EfABI4 Transcription Factor Is Involved in the Regulation of Starch Biosynthesis in Euryale ferox Salisb Seeds
Abstract
:1. Introduction
2. Results
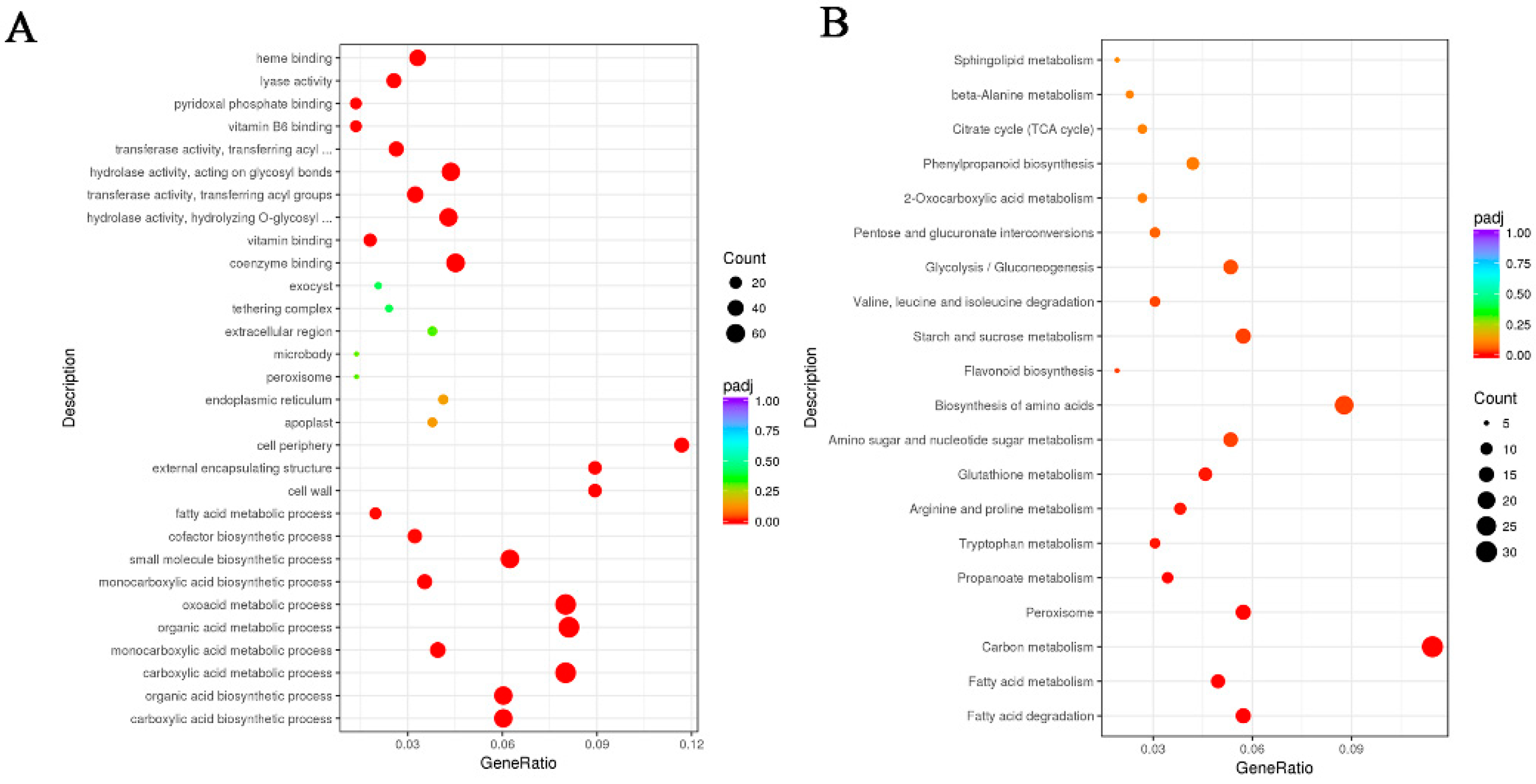
2.1. Transcriptome Analysis of ABA Treated on Euryale ferox Seeds
2.2. DEGs Related to Starch Biosynthesis
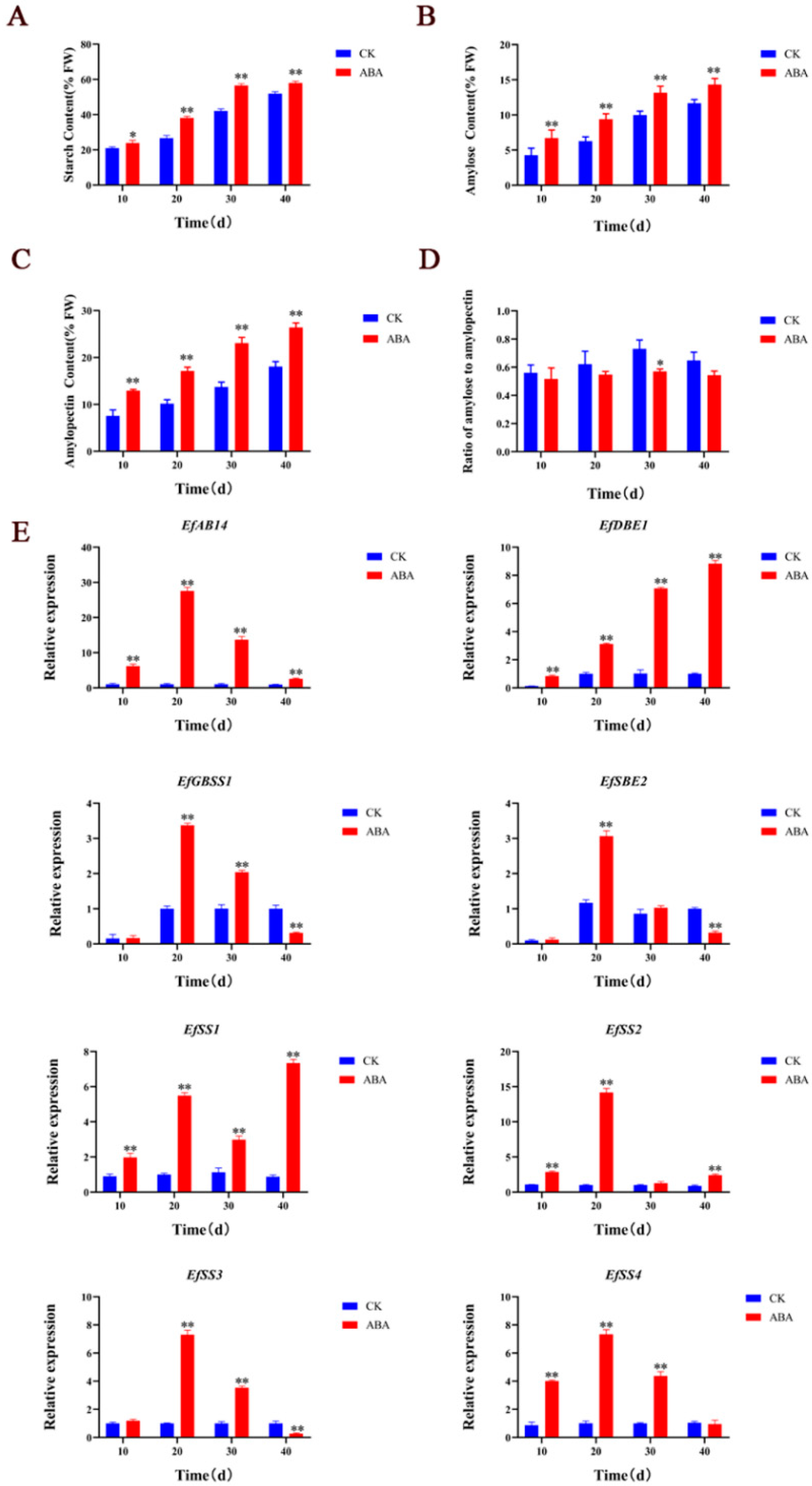
2.3. ABA Treatment on Starch Biosynthesis during Euryale ferox Salisb Development
2.4. EfAB14 Directly Promotes the Expression of EfSS1
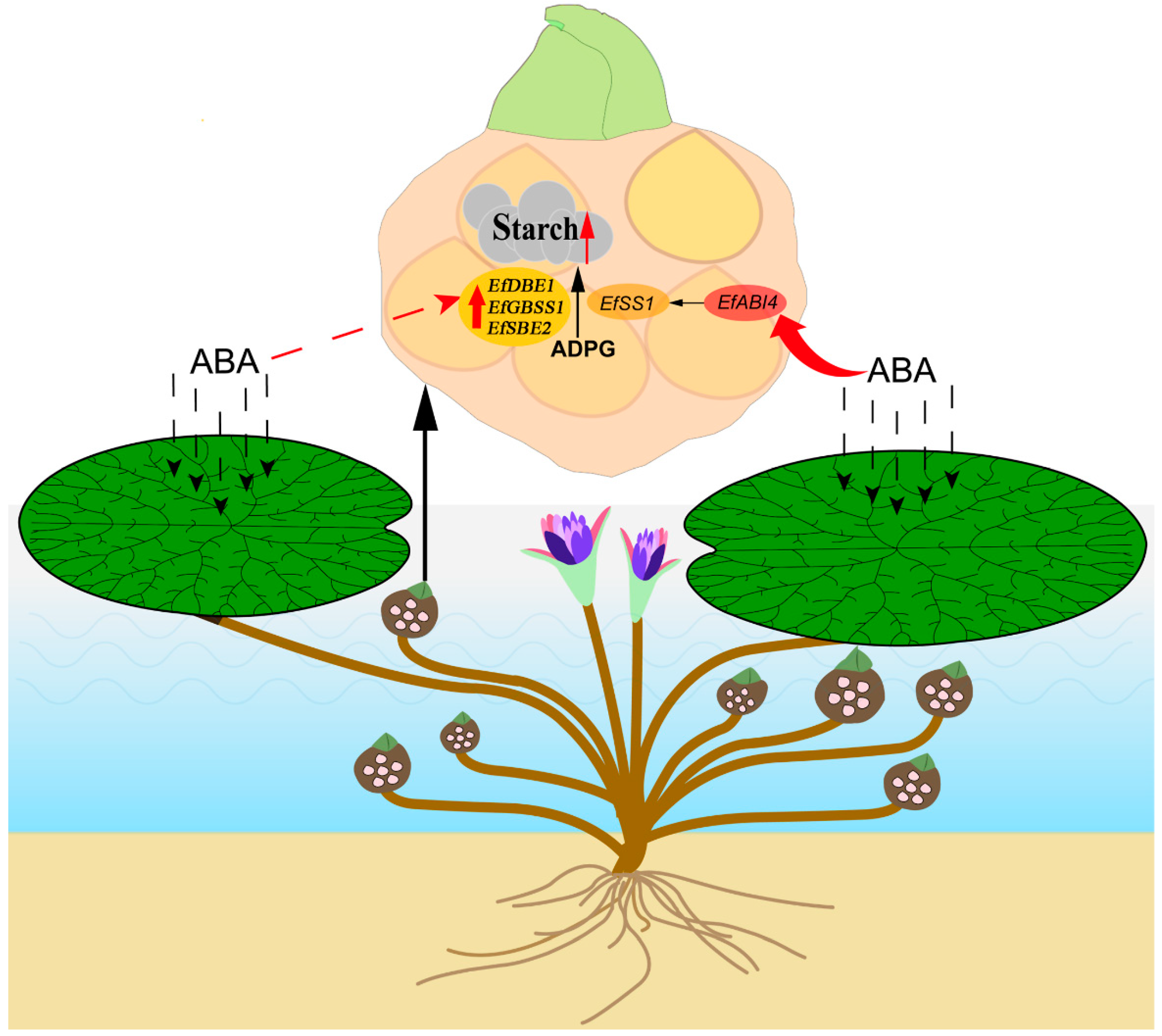
3. Discussion
4. Materials and Methods
4.1. Plant Materials and ABA Treatment
4.2. RNA Extraction and Sequencing
4.3. Reverse Transcription-Quantitative PCR (RT-qPCR)
4.4. Determination of Starch Content in ABA Treated Leaves
4.5. Y1H Assays
4.6. Subcellular Localization
4.7. Transient Dual-Luciferase Detection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hsu, P.K.; Dubeaux, G.; Takahashi, Y.; Schroeder, J.I. Signaling mechanisms in abscisic acid-mediated stomatal closure. Plant J. 2021, 105, 307–321. [Google Scholar] [CrossRef]
