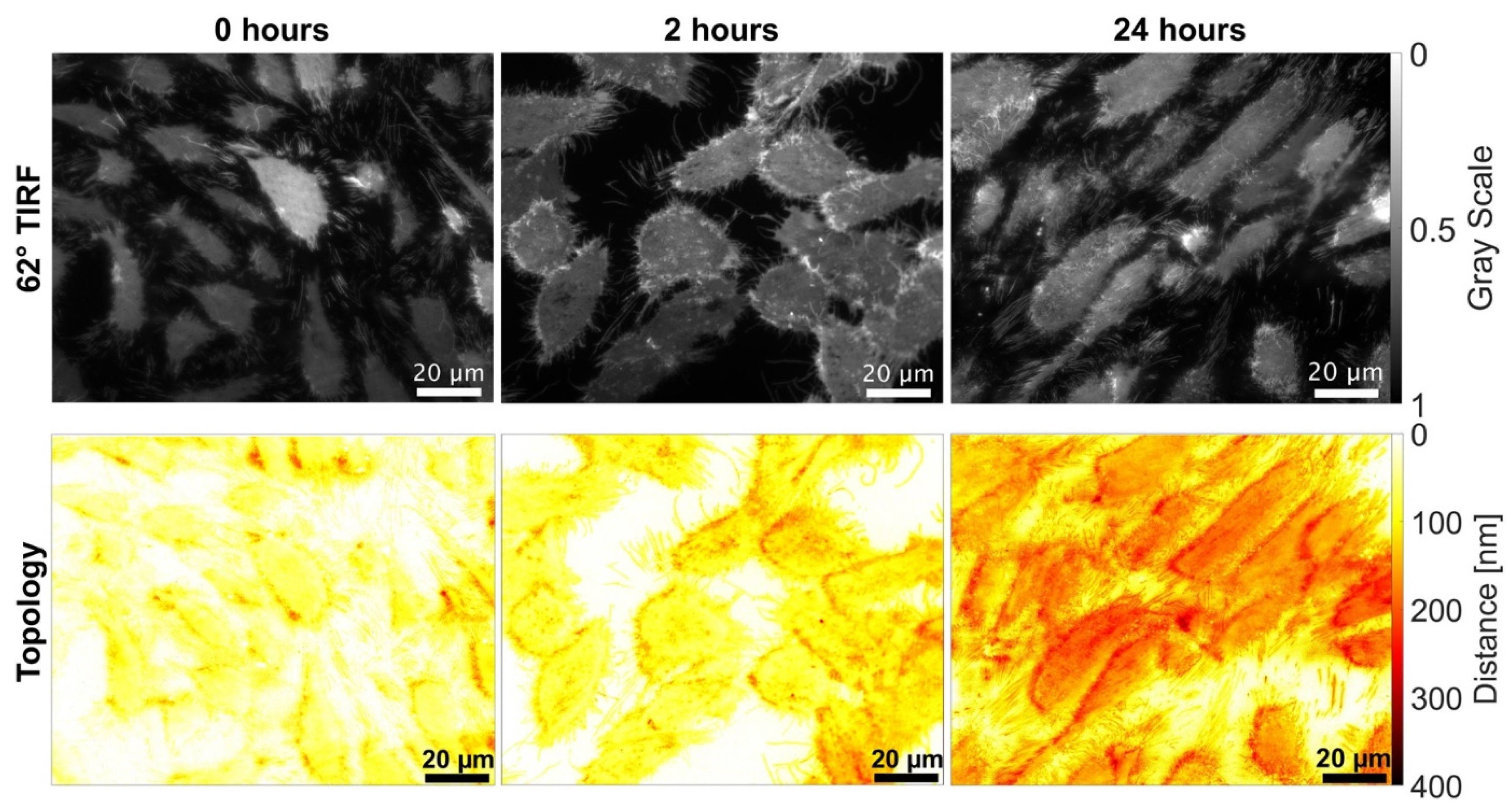
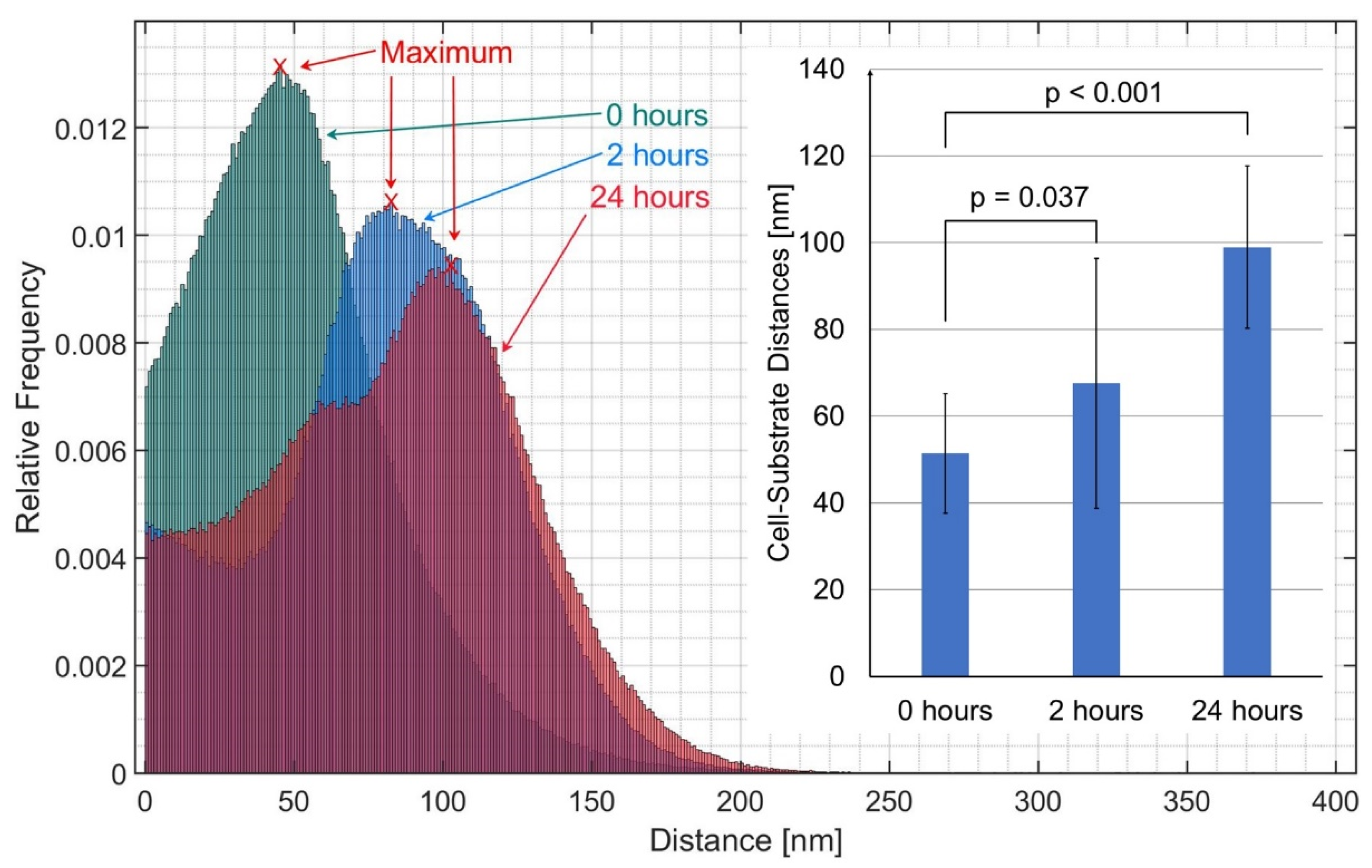
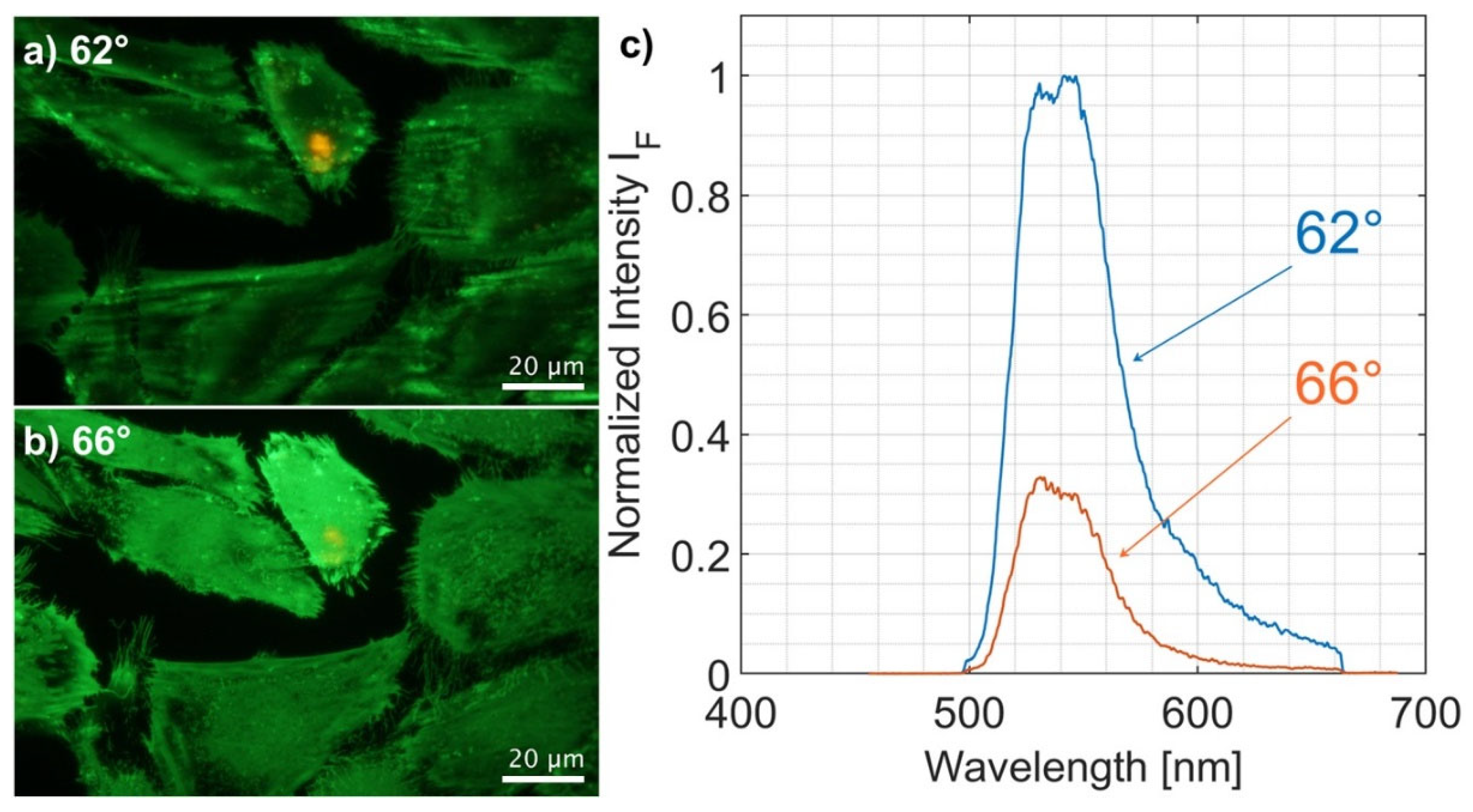
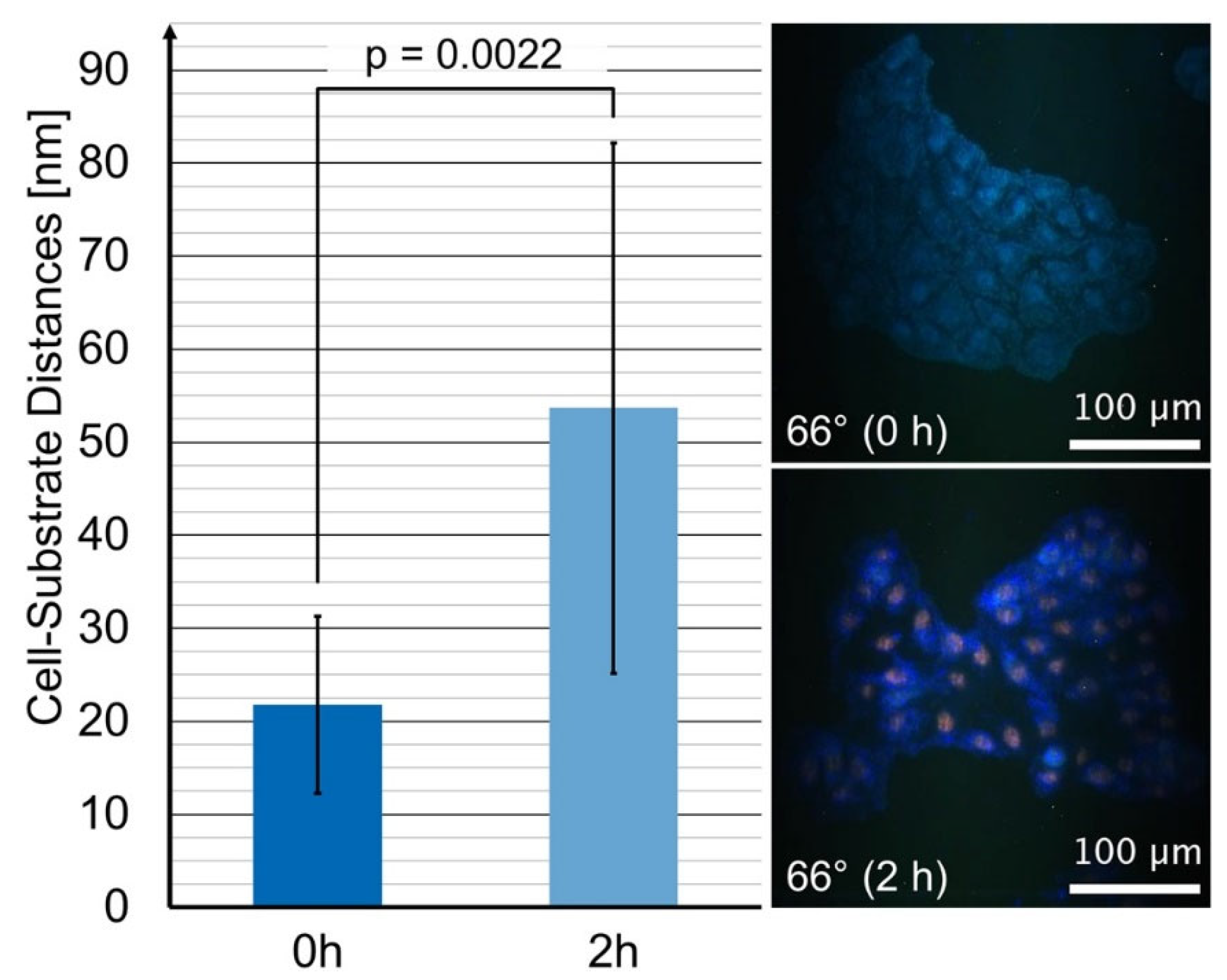
3. Results
4. Discussion
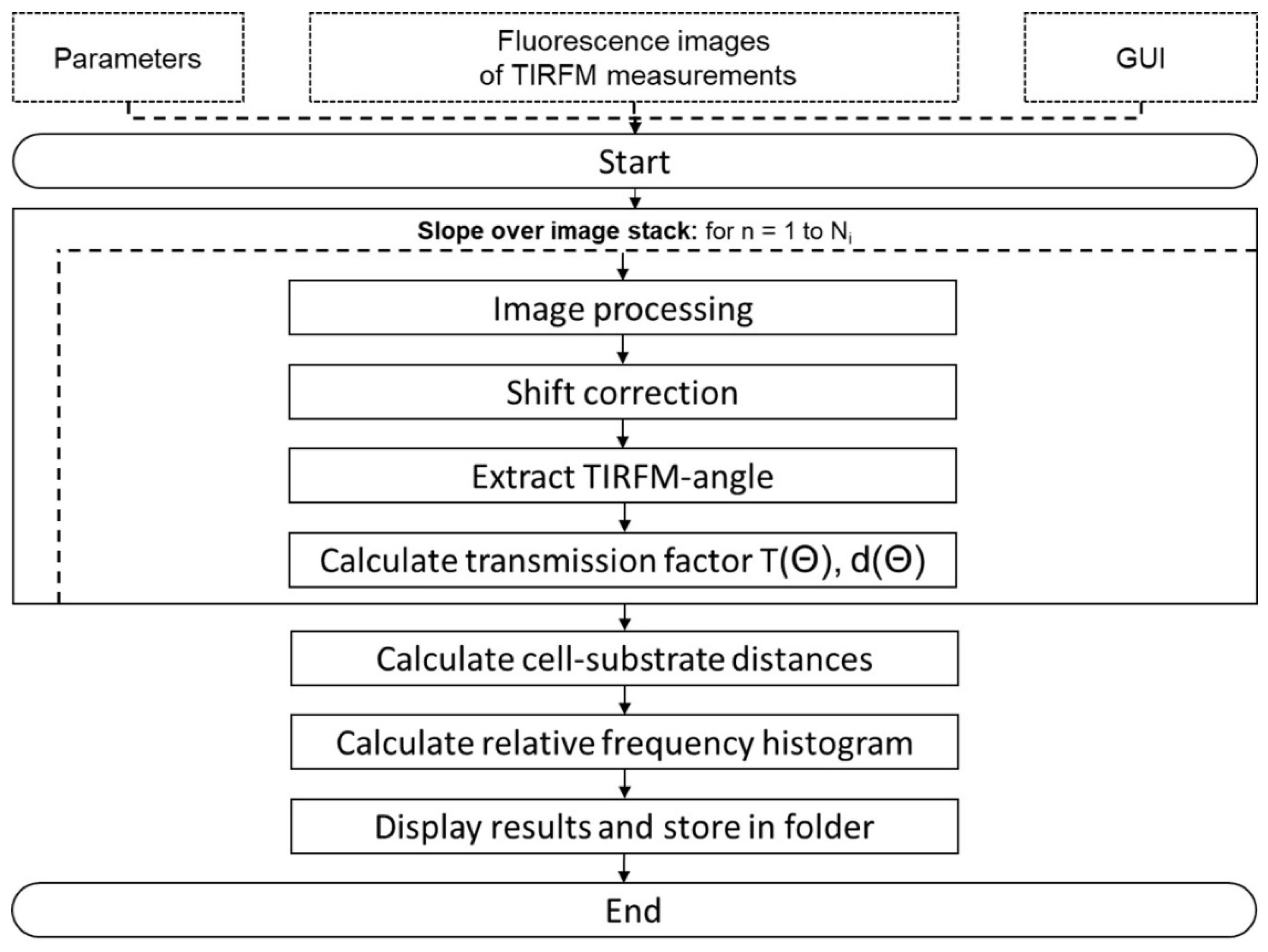
5. Materials and Methods
5.1. Cells
5.2. VA-TIRFM
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carter, S.K.; Blum, R.H. New chemotherapeutic agents—Bleomycin and adriamycin. CA Cancer J. Clin. 1974, 24, 322–331. [Google Scholar] [CrossRef]
- Blum, R.H.; Carter, S.K. Adriamycin. A new anticancer drug with significant clinical activity. Ann. Intern. Med. 1974, 80, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Szachowicz-Petelska, B.; Figaszewski, Z.; Lewandowski, W. Mechanisms of transport across cell membranes of complexes contained in antitumour drugs. Int. J. Pharm. 2001, 222, 169–182. [Google Scholar] [CrossRef]
- Slingerland, M.; Guchelaar, H.J.; Gelderblom, H. Liposomal drug formulations in cancer therapy: 15 years along the road. Drug Discov. Today 2012, 17, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Rivankar, S. An overview of doxorubicin formulations in cancer therapy. J. Cancer Res. Ther. 2014, 10, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.X.; Wang, T.T.; Wu, Y.T.; Xu, C.M.; Dong, M.Y.; Sheng, J.Z.; Huang, H.F. Adriamycin induces H2AX phosphorylation in human spermatozoa. Asian J. Androl. 2008, 10, 749–757. [Google Scholar] [CrossRef]
