Generation of TRIM28 Knockout K562 Cells by CRISPR/Cas9 Genome Editing and Characterization of TRIM28-Regulated Gene Expression in Cell Proliferation and Hemoglobin Beta Subunits
Abstract
:1. Introduction
2. Results
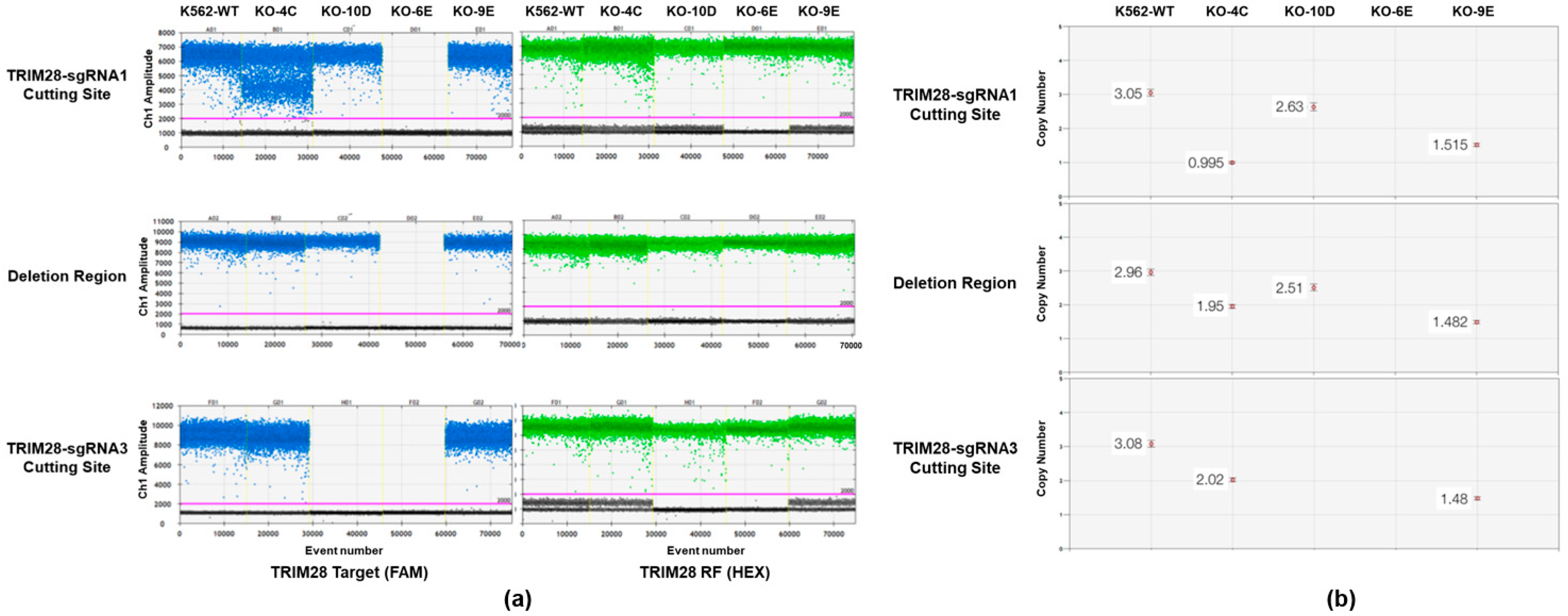
2.1. Knockout of TRIM28 in K562 Using CRISPR/Cas9 Mediated Genome Editing
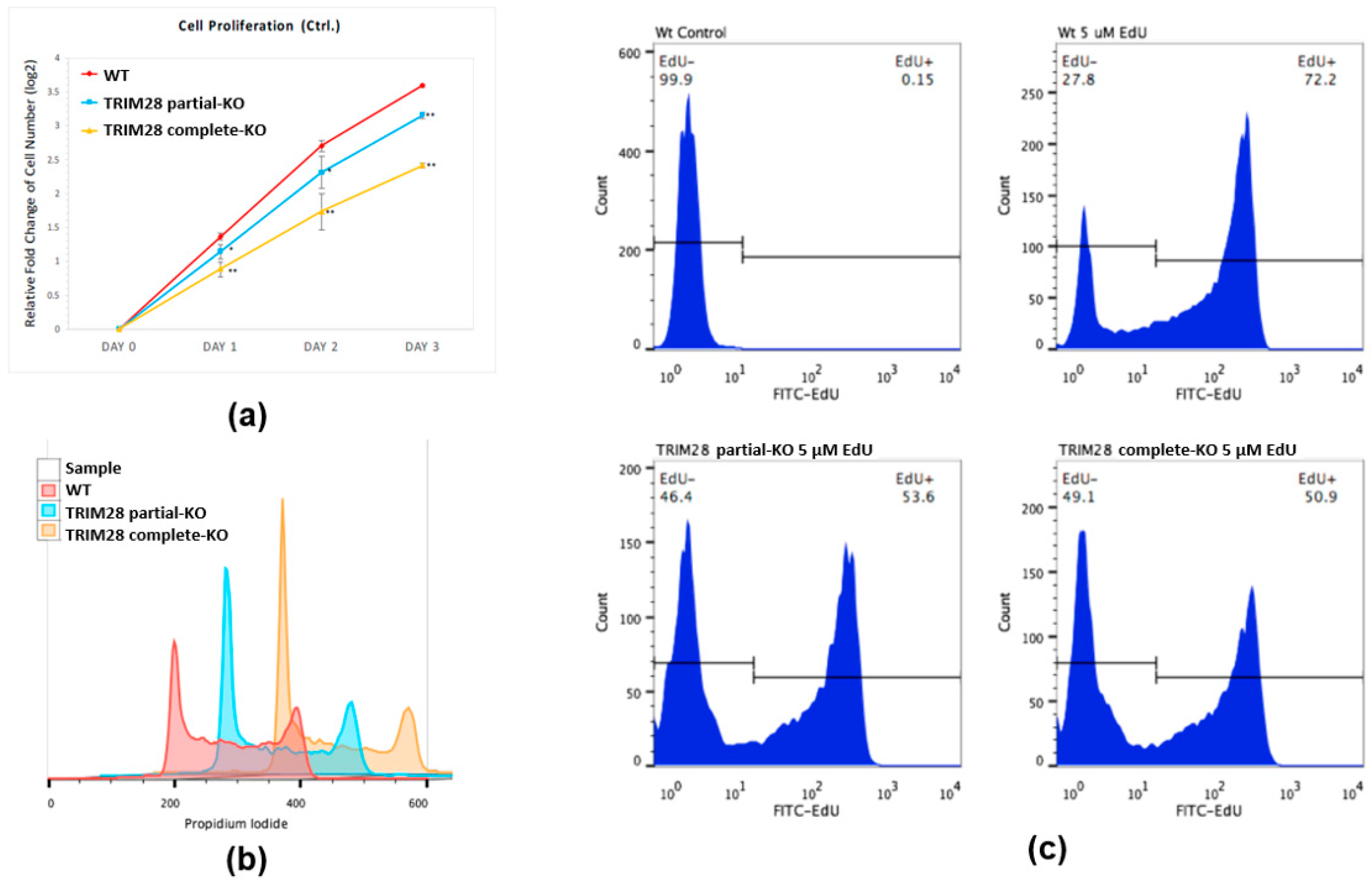
2.2. Knockout of TRIM28 Inhibits the Cell Proliferation
2.3. TRIM28 Regulates Cell Proliferation-Related Gene Expression
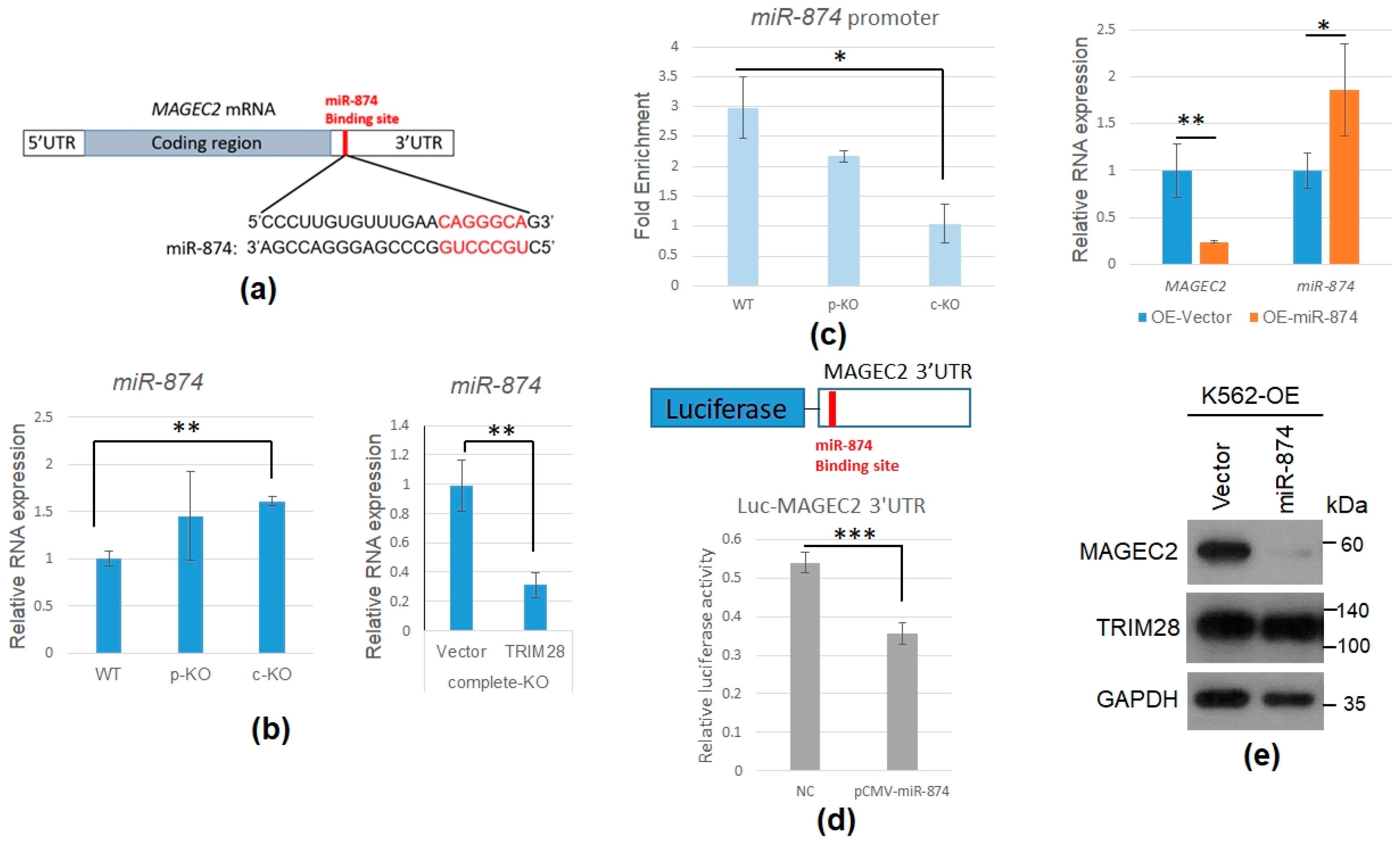
2.4. TRIM28 Involves in MAGEC2 mRNA Expression
2.5. TRIM28 KO Induces miR-874 to Downregulate MAGEC2 Expression
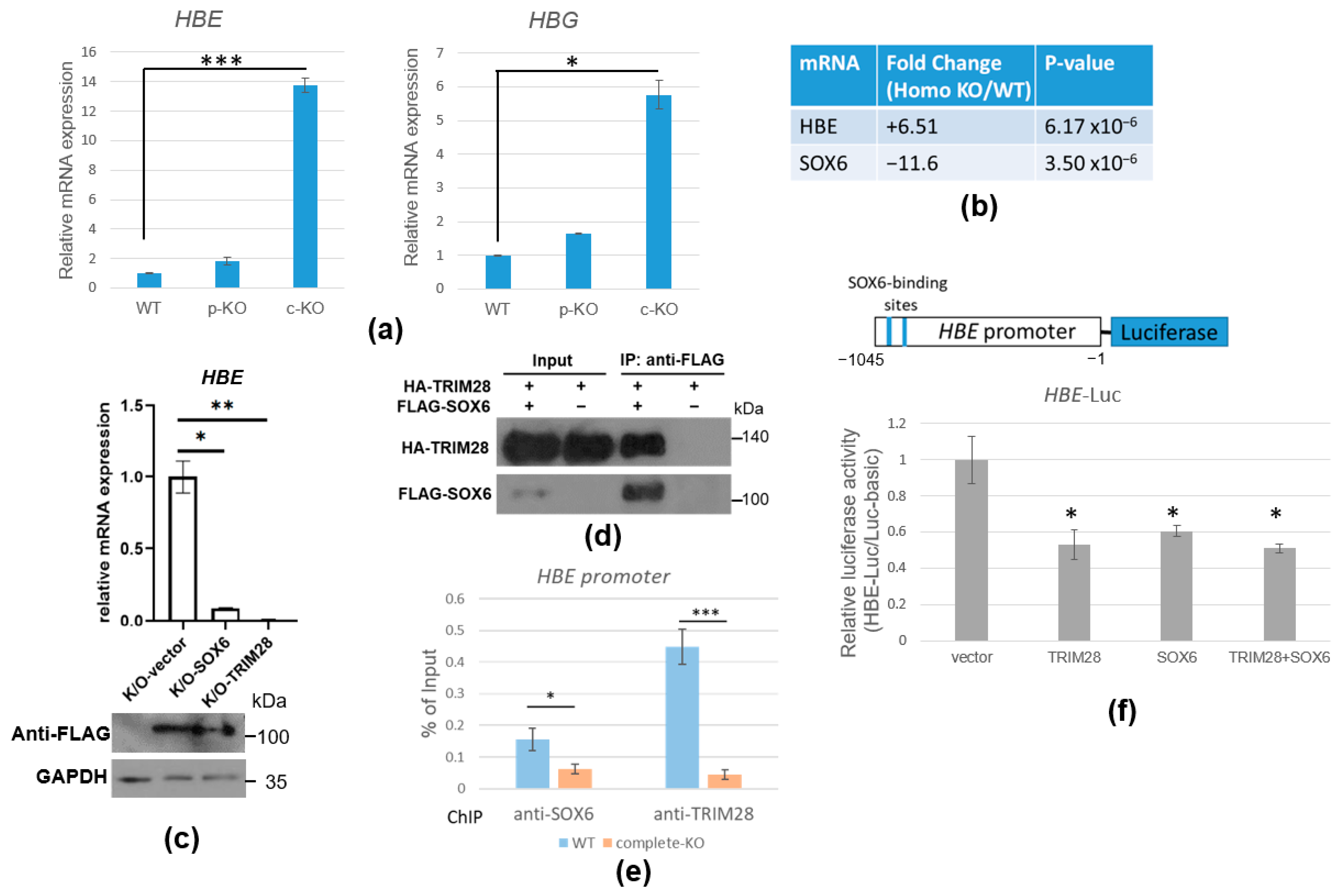
2.6. TRIM28 Regulates HBE mRNA Expression via SOX6 Transcription Repressor
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. Plasmids
4.3. CRISPR/Cas9-Mediated TRIM28 Knockout in K562 Cells
4.4. Analysis of TRIM28 Knockout by Genomic PCR
4.5. Copy Number Variation Detection by Quantitative Droplet Digital PCR (ddPCR)
4.6. RNA Isolation, Reverse Transcription and Quantitative PCR
4.7. Clariom D Whole Transcriptome Microarray Analysis
4.8. Preparation of Whole Cell Extracts and Western Blot Analysis
4.9. Cell Proliferation and Cell Viability Assay
4.10. Cell Cycle Analysis
4.11. Immunoprecipitation Assay
4.12. ChIP Assay
4.13. Overexpression of Trim28, SOX6 or miR-874 in TRIM28 Complete-KO K562 Cells
4.14. Dual Luciferase Reporter Assay
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Friedman, J.R.; Fredericks, W.J.; Jensen, D.E.; Speicher, D.W.; Huang, X.P.; Neilson, E.G.; Rauscher, F.J., 3rd. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996, 10, 2067–2078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, H.; Begg, G.E.; Schultz, D.C.; Friedman, J.R.; Jensen, D.E.; Speicher, D.W.; Rauscher, F.J., 3rd. Reconstitution of the KRAB-KAP-1 repressor complex: A model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J. Mol. Biol. 2000, 295, 1139–1162. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Keown, J.R.; Black, M.M.; Raclot, C.; Demarais, N.; Trono, D.; Turelli, P.; Goldstone, D.C. A Dissection of Oligomerization by the TRIM28 Tripartite Motif and the Interaction with Members of the Krab-ZFP Family. J. Mol. Biol. 2019, 431, 2511–2527. [Google Scholar] [CrossRef] [PubMed]
