Genome Wide Identification and Characterization of Wheat GH9 Genes Reveals Their Roles in Pollen Development and Anther Dehiscence
Abstract
:1. Introduction
2. Results
2.1. Identification of the GH9 Family in Wheat
2.2. Phylogenetic Analysis of the GH9 Protein
2.3. Chromosomal Localization of the TaGH9 Family and Synteny Analysis
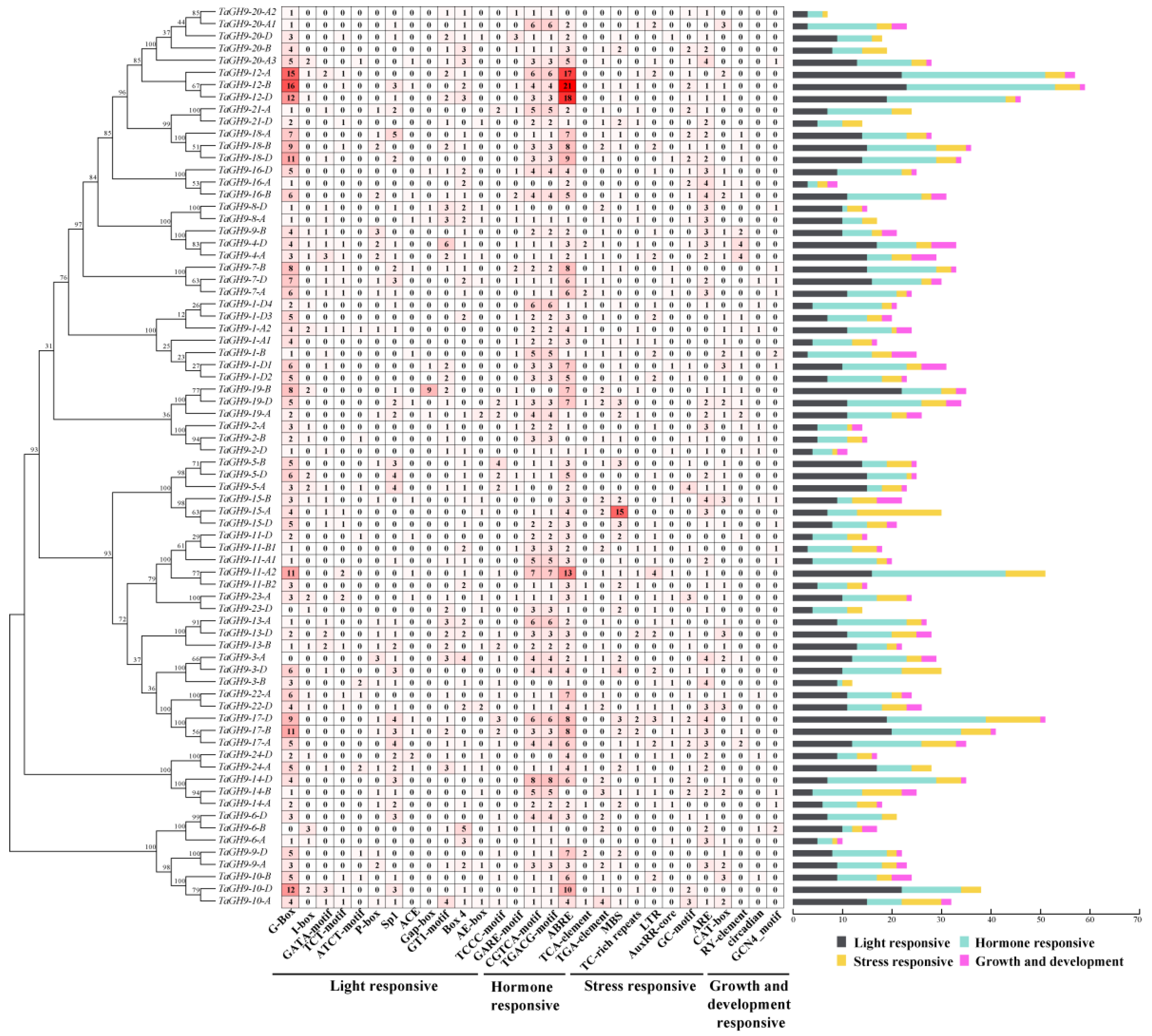
2.4. Structure Analysis and Motif Distribution of the TaGH9 Family
2.5. Cis-Acting Elements in Promoter Have Valuable Information for Analysis of the TaGH9 Family Function
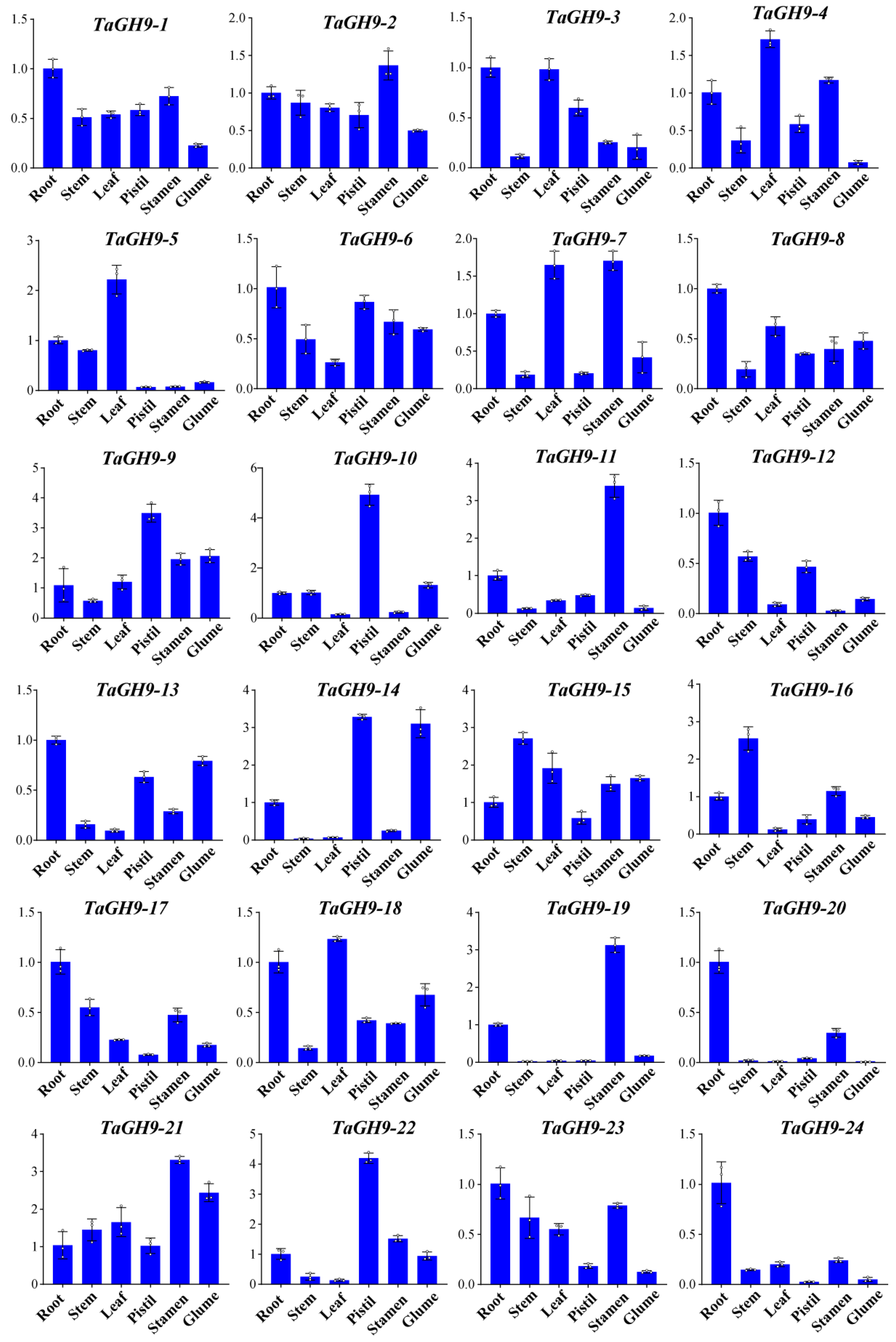
2.6. TaGH9 Genes Were Involved in Stamens Development
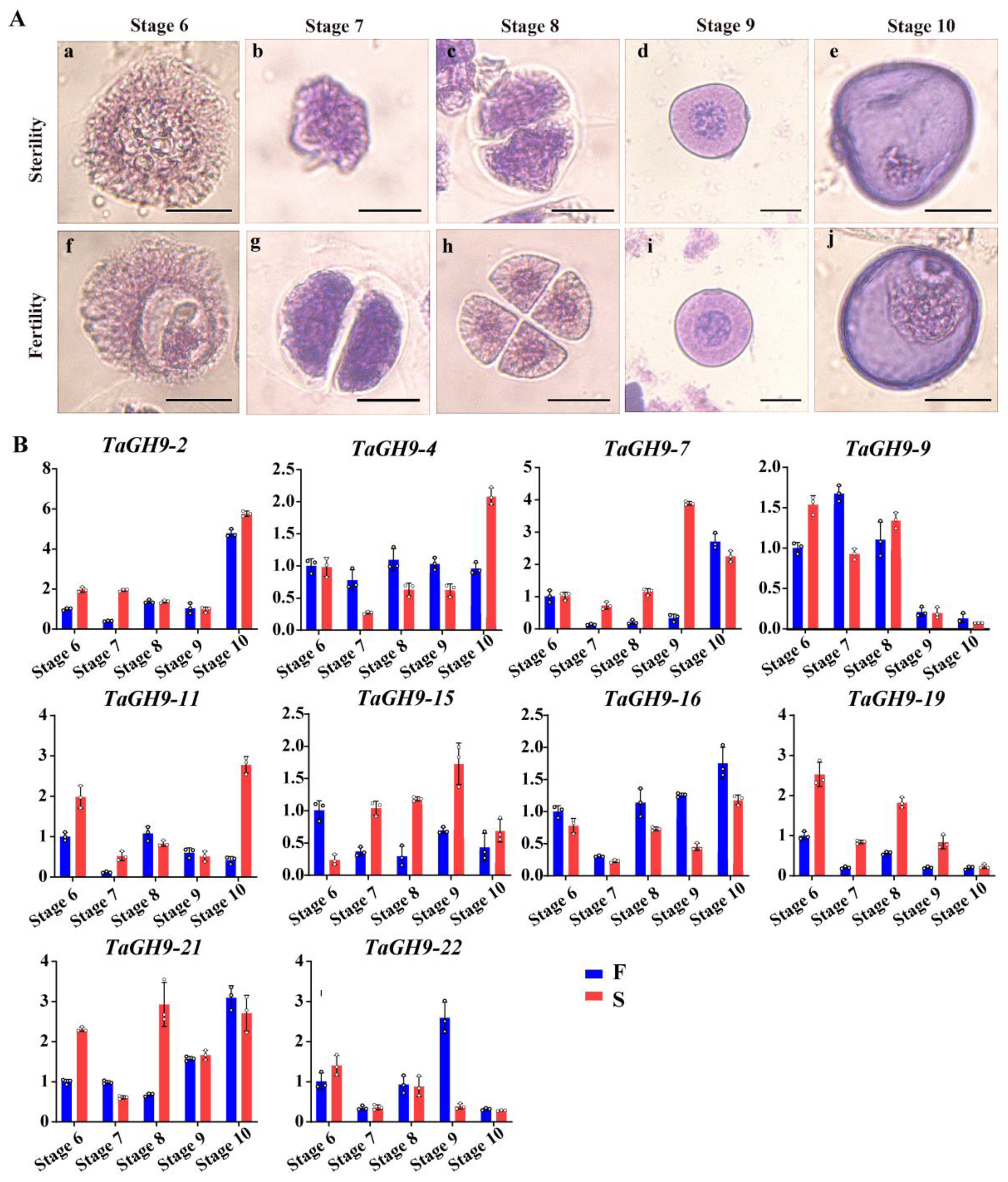
2.7. TaGH9 Involving Pollen Development and Pollen Fertility Conversion
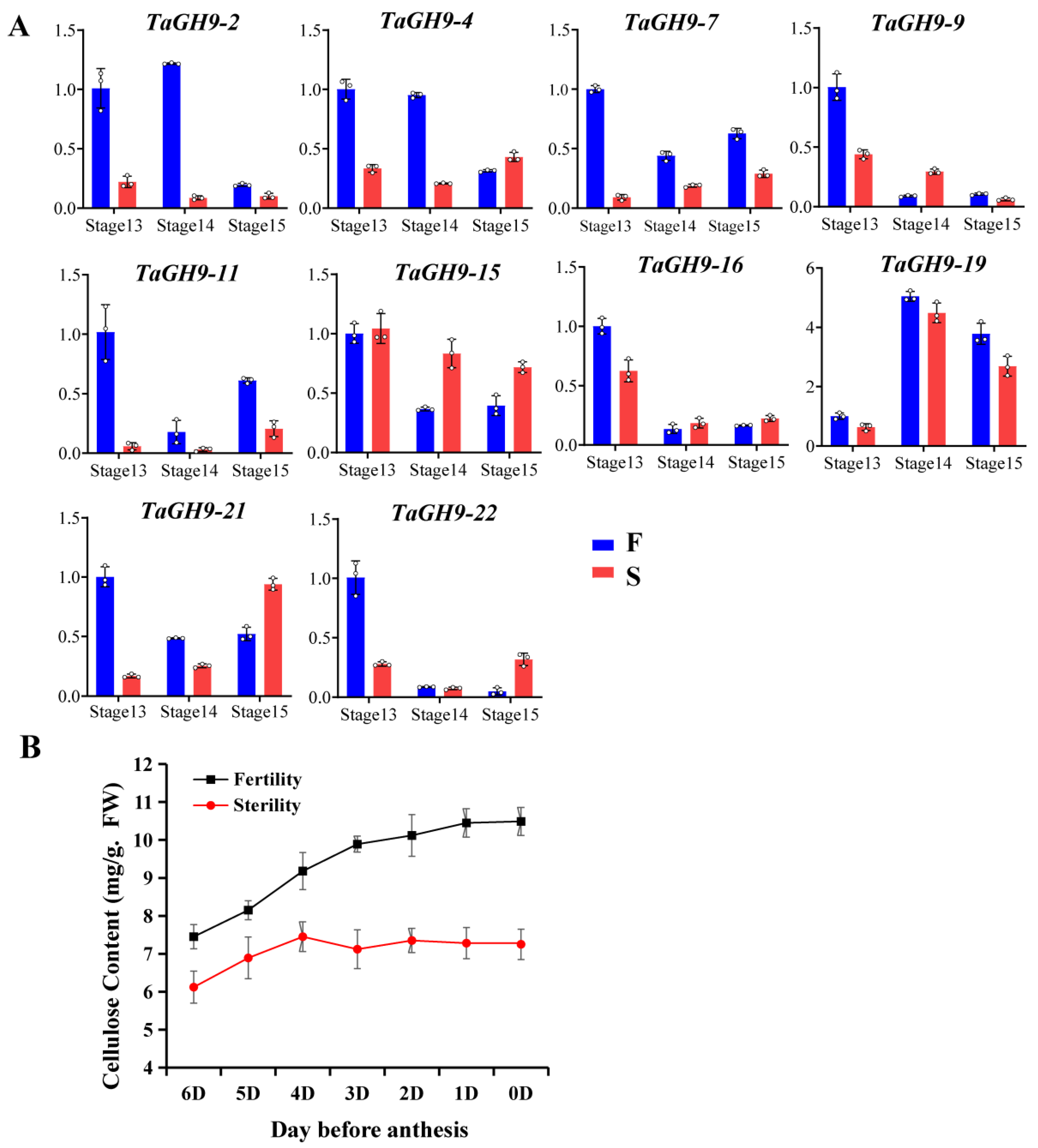
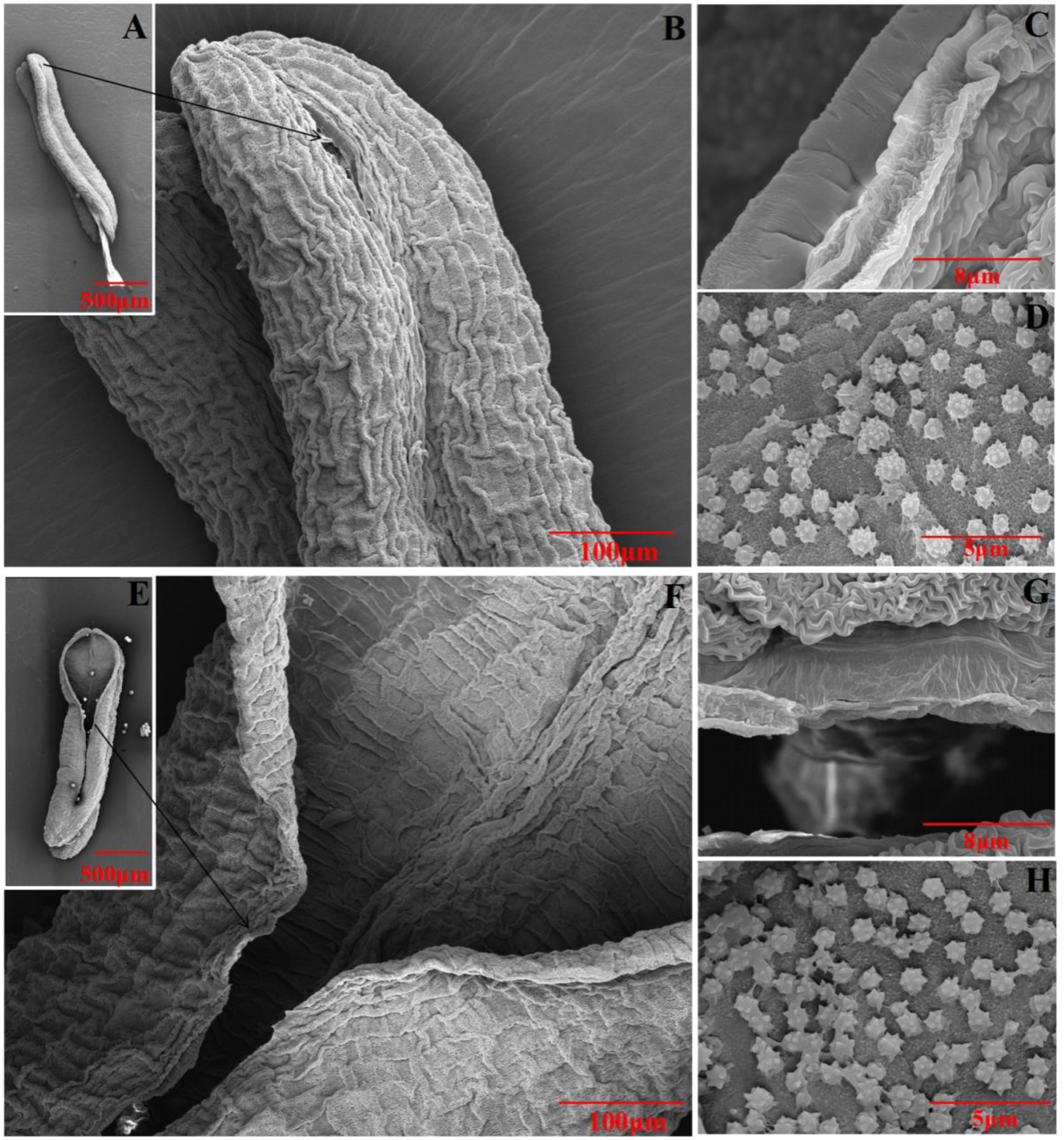
2.8. TaGH9 Involving Anther Dehiscence
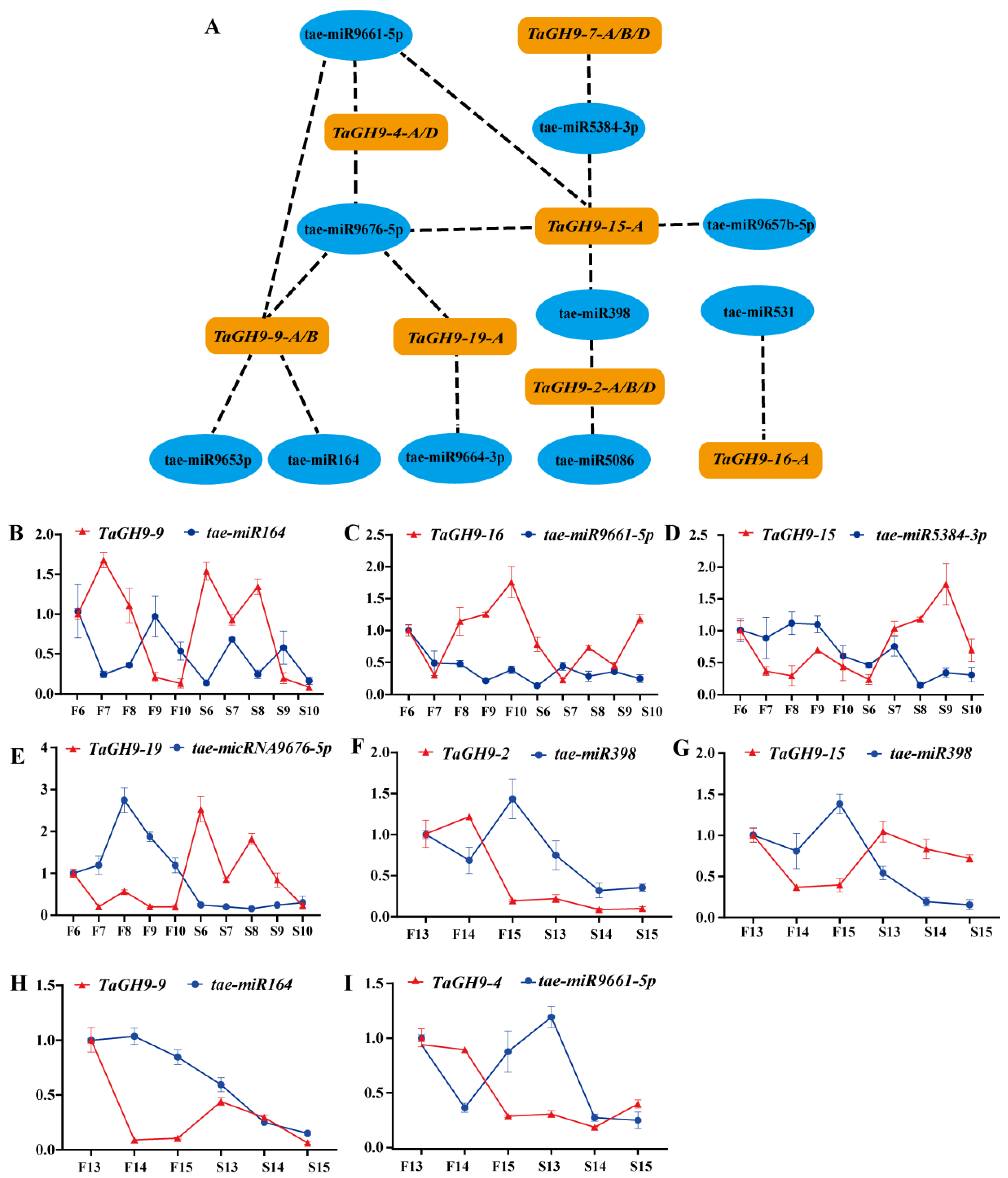
2.9. MicroRNA Targeting Prediction of TaGH9 and Expression Verification
3. Discussion
3.1. TaGH9s Involving Phytohormone Crosstalk-Regulated Pollen Development
3.2. TaGH9 May Be Involved in Anther Growth and Pollen Fertility Conversion
3.3. TaGH9 May Be Involved in Anther Dehiscence
3.4. A Putative Pathway of TaGH9 Regulating Pollen Fertility Conversion and Anther Dehiscence in Wheat
4. Materials and Methods
4.1. Plant Material, Growing Conditions and Sample Collection
4.2. Phenotypic Characterization of BS366
4.3. Data Sources for the GH9 Gene Family
4.4. Identification of Wheat GH9 Family Members
4.5. Chromosomal Locations and Synteny Analysis
4.6. Multiple Sequence Alignment and Phylogenetic Tree Construction
4.7. Structural Analysis of TaGH9 Genes and Proteins
4.8. Analysis of cis-Regulatory Elements of GH9 Genes
4.9. RNA-Seq Data Analysis and Gene Expression Heatmap
4.10. Measurement of Cellulose Content in Anther
4.11. miRNA Targeting Prediction of the TaGH9 Family
4.12. Expression Analysis of TaGH9 and miRNAs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| ABRE | Abscisic acid response element |
| CESA | Cellulose synthase |
| CSC | Cellulose synthase complex |
| EXPB | Expansin precursor protein |
| GA | Gibberellin acid |
| JA | Jasmonic acid |
| MeJA | Methyl Jasmonate |
| PG | Polygalacturonase |
| PTGMS | Photoperiod and thermosensitive genetic male sterile |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| SEM | Scanning electron microscope |
References
