Tissue-Specific Downregulation of Fatty Acid Synthase Suppresses Intestinal Adenoma Formation via Coordinated Reprograming of Transcriptome and Metabolism in the Mouse Model of Apc-Driven Colorectal Cancer
Abstract
:1. Introduction
2. Results
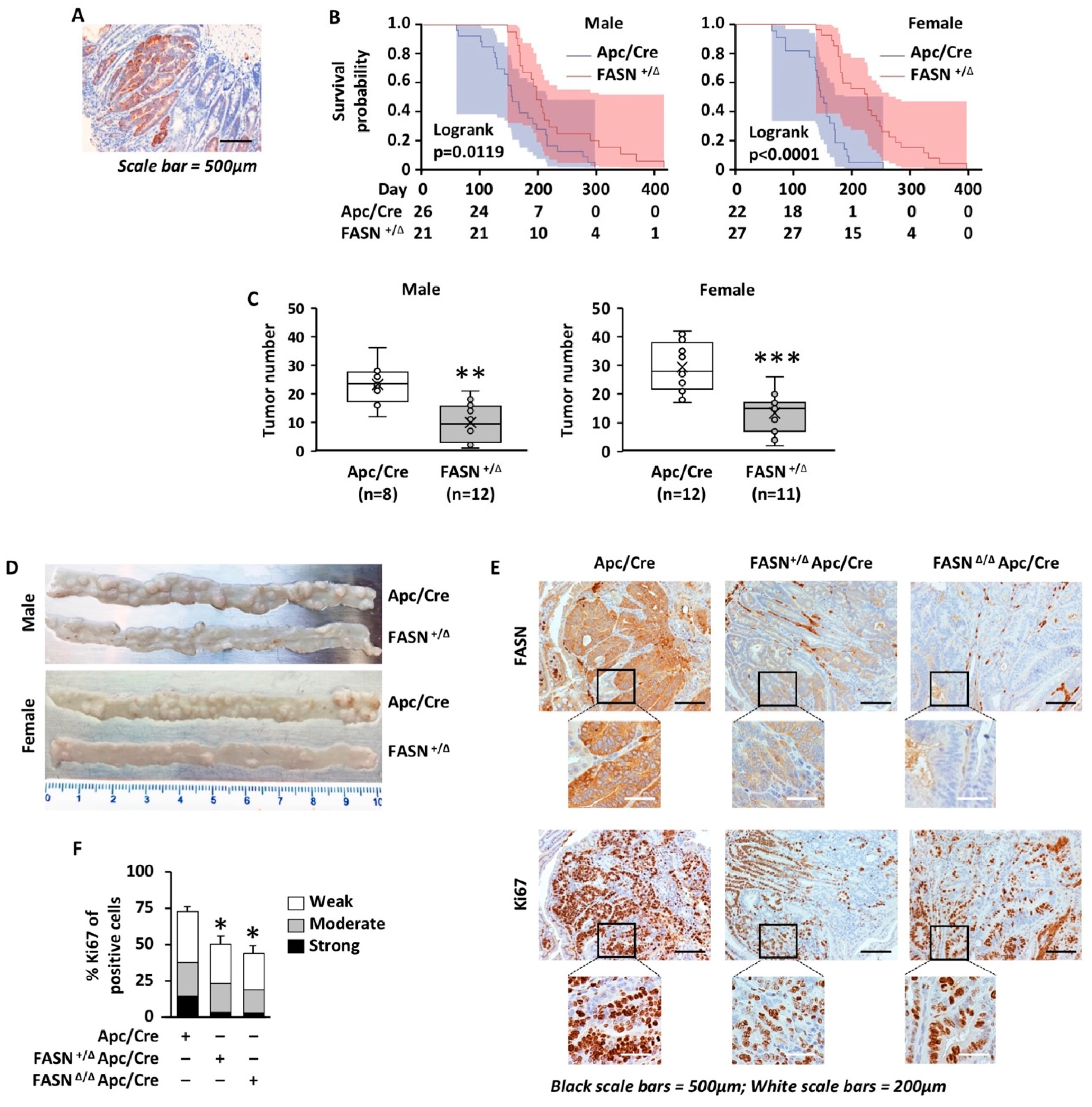
2.1. Heterozygous Deletion of FASN Increases Survival and Decreases the Number of Intestinal Adenomas during Apc-Driven Carcinogenesis
2.2. A Decrease in FASN Expression Is Associated with Downregulation of Pathways Linked to Cellular Proliferation, Energy Production, and Cancer-Associated Signaling
2.3. Heterozygous Deletion of FASN Alters the Levels of Diglycerides, but Does Not Change the Total Levels of Free Fatty Acid and Sphingolipid Species in Mouse Adenomas
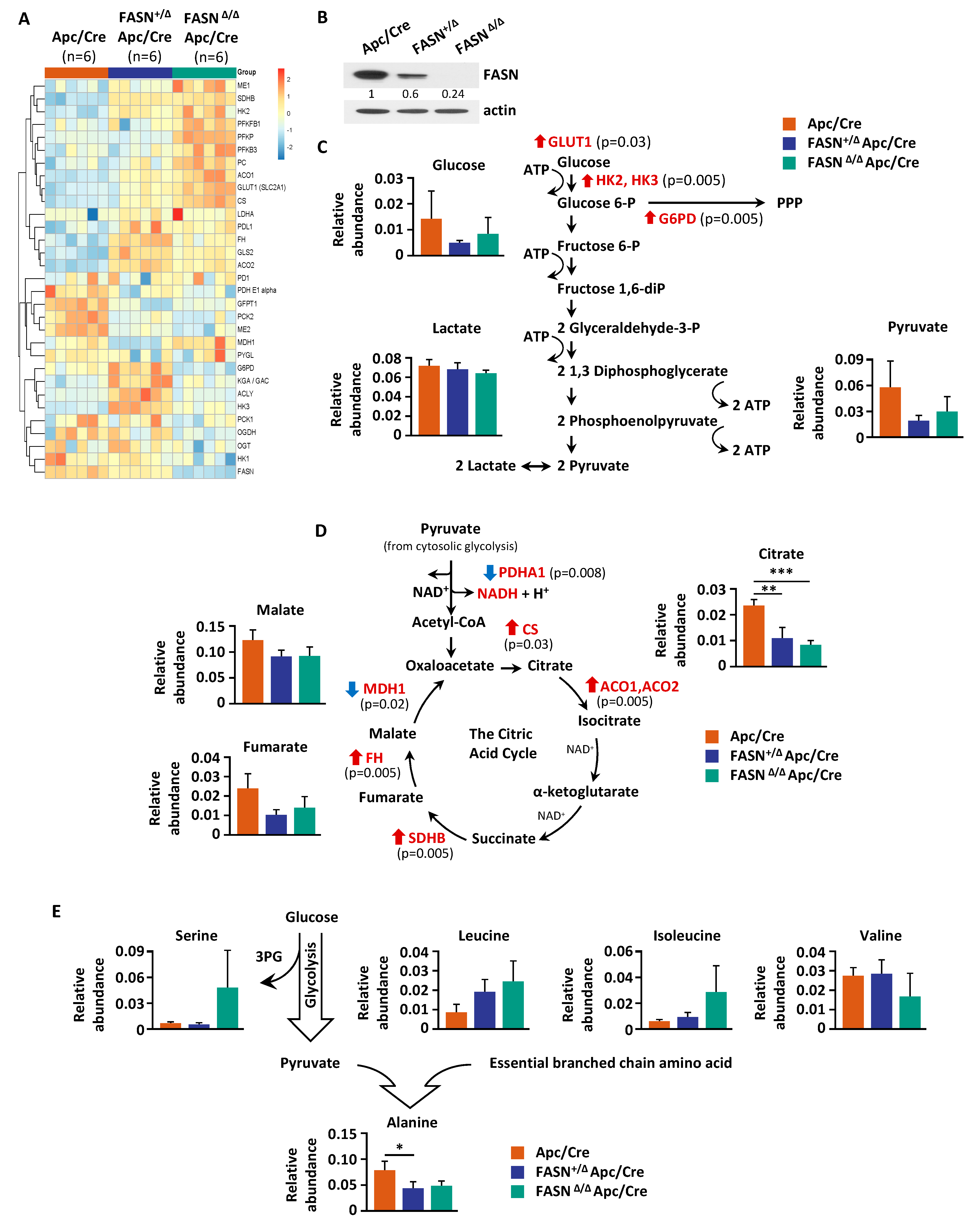
2.4. Downregulation of FASN in Mouse Adenomas Alters the Levels of Cellular Metabolites
2.5. Deletion of FASN Alters Expression of Metabolites and Their Metabolizing Enzymes in Adenomas
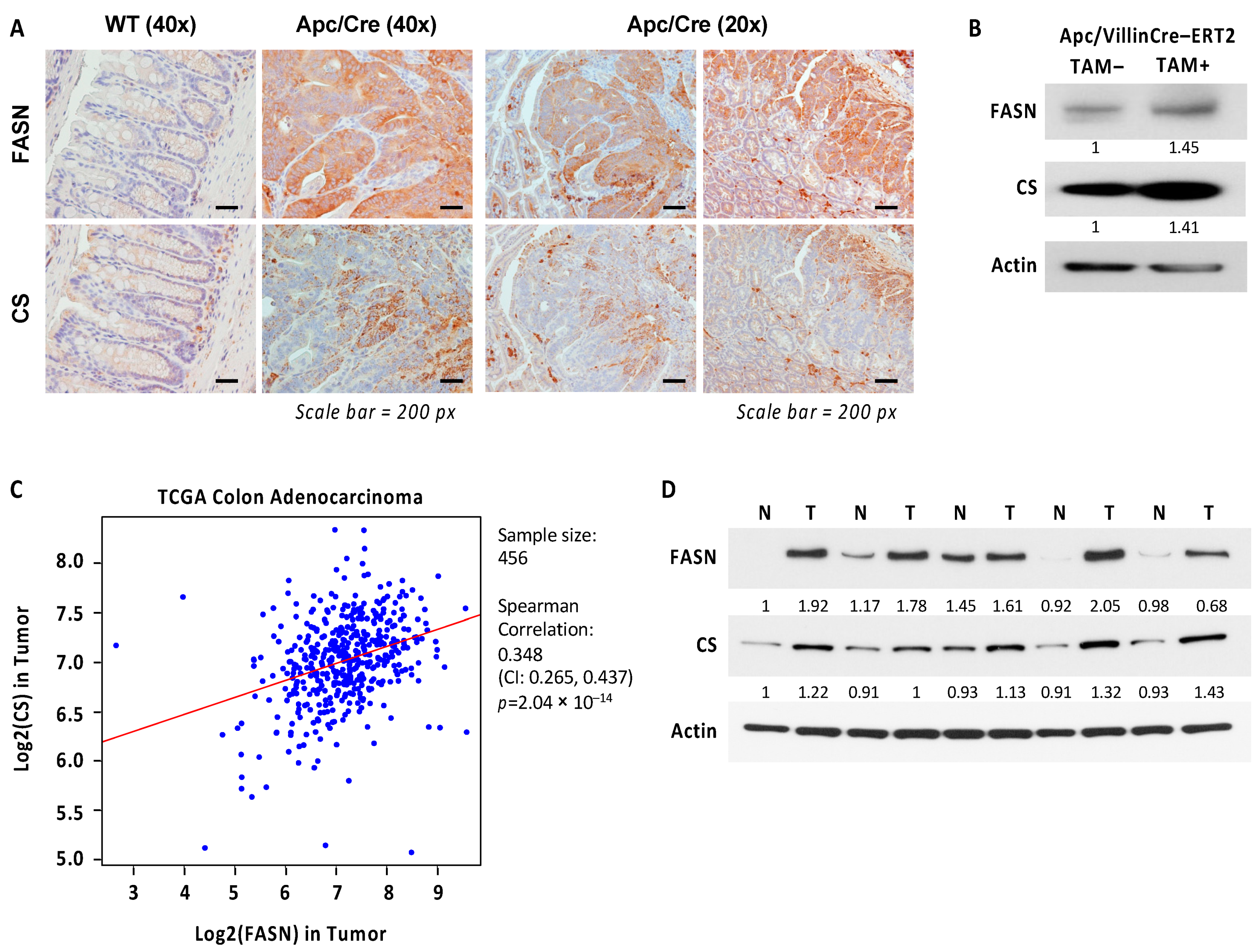
2.6. Expression of CS Is Upregulated and Correlates with Expression of FASN in Colorectal Cancer
3. Discussion
4. Materials and Methods