- Stoll, G.A.; Oda, S.I.; Chong, Z.S.; Yu, M.; McLaughlin, S.H.; Modis, Y. Structure of KAP1 tripartite motif identifies molecular interfaces required for retroelement silencing. Proc. Natl. Acad. Sci. USA 2019, 116, 15042–15051. [Google Scholar] [CrossRef] [Green Version]
- Schultz, D.C.; Friedman, J.R.; Rauscher, F.J., 3rd. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: The PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001, 15, 428–443. [Google Scholar] [CrossRef] [Green Version]
- Schultz, D.C.; Ayyanathan, K.; Negorev, D.; Maul, G.G.; Rauscher, F.J., 3rd. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002, 16, 919–932. [Google Scholar] [CrossRef] [Green Version]
- Ryan, R.F.; Schultz, D.C.; Ayyanathan, K.; Singh, P.B.; Friedman, J.R.; Fredericks, W.J.; Rauscher, F.J., 3rd. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: A potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell Biol. 1999, 19, 4366–4378. [Google Scholar] [CrossRef]
- Quenneville, S.; Turelli, P.; Bojkowska, K.; Raclot, C.; Offner, S.; Kapopoulou, A.; Trono, D. The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Rep. 2012, 2, 766–773. [Google Scholar] [CrossRef] [Green Version]
- Meroni, G.; Diez-Roux, G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays 2005, 27, 1147–1157. [Google Scholar] [CrossRef]
- Lee, A.K.; Potts, P.R. A Comprehensive Guide to the MAGE Family of Ubiquitin Ligases. J. Mol. Biol. 2017, 429, 1114–1142. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.W.; Huang, X.; Lin, W.; Min, J.; Miller, D.J.; Mayasundari, A.; Rodrigues, P.; Griffith, E.C.; Gee, C.T.; Li, L.; et al. Structural basis for substrate recognition and chemical inhibition of oncogenic MAGE ubiquitin ligases. Nat. Commun. 2020, 11, 4931. [Google Scholar] [CrossRef] [PubMed]