- Meredith, A.M.; Dass, C.R. Increasing role of the cancer chemotherapeutic doxorubicin in cellular metabolism. J. Pharm. Pharmacol. 2016, 68, 729–741. [Google Scholar] [CrossRef] [Green Version]
- Minotti, G.; Menna, P.; Salvatorelli, E.; Cairo, G.; Gianni, L. Anthracyclines: Molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol. Rev. 2004, 56, 185–229. [Google Scholar] [CrossRef] [Green Version]
- Cardinale, D.; Colombo, A.; Lamantia, G.; Colombo, N.; Civelli, M.; De Giacomi, G.; Rubino, M.; Veglia, F.; Fiorentini, C.; Cipolla, C.M. Anthracycline-induced cardiomyopathy: Clinical relevance and response to pharmacologic therapy. J. Am. Coll. Cardiol. 2010, 55, 213–220. [Google Scholar] [CrossRef] [Green Version]
- Karukstis, K.K.; Thompson, E.H.; Whiles, J.A.; Rosenfeld, R.J. Deciphering the fluorescence signature of daunomycin and doxorubicin. Biophys. Chem. 1998, 73, 249–263. [Google Scholar] [CrossRef]
- Chen, N.T.; Wu, C.Y.; Chung, C.Y.; Hwu, Y.; Cheng, S.H.; Mou, C.Y.; Lo, L.W. Probing the dynamics of doxorubicin-DNA intercalation during the initial activation of apoptosis by fluorescence lifetime imaging microscopy (FLIM). PLoS ONE 2012, 7, e44947. [Google Scholar] [CrossRef]
- Dai, X.; Yue, Z.; Eccleston, M.E.; Swartling, J.; Slater, N.K.; Kaminski, C.F. Fluorescence intensity and lifetime imaging of free and micellar-encapsulated doxorubicin in living cells. Nanomedicine 2008, 4, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Bakker, G.J.; Andresen, V.; Hoffman, R.M.; Friedl, P. Fluorescence lifetime microscopy of tumor cell invasion, drug delivery, and cytotoxicity. Methods Enzymol. 2012, 504, 109–125. [Google Scholar] [CrossRef]