4.1. Mouse Colonies
4.2. Survival Analysis and Tumor Number Studies
4.3. Histologic Analysis and Immunohistochemical (IHC) Staining
4.4. Western Blot Analysis
4.5. Antibodies for Western Blot and IHC Staining
4.6. RNA-Sequencing and Gene Set Enrichment Analysis
4.7. Metabolite Extraction
4.8. Sample Derivatization and Gas Chromatography-Mass Spectrometry (GC-MS) Quantification
4.9. Metabolite Analysis
4.10. Lipidomic Analysis
4.11. Reverse Phase Protein Analysis
4.12. Analysis of Correlation between FASN and Citrate Synthase (CS)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Menendez, J.A.; Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 2007, 7, 763–777. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Geng, F.; Cheng, X.; Guo, D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. 2018, 38, 27. [Google Scholar] [CrossRef] [PubMed]
- Zaytseva, Y. Lipid Metabolism as a Targetable Metabolic Vulnerability in Colorectal Cancer. Cancers 2021, 13, 301. [Google Scholar] [CrossRef] [PubMed]
- Buckley, D.; Duke, G.; Heuer, T.S.; O’Farrell, M.; Wagman, A.S.; McCulloch, W.; Kemble, G. Fatty acid synthase—Modern tumor cell biology insights into a classical oncology target. Pharmacol. Ther. 2017, 177, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Cheng, C.; Tan, Z.; Li, N.; Tang, M.; Yang, L.; Cao, Y. Emerging roles of lipid metabolism in cancer metastasis. Mol. Cancer 2017, 16, 76. [Google Scholar] [CrossRef] [Green Version]
- Zaytseva, Y.Y.; Rychahou, P.G.; Gulhati, P.; Elliott, V.A.; Mustain, W.C.; O’Connor, K.; Morris, A.J.; Sunkara, M.; Weiss, H.L.; Lee, E.Y.; et al. Inhibition of fatty acid synthase attenuates CD44-associated signaling and reduces metastasis in colorectal cancer. Cancer Res. 2012, 72, 1504–1517. [Google Scholar] [CrossRef] [Green Version]
- Elliott, V.A.; Rychahou, P.; Zaytseva, Y.Y.; Evers, B.M. Activation of c-Met and upregulation of CD44 expression are associated with the metastatic phenotype in the colorectal cancer liver metastasis model. PLoS ONE 2014, 9, e97432. [Google Scholar] [CrossRef]
- Zaytseva, Y.Y.; Harris, J.W.; Mitov, M.I.; Kim, J.T.; Butterfield, D.A.; Lee, E.Y.; Weiss, H.L.; Gao, T.; Evers, B.M. Increased expression of fatty acid synthase provides a survival advantage to colorectal cancer cells via upregulation of cellular respiration. Oncotarget 2015, 6, 18891–18904. [Google Scholar] [CrossRef] [Green Version]