- Lian, Y.; Meng, L.; Ding, P.; Sang, M. Epigenetic regulation of MAGE family in human cancer progression-DNA methylation, histone modification, and non-coding RNAs. Clin. Epigenet. 2018, 10, 115. [Google Scholar] [CrossRef] [PubMed]
- Cammas, F.; Mark, M.; Dolle, P.; Dierich, A.; Chambon, P.; Losson, R. Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development 2000, 127, 2955–2963. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Kim, J.; Xu, Q.; Leng, Y.; Orkin, S.H.; Elledge, S.J. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009, 23, 837–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seki, Y.; Kurisaki, A.; Watanabe-Susaki, K.; Nakajima, Y.; Nakanishi, M.; Arai, Y.; Shiota, K.; Sugino, H.; Asashima, M. TIF1beta regulates the pluripotency of embryonic stem cells in a phosphorylation-dependent manner. Proc. Natl. Acad. Sci. USA 2010, 107, 10926–10931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santoni de Sio, F.R.; Barde, I.; Offner, S.; Kapopoulou, A.; Corsinotti, A.; Bojkowska, K.; Genolet, R.; Thomas, J.H.; Luescher, I.F.; Pinschewer, D.; et al. KAP1 regulates gene networks controlling T-cell development and responsiveness. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2012, 26, 4561–4575. [Google Scholar] [CrossRef] [Green Version]
- Santoni de Sio, F.R.; Massacand, J.; Barde, I.; Offner, S.; Corsinotti, A.; Kapopoulou, A.; Bojkowska, K.; Dagklis, A.; Fernandez, M.; Ghia, P.; et al. KAP1 regulates gene networks controlling mouse B-lymphoid cell differentiation and function. Blood 2012, 119, 4675–4685. [Google Scholar] [CrossRef] [Green Version]
- Barde, I.; Rauwel, B.; Marin-Florez, R.M.; Corsinotti, A.; Laurenti, E.; Verp, S.; Offner, S.; Marquis, J.; Kapopoulou, A.; Vanicek, J.; et al. A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science 2013, 340, 350–353. [Google Scholar] [CrossRef] [Green Version]