- Huang, J.; Xia, T.; Li, G.; Li, X.; Li, Y.; Wang, Y.; Wang, Y.; Chen, Y.; Xie, G.; Bai, F.-W.; et al. Overproduction of native endo-β-1,4-glucanases leads to largely enhanced biomass saccharification and bioethanol production by specific modification of cellulose features in transgenic rice. Biotechnol. Biofuels 2019, 12, 11. [Google Scholar] [CrossRef]
- Yoshida, K.; Komae, K. A Rice Family 9 Glycoside hydrolase isozyme with broad substrate specificity for hemicelluloses in type II cell walls. Plant Cell Physiol. 2006, 47, 1541–1554. [Google Scholar] [CrossRef] [Green Version]
- Woo, M.-H.; Chang, Y.-H.; Lee, H.-S.; Pak, P.J.; Kim, J.-S.; Chung, N. First thermostable endo-β-1,4-glucanase from newly isolated xanthomonas sp. EC102. Protein J. 2014, 33, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Shi, P.; Huang, H.; Li, Z.; Yuan, T.; Yang, P.; Luo, H.; Bai, Y.; Yao, B. A novel thermoacidophilic and thermostable endo-β-1,4-glucanase from Phialophora sp. G5: Its thermostability influenced by a distinct β-sheet and the carbohydrate-binding module. Appl. Microbiol. Biotechnol. 2012, 95, 947–955. [Google Scholar] [CrossRef]
- Kundu, S.; Sharma, R. Origin, evolution, and divergence of plant class C GH9 endoglucanases. BMC Evol. Biol. 2018, 18, 79. [Google Scholar] [CrossRef] [PubMed]
- Libertini, E.; Li, Y.; McQueen-Mason, S.J. Phylogenetic analysis of the plant endo-β-1,4-glucanase gene family. J. Mol. Evol. 2004, 58, 506–515. [Google Scholar] [CrossRef]
- del Campillo, E.; Gaddam, S.; Mettle-Amuah, D.; Heneks, J. A tale of two tissues: AtGH9C1 is an endo-β-1,4-glucanase involved in root hair and endosperm development in Arabidopsis. PLoS ONE 2012, 7, e49363. [Google Scholar] [CrossRef] [Green Version]
- Xie, G.; Yang, B.; Xu, Z.; Li, F.; Guo, K.; Zhang, M.; Wang, L.; Zou, W.; Wang, Y.; Peng, L. Global identification of multiple OsGH9 family members and their involvement in cellulose crystallinity modification in rice. PLoS ONE 2013, 8, e50171. [Google Scholar] [CrossRef]
- Du, Q.; Wang, L.; Yang, X.; Gong, C.; Zhang, D. Populus endo-β-1,4-glucanases gene family: Genomic organization, phylogenetic analysis, expression profiles and association mapping. Planta 2015, 241, 1417–1434. [Google Scholar] [CrossRef]
- Zhao, Q.; Li, X. Genome-wide identification and analysis of the GH9 family genes in cotton. J. Shanxi Agric. Sci. 2018, 46, 1065–1069. [Google Scholar]