- Haaland, D.M.; Jones, H.D.; van Benthem, M.H.; Sinclair, M.B.; Melgaard, D.K.; Stork, C.L.; Pedroso, M.C.; Liu, P.; Brasier, A.R.; Andrews, N.L.; et al. Hyperspectral confocal fluorescence imaging: Exploring alternative multivariate curve resolution approaches. Appl. Spectrosc. 2009, 63, 271–279. [Google Scholar] [CrossRef]
- Weber, P.; Wagner, M.; Schneckenburger, H. Cholesterol dependent uptake and interaction of doxorubicin in MCF-7 breast cancer cells. Int. J. Mol. Sci. 2013, 14, 8358–8366. [Google Scholar] [CrossRef]
- Richter, V.; Weber, P.; Wagner, M.; Schneckenburger, H. 3D visualization of cellular location and cytotoxic reactions of doxorubicin, a chemotherapeutic agent. Med. Res. Arch. 2018, 6, 1–9. [Google Scholar]
- Stock, K.; Sailer, R.; Strauss, W.S.L.; Lyttek, M.; Steiner, R.; Schneckenburger, H. Variable-angle total internal reflection fluorescence microscopy (VA-TIRFM): Realization and application of a compact illumination device. J. Microsc. 2003, 211, 19–29. [Google Scholar] [CrossRef]
- Koutsilieris, M.; Reyes-Moreno, C.; Choki, I.; Sourla, A.; Doillon, C.; Pavlidis, N. Chemotherapy cytotoxicity of human MCF-7 and MDA-MB 231 breast cancer cells is altered by osteoblast-derived growth factors. Mol. Med. 1999, 5, 