- Zaytseva, Y.Y.; Rychahou, P.G.; Le, A.T.; Scott, T.L.; Flight, R.M.; Kim, J.T.; Harris, J.; Liu, J.; Wang, C.; Morris, A.J.; et al. Preclinical evaluation of novel fatty acid synthase inhibitors in primary colorectal cancer cells and a patient-derived xenograft model of colorectal cancer. Oncotarget 2018, 9, 24787–24800. [Google Scholar] [CrossRef] [Green Version]
- Ventura, R.; Mordec, K.; Waszczuk, J.; Wang, Z.; Lai, J.; Fridlib, M.; Buckley, D.; Kemble, G.; Heuer, T.S. Inhibition of de novo Palmitate Synthesis by Fatty Acid Synthase Induces Apoptosis in Tumor Cells by Remodeling Cell Membranes, Inhibiting Signaling Pathways, and Reprogramming Gene Expression. eBioMedicine 2015, 2, 806–822. [Google Scholar] [CrossRef] [Green Version]
- Flavin, R.; Peluso, S.; Nguyen, P.L.; Loda, M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010, 6, 551–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Cancer Institute. Clinical Trials Using FASN Inhibitor TVB-2640. Available online: www.cancer.gov/about-cancer/treatment/clinical-trials/intervention/C118285 (accessed on 31 January 2022).