- Jara, K.; Castro, R.I.; Ramos, P.; Parra-Palma, C.; Valenzuela-Riffo, F.; Morales-Quintana, L. Molecular insights into FaEG1, a strawberry endoglucanase enzyme expressed during strawberry fruit ripening. Plants 2019, 8, 140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brummell, D.A.; Lashbrook, C.C.; Bennett, A.B. Plant endo-1,4-beta-D-glucanases: Structure, properties, and physiological function. ACS Symp. Ser. Am. Chem. Soc. 1994, 566, 100–129. [Google Scholar]
- Rose, J.K.C.; Bennett, A.B. Cooperative disassembly of the cellulose–xyloglucan network of plant cell walls: Parallels between cell expansion and fruit ripening. Trends Plant Sci. 1999, 4, 176–183. [Google Scholar] [CrossRef]
- Roberts, J.A.; Elliott, K.A.; Gonzalez-Carranza, Z.H. Abscission, dehiscence, and other cell sepration processes. Annu. Rev. Plant Biol. 2002, 53, 131–158. [Google Scholar] [CrossRef]
- Kalaitzis, P.; Hong, S.-B.; Solomos, T.; Tucker, M.L. Molecular characterization of a tomato endo-β-1,4-glucanase gene expressed in mature pistils, abscission zones and fruit. Plant Cell Physiol. 1999, 40, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.K.; Lewandowski, D.J.; Nairn, C.J.; Brown, G.E. Endo-1,4-β-glucanase gene expression and cell wall hydrolase activities during abscission in Valencia orange. Physiol. Plant. 1998, 102, 217–225. [Google Scholar] [CrossRef]
- Nicol, F.; His, I.; Jauneau, A.; Vernhettes, S.; Canut, H.; Höfte, H. A plasma membrane-bound putative endo-1,4-β-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 1998, 17, 5563–5576. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.L.; He, S.J.; Cao, Y.R.; Chen, T.; Du, B.X.; Chu, C.C.; Zhang, J.S.; Chen, S.Y. OsGLU1, A putative membrane-bound endo-1,4-β-D-glucanase from Rice, affects plant internode elongation. Plant Mol. Biol. 2006, 60, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, S.; Ahmed, W.; Alatalo, J.M.; Mahmood, M.; Imtiaz, M.; Ditta, A.; Ali, E.F.; Abdelrahman, H.; Slaný, M.; Antoniadis, V.; et al. Herbal plants- and rice straw-derived biochars reduced metal mobilization in fishpond sediments and improved their potential as fertilizers. Sci. Total Environ. 2022, 826, 154043. [Google Scholar] [CrossRef]
- Scott, R.J.; Spielman, M.; Dickinson, H.G. Stamen structure and function. Plant Cell 2004, 16, S46–S60. [Google Scholar] [CrossRef]