86–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aydın, M.; Özdemir, E.; Altun, Z.; Kılıç, S.; Aktaş, S. Evaluation of liposomal and microbubbles Mediated delivery of doxorubicin in two-dimensional (2D) and three-dimensional (3D) models for breast cancer. Eur. J. Breast Health 2021, 17, 274–282. [Google Scholar] [CrossRef]
- Wang, T.; Dong, J.; Yuan, X.; Wen, H.; Wu, L.; Liu, J.; Sui, H.; Deng, W. A new chalcone derivative C49 reverses doxorubicin resistance in MCF-7/DOX cells by inhibiting p-glycoprotein expression. Front. Pharmacol. 2021, 12, 653306. [Google Scholar] [CrossRef]
- Schneckenburger, H.; Weber, P.; Wagner, M.; Schickinger, S.; Richter, V.; Bruns, T.; Strauss, W.S.; Wittig, R. Light exposure and cell viability in fluorescence microscopy. J. Microsc. 2012, 245, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Gingell, D.; Heavens, O.R.; Mello, J.S. General electromagnetic theory of total internal reflection fluorescence: The quantitative basis for mapping cell-substratum topography. J. Cell Sci. 1987, 87, 677–694. [Google Scholar] [CrossRef] [PubMed]
- Reichert, W.M.; Truskey, G.A. Total internal reflection fluorescence (TIRF) microscopy. (I) Modelling cell contact region fluorescence. J. Cell Sci. 1990, 96, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Weber, P.; Baumann, H.; Schneckenburger, H. Nanotopology of cell adhesion upon variable-angle total internal reflection fluorescence microscopy (VA-TIRFM). J. Vis. Exp. 2012, 68, e4133. [Google Scholar] [CrossRef] [Green Version]