- Hosoya, T.; Clifford, M.; Losson, R.; Tanabe, O.; Engel, J.D. TRIM28 is essential for erythroblast differentiation in the mouse. Blood 2013, 122, 3798–3807. [Google Scholar] [CrossRef] [Green Version]
- Miyagi, S.; Koide, S.; Saraya, A.; Wendt, G.R.; Oshima, M.; Konuma, T.; Yamazaki, S.; Mochizuki-Kashio, M.; Nakajima-Takagi, Y.; Wang, C.; et al. The TIF1beta-HP1 system maintains transcriptional integrity of hematopoietic stem cells. Stem Cell Rep. 2014, 2, 145–152. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Park, J.W.; Kang, J.Y.; Seo, S.B. Negative Regulation of Erythroid Differentiation via the CBX8-TRIM28 Axis. Mol. Cells 2021, 44, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Lozzio, C.B.; Lozzio, B.B. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood 1975, 45, 321–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raitano, A.B.; Halpern, J.R.; Hambuch, T.M.; Sawyers, C.L. The Bcr-Abl leukemia oncogene activates Jun kinase and requires Jun for transformation. Proc. Natl. Acad. Sci. USA 1995, 92, 11746–11750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsiftsoglou, A.S.; Pappas, I.S.; Vizirianakis, I.S. Mechanisms involved in the induced differentiation of leukemia cells. Pharmacol. Ther. 2003, 100, 257–290. [Google Scholar] [CrossRef]
- Schechter, A.N. Hemoglobin research and the origins of molecular medicine. Blood 2008, 112, 3927–3938. [Google Scholar] [CrossRef] [Green Version]
- Dean, A.; Ley, T.J.; Humphries, R.K.; Fordis, M.; Schechter, A.N. Inducible transcription of five globin genes in K562 human leukemia cells. Proc. Natl. Acad. Sci. USA 1983, 80, 5515–5519. [Google Scholar] [CrossRef] [Green Version]
- Yi, Z.; Cohen-Barak, O.; Hagiwara, N.; Kingsley, P.D.; Fuchs, D.A.; Erickson, D.T.; Epner, E.M.; Palis, J.; Brilliant, M.H. Sox6 directly silences epsilon globin expression in definitive erythropoiesis. PLoS Genet. 2006, 2, e14. [Google Scholar] [CrossRef]