- Brummell, D.A.; Hall, B.D.; Bennett, A.B. Antisense suppression of tomato endo-1,4-β-glucanase Cel2 mRNA accumulation increases the force required to break fruit abscission zones but does not affect fruit softening. Plant Mol. Biol. 1999, 40, 615–622. [Google Scholar] [CrossRef]
- Cho, H.T.; Cosgrove, D.J. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2000, 97, 9783–9788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, S.; Kim, S.R.; Zhao, G.; Yi, J.; Yoo, Y.; Jin, P.; Lee, S.W.; Jung, K.H.; Zhang, D.; An, G. Rice glycosyltransferase1 encodes a glycosyltransferase essential for pollen wall formation. Plant Physiol. 2013, 161, 663–675. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zhang, P.; Lv, J.; Cheng, Y.; Cui, J.; Zhao, H.; Hu, S. Global dynamic transcriptome programming of rapeseed (Brassica napus L.) anther at different development stages. PLoS ONE 2016, 11, e0154039. [Google Scholar] [CrossRef] [Green Version]
- Zhiliang, Y. Functional analysis of glycoside hydrolase-like gene AtGHL in Arab. Thaliana. Agric. Univ. Hebei 2012, 8, 22–23. [Google Scholar]
- Tan, Z.; Li, Y.M.; Bai, J.F.; Guo, H.Y.; Li, T.T.; Duan, W.J.; Liu, Z.H.; Yuan, S.H.; Zhang, T.B.; Zhang, F.T.; et al. Cloning of TaBG and analysis of its function in anther dehiscence in wheat. Sci. Agric. Sin. 2021, 54, 2710–2723. [Google Scholar]
- Tang, Z.; Zhang, L.; Xu, C.; Yuan, S.; Zhang, F.; Zheng, Y.; Zhao, C. Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing. Plant Physiol. 2012, 159, 721–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, J.-F.; Wang, Y.K.; Wang, P.; Duan, W.J.; Yuan, S.H.; Sun, H.; Yuan, G.L.; Ma, J.X.; Wang, N.; Zhang, F.T.; et al. Uncovering male fertility transition responsive miRNA in a wheat photo-thermosensitive genic male sterile line by deep sequencing and degradome analysis. Front. Plant Sci. 2017, 8, 1370. [Google Scholar] [CrossRef] [Green Version]
- Bai, J.F.; Wang, Y.K.; Guo, L.P.; Guo, X.M.; Guo, H.Y.; Yuan, S.H.; Duan, W.J.; Liu, Z.; Zhao, C.P.; Zhang, F.T.; et al. Genomic identification and characterization of MYC family genes in wheat (Triticum aestivum L.). BMC Genom. 2019, 20, 1032. [Google Scholar] [CrossRef] [Green Version]
- Abbas, A.; Yu, P.; Sun, L.; Yang, Z.; Chen, D.; Cheng, S.; Cao, L. Exploiting genic male sterility in rice: From molecular dissection to breeding applications. Front. Plant Sci. 2021, 12, 629314. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.-J.; Liu, Z.-H.; Bai, J.-F.; Yuan, S.-H.; Li, Y.-M.; Lu, F.-K.; Zhang, T.-B.; Sun, J.-H.; Zhang, F.-T.; Zhao, C.-P.; et al. Comprehensive analysis of formin gene family highlights candidate genes related to pollen cytoskeleton and male fertility in wheat (Triticum aestivum L.). BMC Genom. 2021, 22, 570. [Google Scholar] [CrossRef] [PubMed]