- Satoh, K.; Yachida, S.; Sugimoto, M.; Oshima, M.; Nakagawa, T.; Akamoto, S.; Tabata, S.; Saitoh, K.; Kato, K.; Sato, S.; et al. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc. Natl. Acad. Sci. USA 2017, 114, E7697–E7706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, D.S.; Archer, M.C. Fatty acid synthase is over-expressed in large aberrant crypt foci in rats treated with azoxymethane. Int. J. Cancer 2009, 124, 2750–2753. [Google Scholar] [CrossRef] [PubMed]
- Visca, P.; Alo, P.L.; Del Nonno, F.; Botti, C.; Trombetta, G.; Marandino, F.; Filippi, S.; Di Tondo, U.; Donnorso, R.P. Immunohistochemical expression of fatty acid synthase, apoptotic-regulating genes, proliferating factors, and ras protein product in colorectal adenomas, carcinomas, and adjacent nonneoplastic mucosa. Clin. Cancer Res. 1999, 5, 4111–4118. [Google Scholar]
- Kearney, K.E.; Pretlow, T.G.; Pretlow, T.P. Increased expression of fatty acid synthase in human aberrant crypt foci: Possible target for colorectal cancer prevention. Int. J. Cancer 2009, 125, 249–252. [Google Scholar] [CrossRef]
- Cruz, M.D.; Wali, R.K.; Bianchi, L.K.; Radosevich, A.J.; Crawford, S.E.; Jepeal, L.; Goldberg, M.J.; Weinstein, J.; Momi, N.; Roy, P.; et al. Colonic mucosal fatty acid synthase as an early biomarker for colorectal neoplasia: Modulation by obesity and gender. Cancer Epidemiol. Biomark. Prev. 2014, 23, 2413–2421. [Google Scholar] [CrossRef] [Green Version]
- Bueno, M.J.; Jimenez-Renard, V.; Samino, S.; Capellades, J.; Junza, A.; Lopez-Rodriguez, M.L.; Garcia-Carceles, J.; Lopez-Fabuel, I.; Bolanos, J.P.; Chandel, N.S.; et al. Essentiality of fatty acid synthase in the 2D to anchorage-independent growth transition in transforming cells. Nat. Commun. 2019, 10, 5011. [Google Scholar] [CrossRef]
- Yang, J.; Yu, J. The association of diet, gut microbiota and colorectal cancer: What we eat may imply what we get. Protein Cell 2018, 9, 474–487. [Google Scholar] [CrossRef] [Green Version]
- Tabung, F.K.; Liu, L.; Wang, W.; Fung, T.T.; Wu, K.; Smith-Warner, S.A.; Cao, Y.; Hu, F.B.; Ogino, S.; Fuchs, C.S.; et al. Association of Dietary Inflammatory Potential with Colorectal Cancer Risk in Men and Women. JAMA Oncol. 2018, 4, 366–373. [Google Scholar] [CrossRef] [Green Version]
- Drury, J.; Rychahou, P.G.; He, D.; Jafari, N.; Wang, C.; Lee, E.Y.; Weiss, H.L.; Evers, B.M.; Zaytseva, Y.Y. Inhibition of Fatty Acid Synthase Upregulates Expression of CD36 to Sustain Proliferation of Colorectal Cancer Cells. Front. Oncol. 2020, 10, 1185. [Google Scholar] [CrossRef]
- McCart, A.E.; Vickaryous, N.K.; Silver, A. Apc mice: Models, modifiers and mutants. Pathol. Res. Pract. 2008, 204, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, M.V.; Pan, Z.; Zhu, Y.; Tordjman, K.; Schneider, J.G.; Coleman, T.; Turk, J.; Semenkovich, C.F. “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 2005, 1, 309–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, X.; Yang, Z.; Rey, F.E.; Ridaura, V.K.; Davidson, N.O.; Gordon, J.I.; Semenkovich, C.F. Fatty acid synthase modulates intestinal barrier function through palmitoylation of mucin 2. Cell Host Microbe 2012, 11, 140–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagy, A. Cre recombinase: The universal reagent for genome tailoring. Genesis 2000, 26, 99–109. [Google Scholar] [CrossRef]
- Li, L.T.; Jiang, G.; Chen, Q.; Zheng, J.N. Ki67 is a promising molecular target in the diagnosis of cancer (review). Mol. Med. Rep. 2015, 11, 1566–1572. [Google Scholar] [CrossRef] [Green Version]
- Jafari, N.; Drury, J.; Morris, A.J.; Onono, F.O.; Stevens, P.D.; Gao, T.; Liu, J.; Wang, C.; Lee, E.Y.; Weiss, H.L.; et al. De Novo Fatty Acid Synthesis-Driven Sphingolipid Metabolism Promotes Metastatic Potential of Colorectal Cancer. Mol. Cancer Res. 2019, 17, 140–152. [Google Scholar] [CrossRef] [Green Version]