- Li, J.; Lai, Y.; Luo, J.; Luo, L.; Liu, R.; Liu, Z.; Zhao, W. SOX6 Downregulation Induces gamma-Globin in Human beta-Thalassemia Major Erythroid Cells. BioMed Res. Int. 2017, 2017, 9496058. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Sankaran, V.G.; Ni, M.; Menne, T.F.; Puram, R.V.; Kim, W.; Orkin, S.H. Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 2010, 24, 783–798. [Google Scholar] [CrossRef] [Green Version]
- Cui, S.; Kolodziej, K.E.; Obara, N.; Amaral-Psarris, A.; Demmers, J.; Shi, L.; Engel, J.D.; Grosveld, F.; Strouboulis, J.; Tanabe, O. Nuclear receptors TR2 and TR4 recruit multiple epigenetic transcriptional corepressors that associate specifically with the embryonic beta-type globin promoters in differentiated adult erythroid cells. Mol. Cell Biol. 2011, 31, 3298–3311. [Google Scholar] [CrossRef] [Green Version]
- Naumann, S.; Reutzel, D.; Speicher, M.; Decker, H.J. Complete karyotype characterization of the K562 cell line by combined application of G-banding, multiplex-fluorescence in situ hybridization, fluorescence in situ hybridization, and comparative genomic hybridization. Leuk Res. 2001, 25, 313–322. [Google Scholar] [CrossRef]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedlazeck, F.J.; Rescheneder, P.; Smolka, M.; Fang, H.; Nattestad, M.; von Haeseler, A.; Schatz, M.C. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 2018, 15, 461–468. [Google Scholar] [CrossRef] [Green Version]
- Corbisier, P.; Pinheiro, L.; Mazoua, S.; Kortekaas, A.M.; Chung, P.Y.; Gerganova, T.; Roebben, G.; Emons, H.; Emslie, K. DNA copy number concentration measured by digital and droplet digital quantitative PCR using certified reference materials. Anal. Bioanal. Chem. 2015, 407, 1831–1840. [Google Scholar] [CrossRef] [Green Version]
- Deprez, L.; Corbisier, P.; Kortekaas, A.M.; Mazoua, S.; Beaz Hidalgo, R.; Trapmann, S.; Emons, H. Validation of a digital PCR method for quantification of DNA copy number concentrations by using a certified reference material. Biomol. Detect. Quantif. 2016, 9, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.E.; Hakim, C.H.; Ousterout, D.G.; Thakore, P.I.; Moreb, E.A.; Castellanos Rivera, R.M.; Madhavan, S.; Pan, X.; Ran, F.A.; Yan, W.X.; et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016, 351, 403–407. [Google Scholar] [CrossRef] [Green Version]