- Browne, R.G.; Iacuone, S.; Li, S.F.; Dolferus, R.; Parish, R.W. Anther morphological development and stage determination in Triticum aestivum. Front. Plant Sci. 2018, 9, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Khanna, K.; Ruan, S. Expression of microRNAs and its regulation in plants. Semin. Cell Dev. Biol. 2010, 21, 790–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Shen, Y.; Zhu, J.; Liu, S.; Zeng, N.; Zhan, X. miR398 is involved in the relief of phenanthrene-induced oxidative toxicity in wheat roots. Environ. Pollut. 2020, 258, 113701. [Google Scholar] [CrossRef]
- Zhu, C.; Ding, Y.; Liu, H. MiR398 and plant stress responses. Physiol. Plant. 2011, 143, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [Green Version]
- Bai, J.f.; Wang, Y.k.; Wang, P.; Yuan, S.h.; Gao, J.g.; Duan, W.j.; Wang, N.; Zhang, F.t.; Zhang, W.j.; Qin, M.y.; et al. Genome-wide identification and analysis of the COI gene family in wheat (Triticum aestivum L.). BMC Genom. 2018, 19, 754. [Google Scholar] [CrossRef]
- Marciniak, K.; Przedniczek, K. Comprehensive insight into gibberellin and jasmonate-mediated stamen development. Genes (Basel) 2019, 10, 811. [Google Scholar] [CrossRef] [Green Version]
- Cheng, H.; Song, S.; Xiao, L.; Soo, H.M.; Cheng, Z.; Xie, D.; Peng, J. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet 2009, 5, e1000440. [Google Scholar] [CrossRef] [Green Version]
- Keijzer, C.J. The processes of anther dehiscence and pollen dispersal. New Phytol. 1987, 105, 487–498. [Google Scholar] [CrossRef]
- Wang, Y.; Qiao, L.; Bai, J.; Wang, P.; Duan, W.; Yuan, S.; Yuan, G.; Zhang, F.; Zhang, L.; Zhao, C. Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.). BMC Genom. 2017, 18, 152. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Wu, Y.; Zhang, G. Formation pattern and regulatory mechanisms of pollen wall in Arabidopsis. J. Plant Physiol. 2021, 260, 153388. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, Z.; Cao, J. Pollen wall development: The associated enzymes and metabolic pathways. Plant Biol. 2013, 15, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Radja, A.; Horsley, E.M.; Lavrentovich, M.O.; Sweeney, A.M. Pollen cell wall patterns form from modulated phases. Cell 2019, 176, 856–868.e810. [Google Scholar] [CrossRef] [Green Version]