- Hovorka, O.; Šubr, V.; Vetvicka, D.; Kovar, L.; Strohalm, J.; Strohalm, M.; Benda, A.; Hof, M.; Ulbrich, K.; Rihova, B. Spectral analysis of doxorubicin accumulation and the indirect quantification of its DNA intercalation. Eur. J. Pharm. Biopharm. 2010, 76, 514–524. [Google Scholar] [CrossRef]
- Kubiak, A.; Chighizola, M.; Schulte, C.; Bryniarska, N.; Wesołowska, J.; Pudełek, M.; Lasota, M.; Ryszawy, D.; Basta-Kaim, A.; Laidler, P.; et al. Stiffening of DU145 prostate cancer cells driven by actin filaments-microtubule crosstalk conferring resistance to microtubule-targeting drugs. Nanoscale 2021, 13, 6212–6226. [Google Scholar] [CrossRef]
- Mulligan, J.A.; Feng, X.; Adie, S.G. Quantitative reconstruction of time-varying 3D cell forces with traction force optical coherence microscopy. Sci. Rep. 2019, 9, 4086. [Google Scholar] [CrossRef] [Green Version]
- Richter, V.; Lanzerstorfer, P.; Weghuber, J.; Schneckenburger, H. Super-resolution live cell microscopy of membrane-proximal fluorophores. Int. J. Mol. Sci. 2020, 21, 7099. [Google Scholar] [CrossRef]
- Martino, F.; Perestrelo, A.R.; Vinarský, V.; Pagliari, S.; Forte, G. Cellular Mechanotransduction: From Tension to Function. Front. Physiol. 2018, 9, 824. [Google Scholar] [CrossRef]
- Weber, P.; Wagner, M.; Schneckenburger, H. Fluorescence Imaging of Membrane Dynamics in Living Cells. J. Biomed. Opt. 2010, 15, 046017. [Google Scholar] [CrossRef] [Green Version]