- Bhatia, N.; Xiao, T.Z.; Rosenthal, K.A.; Siddiqui, I.A.; Thiyagarajan, S.; Smart, B.; Meng, Q.; Zuleger, C.L.; Mukhtar, H.; Kenney, S.C.; et al. MAGE-C2 promotes growth and tumorigenicity of melanoma cells, phosphorylation of KAP1, and DNA damage repair. J. Investig. Derm. 2013, 133, 759–767. [Google Scholar] [CrossRef] [Green Version]
- Weon, J.L.; Potts, P.R. The MAGE protein family and cancer. Curr. Opin. Cell Biol. 2015, 37, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Lajmi, N.; Luetkens, T.; Yousef, S.; Templin, J.; Cao, Y.; Hildebrandt, Y.; Bartels, K.; Kroger, N.; Atanackovic, D. Cancer-testis antigen MAGEC2 promotes proliferation and resistance to apoptosis in Multiple Myeloma. Br. J. Haematol. 2015, 171, 752–762. [Google Scholar] [CrossRef]
- Song, X.; Guo, C.; Zheng, Y.; Wang, Y.; Jin, Z.; Yin, Y. Post-transcriptional regulation of cancer/testis antigen MAGEC2 expression by TRIM28 in tumor cells. BMC Cancer 2018, 18, 971. [Google Scholar] [CrossRef]
- Song, X.; Song, W.; Wang, Y.; Wang, J.; Li, Y.; Qian, X.; Pang, X.; Zhang, Y.; Yin, Y. MicroRNA-874 Functions as a Tumor Suppressor by Targeting Cancer/Testis Antigen HCA587/MAGE-C2. J. Cancer 2016, 7, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czerwinska, P.; Mazurek, S.; Wiznerowicz, M. The complexity of TRIM28 contribution to cancer. J. Biomed. Sci. 2017, 24, 63. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Yoon, B.H.; Kim, S.K.; Kim, S.Y. GENT2: An updated gene expression database for normal and tumor tissues. BMC Med. Genom. 2019, 12, 101. [Google Scholar] [CrossRef] [PubMed]
- Iyengar, S.; Ivanov, A.V.; Jin, V.X.; Rauscher, F.J., 3rd; Farnham, P.J. Functional analysis of KAP1 genomic recruitment. Mol. Cell Biol. 2011, 31, 1833–1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Florke Gee, R.R.; Chen, H.; Lee, A.K.; Daly, C.A.; Wilander, B.A.; Fon Tacer, K.; Potts, P.R. Emerging roles of the MAGE protein family in stress response pathways. J. Biol. Chem. 2020, 295, 16121–16155. [Google Scholar] [CrossRef]