- Mueller, S.C.; Brown, R.M., Jr. Evidence for an intramembrane component associated with a cellulose microfibril-synthesizing complex in higher plants. J. Cell Biol. 1980, 84, 315–326. [Google Scholar] [CrossRef]
- Persson, S.; Paredez, A.; Carroll, A.; Palsdottir, H.; Doblin, M.; Poindexter, P.; Khitrov, N.; Auer, M.; Somerville, C.R. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 15566–15571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shim, S.-H.; Mahong, B.; Lee, S.-K.; Kongdin, M.; Lee, C.; Kim, Y.J.; Qu, G.; Zhang, D.; Ketudat Cairns, J.R.; Jeon, J.S. Rice β-glucosidase Os12BGlu38 is required for synthesis of intine cell wall and pollen fertility. J. Exp. Bot. 2022, 73, 784–800. [Google Scholar] [CrossRef]
- Xie, X.-J.; Huang, J.J.; Gao, H.-H.; Guo, G.-Q. Expression patterns of two Arabidopsis endo-β-1,4-glucanase genes (At3g43860, At4g39000) in reproductive development. Mol. Biol. 2011, 45, 458–465. [Google Scholar] [CrossRef]
- Kim Jin, H.; Woo Hye, R.; Kim, J.; Lim Pyung, O.; Lee In, C.; Choi Seung, H.; Hwang, D.; Nam Hong, G. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 2009, 323, 1053–1057. [Google Scholar] [CrossRef] [Green Version]
- Laufs, P.; Peaucelle, A.; Morin, H.; Traas, J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 2004, 131, 4311–4322. [Google Scholar] [CrossRef] [Green Version]
- Mallory, A.C.; Dugas, D.V.; Bartel, D.P.; Bartel, B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 2004, 14, 1035–1046. [Google Scholar] [CrossRef] [Green Version]
- Casati, P. Analysis of UV-B regulated miRNAs and their targets in maize leaves. Plant Signal. Behav. 2013, 8, psb.26758. [Google Scholar] [CrossRef] [Green Version]
- Xiang, X.; Zhang, P.; Yu, P.; Zhang, Y.; Yang, Z.; Sun, L.; Wu, W.; Khan, R.M.; Abbas, A.; Cheng, S.; et al. LSSR1 facilitates seed setting rate by promoting fertilization in rice. Rice 2019, 12, 31. [Google Scholar] [CrossRef] [Green Version]