- Burmeister, J.S.; Truskey, G.A.; Reichert, W.M. Quantitative analysis of variable-angle total internal reflection fluorescence microscopy (VA-TIRFM) of cell/substrate contacts. J. Microsc. 1994, 173, 39–51. [Google Scholar] [CrossRef]
- Lassalle, H.P.; Baumann, H.; Strauss, W.S.; Schneckenburger, H. Cell-substrate topology upon ALA-PDT using variable-angle total internal reflection fluorescence microscopy (VA-TIRFM). J. Environ. Pathol. Toxicol. Oncol. 2007, 26, 83–88. [Google Scholar] [CrossRef]
- Parasassi, T.; de Stasio, G.; d’Ubaldo, A.; Gratton, E. Phase fluctuation in phospholipid membranes revealed by laurdan fluorescence. Biophys. J. 1990, 57, 1179–1186. [Google Scholar] [CrossRef] [Green Version]
- Van Cruchten, S.; Van Den Broeck, W. Morphological and biochemical aspects of apoptosis, oncosis and necrosis. Anat. Histol. Embryol. 2002, 31, 214–223. [Google Scholar] [CrossRef]
- Bortner, C.D.; Cidlowski, J.A. Apoptotic volume decrease and the incredible shrinking cell. Cell Death Differ. 2002, 9, 1307–1310. [Google Scholar] [CrossRef]
- Mulvey, C.S.; Sherwood, C.A.; Bigio, I.J. Wavelength-dependent backscattering measurements for quantitative real-time monitoring of apoptosis in living cells. J. Biomed. Opt. 2009, 14, 064013. [Google Scholar] [CrossRef]
- Richter, V.; Voit, F.; Kienle, A.; Schneckenburger, H. Light scattering microscopy with angular resolution and its possible application to apoptosis. J. Microsc. 2015, 257, 1–7. [Google Scholar] [CrossRef]