- Fan, T.W.M.; Bruntz, R.C.; Yang, Y.; Song, H.; Chernyavskaya, Y.; Deng, P.; Zhang, Y.; Shah, P.P.; Beverly, L.J.; Qi, Z.; et al. De novo synthesis of serine and glycine fuels purine nucleotide biosynthesis in human lung cancer tissues. J. Biol. Chem. 2019, 294, 13464–13477. [Google Scholar] [CrossRef]
- Liu, Z.; Yu, M.; Fei, B.; Fang, X.; Ma, T.; Wang, D. miR215p targets PDHA1 to regulate glycolysis and cancer progression in gastric cancer. Oncol. Rep. 2018, 40, 2955–2963. [Google Scholar] [CrossRef]
- Murai, S.; Ando, A.; Ebara, S.; Hirayama, M.; Satomi, Y.; Hara, T. Inhibition of malic enzyme 1 disrupts cellular metabolism and leads to vulnerability in cancer cells in glucose-restricted conditions. Oncogenesis 2017, 6, e329. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef] [Green Version]
- Balsa-Martinez, E.; Puigserver, P. Cancer Cells Hijack Gluconeogenic Enzymes to Fuel Cell Growth. Mol. Cell 2015, 60, 509–511. [Google Scholar] [CrossRef] [PubMed]
- Lieu, E.L.; Nguyen, T.; Rhyne, S.; Kim, J. Amino acids in cancer. Exp. Mol. Med. 2020, 52, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Liu, X.; Cheng, C.; Yu, W.; Yi, P. Metabolism of Amino Acids in Cancer. Front. Cell Dev. Biol. 2020, 8, 603837. [Google Scholar] [CrossRef]
- Galim, E.B.; Hruska, K.; Bier, D.M.; Matthews, D.E.; Haymond, M.W. Branched-chain amino acid nitrogen transfer to alamine in vivo in dogs. Direct isotopic determination with [15N]leucine. J. Clin. Investig. 1980, 66, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.M.; Biancur, D.E.; Wang, X.; Halbrook, C.J.; Sherman, M.H.; Zhang, L.; Kremer, D.; Hwang, R.F.; Witkiewicz, A.K.; Ying, H.; et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 2016, 536, 479–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlichtholz, B.; Turyn, J.; Goyke, E.; Biernacki, M.; Jaskiewicz, K.; Sledzinski, Z.; Swierczynski, J. Enhanced citrate synthase activity in human pancreatic cancer. Pancreas 2005, 30, 99–104. [Google Scholar] [CrossRef]
- Kridel, S.J.; Axelrod, F.; Rozenkrantz, N.; Smith, J.W. Orlistat is a novel inhibitor of fatty acid synthase with antitumor activity. Cancer Res. 2004, 64, 2070–2075. [Google Scholar] [CrossRef] [Green Version]
- Dowling, S.; Cox, J.; Cenedella, R.J. Inhibition of fatty acid synthase by Orlistat accelerates gastric tumor cell apoptosis in culture and increases survival rates in gastric tumor bearing mice in vivo. Lipids 2009, 44, 489–498. [Google Scholar] [CrossRef]
- Notarnicola, M.; Barone, M.; Francavilla, A.; Tutino, V.; Bianco, G.; Tafaro, A.; Minoia, M.; Polimeno, L.; Napoli, A.; Scavo, M.P.; et al. Lovastatin, but not orlistat, reduces intestinal polyp volume in an ApcMin/+ mouse model. Oncol. Rep. 2016, 36, 893–899. [Google Scholar] [CrossRef] [Green Version]
- Migita, T.; Ruiz, S.; Fornari, A.; Fiorentino, M.; Priolo, C.; Zadra, G.; Inazuka, F.; Grisanzio, C.; Palescandolo, E.; Shin, E.; et al. Fatty acid synthase: A metabolic enzyme and candidate oncogene in prostate cancer. J. Natl. Cancer Inst. 2009, 101, 519–532. [Google Scholar] [CrossRef]
- Alwarawrah, Y.; Hughes, P.; Loiselle, D.; Carlson, D.A.; Darr, D.B.; Jordan, J.L.; Xiong, J.; Hunter, L.M.; Dubois, L.G.; Thompson, J.W.; et al. Fasnall, a Selective FASN Inhibitor, Shows Potent Anti-tumor Activity in the MMTV-Neu Model of HER2(+) Breast Cancer. Cell Chem. Biol. 2016, 23, 678–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alli, P.M.; Pinn, M.L.; Jaffee, E.M.; McFadden, J.M.; Kuhajda, F.P. Fatty acid synthase inhibitors are chemopreventive for mammary cancer in neu-N transgenic mice. Oncogene 2005, 24, 39–46. [Google Scholar] [CrossRef] [Green Version]