- Yang, B.; O’Herrin, S.M.; Wu, J.; Reagan-Shaw, S.; Ma, Y.; Bhat, K.M.; Gravekamp, C.; Setaluri, V.; Peters, N.; Hoffmann, F.M.; et al. MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apoptosis in MAGE-positive cell lines. Cancer Res. 2007, 67, 9954–9962. [Google Scholar] [CrossRef] [Green Version]
- Doyle, J.M.; Gao, J.; Wang, J.; Yang, M.; Potts, P.R. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol. Cell 2010, 39, 963–974. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Pan, Y.; Wang, L.; Zhang, L.; Ravichandran, R.; Potts, P.R.; Jiang, J.; Wu, H.; Huang, H. MAGE-TRIM28 complex promotes the Warburg effect and hepatocellular carcinoma progression by targeting FBP1 for degradation. Oncogenesis 2017, 6, e312. [Google Scholar] [CrossRef]
- Xiao, T.Z.; Suh, Y.; Longley, B.J. MAGE proteins regulate KRAB zinc finger transcription factors and KAP1 E3 ligase activity. Arch. Biochem. Biophys. 2014, 563, 136–144. [Google Scholar] [CrossRef] [Green Version]
- Pineda, C.T.; Ramanathan, S.; Fon Tacer, K.; Weon, J.L.; Potts, M.B.; Ou, Y.H.; White, M.A.; Potts, P.R. Degradation of AMPK by a cancer-specific ubiquitin ligase. Cell 2015, 160, 715–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Yan, D.L.; Yang, F.; Wang, D.D.; Chen, X.; Wu, J.Z.; Tang, J.H.; Xia, W.J. DNA methylation mediated silencing of microRNA-874 is a promising diagnosis and prognostic marker in breast cancer. Oncotarget 2017, 8, 45496–45505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.; Wang, W.; Zhao, Y.; Zhao, Z. MicroRN.NA-874 inhibits cell proliferation and invasion by targeting cyclin-dependent kinase 9 in osteosarcoma. Oncol. Lett. 2018, 15, 7649–7654. [Google Scholar] [CrossRef] [PubMed]