- Drakakaki, G. Polysaccharide deposition during cytokinesis: Challenges and future perspectives. Plant Sci. 2015, 236, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Samuels, A.L.; Giddings, T.H., Jr.; Staehelin, L.A. Cytokinesis in tobacco BY-2 and root tip cells: A new model of cell plate formation in higher plants. J. Cell Biol. 1995, 130, 1345–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, J.; Niu, Q.W.; Nishizawa, N.; Wu, Y.; Kost, B.; Chua, N.H. KORRIGAN, an Arabidopsis endo-1,4-beta-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 2000, 12, 1137–1152. [Google Scholar] [CrossRef] [PubMed]
- Jaffri, S.R.F.; MacAlister, C.A. Sequential deposition and remodeling of cell wall polymers during tomato pollen development. Front. Plant Sci. 2021, 12, 703713. [Google Scholar] [CrossRef] [PubMed]
- Abramova, L.; Avalkina, N.; Golubeva, E.; Pyzhenkova, Z.; Golubovskaya, I. synthesis and deposition of callose in anthers and ovules of meiotic mutants of maize (Zea mays). Russ. J. Plant Physiol. 2003, 50, 324–329. [Google Scholar] [CrossRef]
- Schindfessel, C.; Drozdowska, Z.; De Mooij, L.; Geelen, D. Loss of obligate crossovers, defective cytokinesis and male sterility in barley caused by short-term heat stress. Plant Reprod. 2021, 34, 243–253. [Google Scholar] [CrossRef]
- Sexton, R.; Del Campillo, E.; Duncan, D.; Lewis, L.N. The purification of an anther cellulase (β(1:4)4-glucan hydrolase) from Lathyrus odoratus L. and its relationship to the similar enzyme found in abscission zones. Plant Sci. 1990, 67, 169–176. [Google Scholar] [CrossRef]
- Lane, D.R.; Wiedemeier, A.; Peng, L.; Höfte, H.; Vernhettes, S.; Desprez, T.; Hocart, C.H.; Birch, R.J.; Baskin, T.I.; Burn, J.E.; et al. Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 2001, 126, 278–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Sun, J.; Li, L. PtrCel9A6, an Endo-1,4-β-Glucanase, is required for cell wall formation during xylem differentiation in populus. Mol. Plant 2013, 6, 1904–1917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonner, L.J.; Dickinson, H.G. Anther dehiscence in Lycopersicon esculentum. New Phytol. 1990, 115, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Bots, M.; Vergeldt, F.; Mieke, W.-A.; Weterings, K.; van Henk, A.; Mariani, C. Aquaporins of the PIP2 class are required for efficient anther dehiscence in tobacco. Plant Physiol. 2005, 137, 1049–1056. [Google Scholar] [CrossRef] [Green Version]
- Stadler, R.; Truernit, E.; Gahrtz, M.; Sauer, N. The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. Plant J. 1999, 19, 269–278. [Google Scholar] [CrossRef]