- Luo, Z.W.; Zhu, M.G.; Zhang, Z.Q.; Ye, F.J.; Huang, W.H.; Luo, X.Z. Increased expression of Ki-67 is a poor prognostic marker for colorectal cancer patients: A meta analysis. BMC Cancer 2019, 19, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falchook, G.; Infante, J.; Arkenau, H.T.; Patel, M.R.; Dean, E.; Borazanci, E.; Brenner, A.; Cook, N.; Lopez, J.; Pant, S.; et al. First-in-human study of the safety, pharmacokinetics, and pharmacodynamics of first-in-class fatty acid synthase inhibitor TVB-2640 alone and with a taxane in advanced tumors. eClinicalMedicine 2021, 34, 100797. [Google Scholar] [CrossRef] [PubMed]
- Jensen-Urstad, A.P.; Semenkovich, C.F. Fatty acid synthase and liver triglyceride metabolism: Housekeeper or messenger? Biochim. Biophys. Acta 2012, 1821, 747–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nenkov, M.; Ma, Y.; Gassler, N.; Chen, Y. Metabolic Reprogramming of Colorectal Cancer Cells and the Microenvironment: Implication for Therapy. Int. J. Mol. Sci. 2021, 22, 6262. [Google Scholar] [CrossRef] [PubMed]
- Bruning, U.; Morales-Rodriguez, F.; Kalucka, J.; Goveia, J.; Taverna, F.; Queiroz, K.C.S.; Dubois, C.; Cantelmo, A.R.; Chen, R.; Loroch, S.; et al. Impairment of Angiogenesis by Fatty Acid Synthase Inhibition Involves mTOR Malonylation. Cell Metab. 2018, 28, 866–880.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Browne, C.D.; Hindmarsh, E.J.; Smith, J.W. Inhibition of endothelial cell proliferation and angiogenesis by orlistat, a fatty acid synthase inhibitor. FASEB J. 2006, 20, 2027–2035. [Google Scholar] [CrossRef]
- Xiong, W.; Sun, K.Y.; Zhu, Y.; Zhang, X.; Zhou, Y.H.; Zou, X. Metformin alleviates inflammation through suppressing FASN-dependent palmitoylation of Akt. Cell Death Dis. 2021, 12, 934. [Google Scholar] [CrossRef]
- Qian, X.; Yang, Z.; Mao, E.; Chen, E. Regulation of fatty acid synthesis in immune cells. Scand. J. Immunol. 2018, 88, e12713. [Google Scholar] [CrossRef]
- Gong, J.; Lin, Y.; Zhang, H.; Liu, C.; Cheng, Z.; Yang, X.; Zhang, J.; Xiao, Y.; Sang, N.; Qian, X.; et al. Reprogramming of lipid metabolism in cancer-associated fibroblasts potentiates migration of colorectal cancer cells. Cell Death Dis. 2020, 11, 267. [Google Scholar] [CrossRef] [PubMed]
- Icard, P.; Coquerel, A.; Wu, Z.; Gligorov, J.; Fuks, D.; Fournel, L.; Lincet, H.; Simula, L. Understanding the Central Role of Citrate in the Metabolism of Cancer Cells and Tumors: An Update. Int. J. Mol. Sci. 2021, 22, 6587. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, T.; Zhou, J.; Wang, Y.; Wang, X.; Di, W.; Zhang, S. Citrate synthase expression affects tumor phenotype and drug resistance in human ovarian carcinoma. PLoS ONE 2014, 9, e115708. [Google Scholar] [CrossRef] [Green Version]
- Haferkamp, S.; Drexler, K.; Federlin, M.; Schlitt, H.J.; Berneburg, M.; Adamski, J.; Gaumann, A.; Geissler, E.K.; Ganapathy, V.; Parkinson, E.K.; et al. Extracellular Citrate Fuels Cancer Cell Metabolism and Growth. Front. Cell Dev. Biol. 2020, 8, 602476. [Google Scholar] [CrossRef]
- Drexler, K.; Schmidt, K.M.; Jordan, K.; Federlin, M.; Milenkovic, V.M.; Liebisch, G.; Artati, A.; Schmidl, C.; Madej, G.; Tokarz, J.; et al. Cancer-associated cells release citrate to support tumour metastatic progression. Life Sci. Alliance 2021, 4, e202000903. [Google Scholar] [CrossRef] [PubMed]
- Salas, M.L.; Vinuela, E.; Salas, M.; Sols, A. Citrate Inhibition of Phosphofructokinase and the Pasteur Effect. Biochem. Biophys. Res. Commun. 1965, 19, 371–376. [Google Scholar] [CrossRef]
- Ren, M.; Yang, X.; Bie, J.; Wang, Z.; Liu, M.; Li, Y.; Shao, G.; Luo, J. Citrate synthase desuccinylation by SIRT5 promotes colon cancer cell proliferation and migration. Biol. Chem. 2020, 401, 1031–1039. [Google Scholar] [CrossRef]
- Denkert, C.; Budczies, J.; Weichert, W.; Wohlgemuth, G.; Scholz, M.; Kind, T.; Niesporek, S.; Noske, A.; Buckendahl, A.; Dietel, M.; et al. Metabolite profiling of human colon carcinoma–deregulation of TCA cycle and amino acid turnover. Mol. Cancer 2008, 7, 72. [Google Scholar] [CrossRef] [Green Version]