- Que, K.; Tong, Y.; Que, G.; Li, L.; Lin, H.; Huang, S.; Wang, R.; Tang, L. Downregulation of miR-874-3p promotes chemotherapeutic resistance in colorectal cancer via inactivation of the Hippo signaling pathway. Oncol. Rep. 2017, 38, 3376–3386. [Google Scholar] [CrossRef]
- Leong, K.W.; Cheng, C.W.; Wong, C.M.; Ng, I.O.; Kwong, Y.L.; Tse, E. miR-874-3p is down-regulated in hepatocellular carcinoma and negatively regulates PIN1 expression. Oncotarget 2017, 8, 11343–11355. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.W.; Chou, H.Y.; Lin, Y.S.; Huang, K.H.; Chang, C.J.; Hsu, T.C.; Lee, S.C. Phosphorylation at Ser473 regulates heterochromatin protein 1 binding and corepressor function of TIF1beta/KAP1. BMC Mol. Biol. 2008, 9, 61. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Staahl, B.T.; Alla, R.K.; Doudna, J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 2014, 3, e04766. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Y.-J.; Kang, Z.; Bei, J.; Chou, S.-J.; Lu, M.-Y.J.; Su, Y.-L.; Lin, S.-W.; Wang, H.-H.; Lin, S.; Chang, C.-J. Generation of TRIM28 Knockout K562 Cells by CRISPR/Cas9 Genome Editing and Characterization of TRIM28-Regulated Gene Expression in Cell Proliferation and Hemoglobin Beta Subunits. Int. J. Mol. Sci. 2022, 23, 6839. https://doi.org/10.3390/ijms23126839
Chang Y-J, Kang Z, Bei J, Chou S-J, Lu M-YJ, Su Y-L, Lin S-W, Wang H-H, Lin S, Chang C-J. Generation of TRIM28 Knockout K562 Cells by CRISPR/Cas9 Genome Editing and Characterization of TRIM28-Regulated Gene Expression in Cell Proliferation and Hemoglobin Beta Subunits. International Journal of Molecular Sciences. 2022; 23(12):6839. https://doi.org/10.3390/ijms23126839
Chicago/Turabian StyleChang, Yao-Jen, Zhifu Kang, Jiayuan Bei, Shu-Jen Chou, Mei-Yeh Jade Lu, Yu-Lun Su, Sheng-Wei Lin, Hsin-Hui Wang, Steven Lin, and Ching-Jin Chang. 2022. "Generation of TRIM28 Knockout K562 Cells by CRISPR/Cas9 Genome Editing and Characterization of TRIM28-Regulated Gene Expression in Cell Proliferation and Hemoglobin Beta Subunits" International Journal of Molecular Sciences 23, no. 12: 6839. https://doi.org/10.3390/ijms23126839