- Vagner, T.; Mouravlev, A.; Young, D. A novel bicistronic sensor vector for detecting caspase-3 activation. J. Pharmacol. Toxicol. Methods 2015, 72, 11–18. [Google Scholar] [CrossRef]
- Sergeeva, T.F.; Shirmanova, M.V.; Zlobovskaya, O.A.; Gavrina, A.I.; Dudenkova, V.V.; Lukina, M.M.; Lukyanov, K.A.; Zagaynova, E.V. Relationship between intracellular pH, metabolic co-factors and caspase-3 activation in cancer cells during apoptosis. Biochim. Biophys. Acta Mol. Cell. Res. 2017, 1864, 604–611. [Google Scholar] [CrossRef]
- Savitsky, A.P.; Rusanov, A.L.; Zherdeva, V.V.; Gorodnicheva, T.V.; Khrenova, M.G.; Nemukhin, A.V. FLIM-FRET Imaging of Caspase-3 Activity in Live Cells Using Pair of Red Fluorescent Proteins. Theranostics 2012, 2, 215–226. [Google Scholar] [CrossRef] [Green Version]
- Luo, K.Q.; Yu, V.C.; Pu, Y.; Chang, D.C. Application of the fluorescence resonance energy transfer method for studying the dynamics of caspase-3 activation during UV-induced apoptosis in living HeLa cells. Biochem. Biophys. Res. Commun. 2001, 283, 1054–1060. [Google Scholar] [CrossRef]
- Förster, T. Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann. Physik 1948, 2, 55–75. [Google Scholar] [CrossRef]
- Angres, B.; Steuer, H.; Weber, P.; Wagner, M.; Schneckenburger, H. A membrane-bound FRET-based caspase sensor for detection of apoptosis using fluorescence lifetime and total internal reflection microscopy. Cytom. A 2009, 75, 420–427. [Google Scholar] [CrossRef]