- Vettore, L.; Westbrook, R.L.; Tennant, D.A. New aspects of amino acid metabolism in cancer. Br. J. Cancer 2020, 122, 150–156. [Google Scholar] [CrossRef]
- Choi, B.H.; Coloff, J.L. The Diverse Functions of Non-Essential Amino Acids in Cancer. Cancers 2019, 11, 675. [Google Scholar] [CrossRef] [Green Version]
- Parker, S.J.; Amendola, C.R.; Hollinshead, K.E.R.; Yu, Q.; Yamamoto, K.; Encarnacion-Rosado, J.; Rose, R.E.; LaRue, M.M.; Sohn, A.S.W.; Biancur, D.E.; et al. Selective Alanine Transporter Utilization Creates a Targetable Metabolic Niche in Pancreatic Cancer. Cancer Discov. 2020, 10, 1018–1037. [Google Scholar] [CrossRef] [PubMed]
- Mika, A.; Duzowska, K.; Halinski, L.P.; Pakiet, A.; Czumaj, A.; Rostkowska, O.; Dobrzycka, M.; Kobiela, J.; Sledzinski, T. Rearrangements of Blood and Tissue Fatty Acid Profile in Colorectal Cancer-Molecular Mechanism and Diagnostic Potential. Front. Oncol. 2021, 11, 689701. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mootha, V.; Lindgren, C.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.; Han, Y.; He, Q. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Andres, D.A.; Young, L.E.A.; Veeranki, S.; Hawkinson, T.R.; Levitan, B.M.; He, D.; Wang, C.; Satin, J.; Sun, R.C. Improved workflow for mass spectrometry-based metabolomics analysis of the heart. J. Biol. Chem. 2020, 295, 2676–2686. [Google Scholar] [CrossRef]
- Brewer, M.K.; Uittenbogaard, A.; Austin, G.L.; Segvich, D.M.; DePaoli-Roach, A.; Roach, P.J.; McCarthy, J.J.; Simmons, Z.R.; Brandon, J.A.; Zhou, Z. Targeting pathogenic Lafora bodies in Lafora disease using an antibody-enzyme fusion. Cell Metab. 2019, 30, 689–705.e6. [Google Scholar] [CrossRef]
- Sun, R.C.; Dukhande, V.V.; Zhou, Z.; Young, L.E.; Emanuelle, S.; Brainson, C.F.; Gentry, M.S. Nuclear glycogenolysis modulates histone acetylation in human non-small cell lung cancers. Cell Metab. 2019, 30, 903–916.e7. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics by gas chromatography–mass spectrometry: Combined targeted and untargeted profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30–34. [Google Scholar] [CrossRef]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wei, Q.; Mao, L.; Liu, W.; Mills, G.B.; Coombes, K. Serial dilution curve: A new method for analysis of reverse phase protein array data. Bioinformatics 2009, 25, 650–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neeley, E.S.; Kornblau, S.M.; Coombes, K.R.; Baggerly, K.A. Variable slope normalization of reverse phase protein arrays. Bioinformatics 2009, 25, 1384–1389. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drury, J.; Young, L.E.A.; Scott, T.L.; Kelson, C.O.; He, D.; Liu, J.; Wu, Y.; Wang, C.; Weiss, H.L.; Fan, T.; et al. Tissue-Specific Downregulation of Fatty Acid Synthase Suppresses Intestinal Adenoma Formation via Coordinated Reprograming of Transcriptome and Metabolism in the Mouse Model of Apc-Driven Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 6510. https://doi.org/10.3390/ijms23126510
Drury J, Young LEA, Scott TL, Kelson CO, He D, Liu J, Wu Y, Wang C, Weiss HL, Fan T, et al. Tissue-Specific Downregulation of Fatty Acid Synthase Suppresses Intestinal Adenoma Formation via Coordinated Reprograming of Transcriptome and Metabolism in the Mouse Model of Apc-Driven Colorectal Cancer. International Journal of Molecular Sciences. 2022; 23(12):6510. https://doi.org/10.3390/ijms23126510
Chicago/Turabian StyleDrury, James, Lyndsay E. A. Young, Timothy L. Scott, Courtney O. Kelson, Daheng He, Jinpeng Liu, Yuanyan Wu, Chi Wang, Heidi L. Weiss, Teresa Fan, and et al. 2022. "Tissue-Specific Downregulation of Fatty Acid Synthase Suppresses Intestinal Adenoma Formation via Coordinated Reprograming of Transcriptome and Metabolism in the Mouse Model of Apc-Driven Colorectal Cancer" International Journal of Molecular Sciences 23, no. 12: 6510. https://doi.org/10.3390/ijms23126510