- Thompson, E.P.; Wilkins, C.; Demidchik, V.; Davies, J.M.; Glover, B.J. An Arabidopsis flavonoid transporter is required for anther dehiscence and pollen development. J. Exp. Bot. 2010, 61, 439–451. [Google Scholar] [CrossRef] [Green Version]
- Steiner-Lange, S.; Unte, U.S.; Eckstein, L.; Yang, C.; Wilson, Z.A.; Schmelzer, E.; Dekker, K.; Saedler, H. Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J. 2003, 34, 519–528. [Google Scholar] [CrossRef]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. Squamosa promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Geng, X.; Liu, Z.; Ye, J.; Xu, M.; Zhang, L.; Song, X. A Sterility induction trait in the genic male sterility wheat line 4110S induced by high temperature and its cytological response. Crop Sci. 2018, 58, 1866–1876. [Google Scholar] [CrossRef]
- Lee, T.-H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2013, 41, D1152–D1158. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Li, M.; Bahn, S.C.; Guo, L.; Musgrave, W.; Berg, H.; Welti, R.; Wang, X. Patatin-related phospholipase pPLAIIIβ-induced changes in lipid metabolism alter cellulose content and cell elongation in Arabidopsis. Plant Cell 2011, 23, 1107–1123. [Google Scholar] [CrossRef] [Green Version]
- Duan, W.; Lu, F.; Cui, Y.; Zhang, J.; Du, X.; Hu, Y.; Yan, Y. Genome-wide identification and characterisation of wheat MATE genes reveals their roles in aluminium tolerance. Int. J. Mol. Sci. 2022, 23, 4418. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, L.; Bai, J.; Yuan, S.; Guo, L.; Liu, Z.; Guo, H.; Zhang, T.; Duan, W.; Li, Y.; Zhao, C.; et al. Genome Wide Identification and Characterization of Wheat GH9 Genes Reveals Their Roles in Pollen Development and Anther Dehiscence. Int. J. Mol. Sci. 2022, 23, 6324. https://doi.org/10.3390/ijms23116324
Luo L, Bai J, Yuan S, Guo L, Liu Z, Guo H, Zhang T, Duan W, Li Y, Zhao C, et al. Genome Wide Identification and Characterization of Wheat GH9 Genes Reveals Their Roles in Pollen Development and Anther Dehiscence. International Journal of Molecular Sciences. 2022; 23(11):6324. https://doi.org/10.3390/ijms23116324
Chicago/Turabian StyleLuo, Liqing, Jianfang Bai, Shaohua Yuan, Liping Guo, Zihan Liu, Haoyu Guo, Tianbao Zhang, Wenjing Duan, Yanmei Li, Changping Zhao, and et al. 2022. "Genome Wide Identification and Characterization of Wheat GH9 Genes Reveals Their Roles in Pollen Development and Anther Dehiscence" International Journal of Molecular Sciences 23, no. 11: 6324. https://doi.org/10.3390/ijms23116324