- Weber, P.; Schickinger, S.; Wagner, M.; Angres, B.; Bruns, T.; Schneckenburger, H. Monitoring of apoptosis in 3D cell cultures by FRET and light sheet fluorescence microscopy. Int. J. Mol. Sci. 2015, 16, 5375–5385. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, X.; Gueydan, C.; Han, J. Plasma membrane changes during programmed cell deaths. Cell Res. 2018, 28, 9–21. [Google Scholar] [CrossRef]
- Dimova, R.; Pouligny, B. Optical dynamometry to study phase transitions in lipid membranes. Methods Mol. Biol. 2007, 400, 227–236. [Google Scholar] [CrossRef]
- Leonhart, R. Lehrbuch Statistik; Hogrefe: Bern, Switzerland, 2017; ISBN 9783456957975. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krecsir, A.; Richter, V.; Wagner, M.; Schneckenburger, H. Impact of Doxorubicin on Cell-Substrate Topology. Int. J. Mol. Sci. 2022, 23, 6277. https://doi.org/10.3390/ijms23116277
Krecsir A, Richter V, Wagner M, Schneckenburger H. Impact of Doxorubicin on Cell-Substrate Topology. International Journal of Molecular Sciences. 2022; 23(11):6277. https://doi.org/10.3390/ijms23116277
Chicago/Turabian StyleKrecsir, Andreas, Verena Richter, Michael Wagner, and Herbert Schneckenburger. 2022. "Impact of Doxorubicin on Cell-Substrate Topology" International Journal of Molecular Sciences 23, no. 11: 6277. https://doi.org/10.3390/ijms23116277