- Ackerson, R.C. Invertase Activity and Abscisic Acid in Relation to Carbohydrate Status in Developing Soybean Reproductive Structures. Crop Sci. 1985, 25, 615–618. [Google Scholar] [CrossRef]
- Saftner, R.A.; Wyse, R.E. Effect of Plant Hormones on Sucrose Uptake by Sugar Beet Root Tissue Discs. Plant Physiol. 1984, 74, 951–955. [Google Scholar] [CrossRef] [PubMed]
- Rainbird, T.R.M. An In Vivo Technique for the Study of Phloem Unloading in Seed Coats of Developing Soybean Seeds. Plant Physiol. 1983, 72, 268–271. [Google Scholar]
- Karssen, C.M.; Swan, B.V.D.; Breekland, A.E.; Koornneef, M. Induction of dormancy during seed development by endogenous abscisic acid: Studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 1983, 157, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Tanner, D.W. On the Possible Role of ABA on Phloem Unloading. Berichte Der. Deutschen Botanischen. 1980, 93, 349–351. [Google Scholar]
- Li, J.; Seng, S.; Li, D.; Zhang, F.; Wu, J. Antagonism between abscisic acid and gibberellin regulates starch synthesis and corm development in Gladiolus hybridus. Hortic. Res. 2021, 8, 2023–2034. [Google Scholar] [CrossRef]
- Yu, Y. Studies on Main Pigment Formation and Physiological Mechanism in Tomato by Exogenous GA3 and ABA; Shenyang Agricultural University: Shenyang, China, 2016. [Google Scholar]
- Soderman, E.M.; Brocard, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and function of the arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 2000, 124, 1752–1765. [Google Scholar] [CrossRef] [Green Version]
- Miao, Y.; Zhao, D.; Zhang, K.; Miao, H.; Liu, J.; Jin, Z.; Xu, B. Effects of ABA Treatment on Starch Metabolism in Banana Fruit. Mol. Plant Breeding. 2018, 16, 1807–1811. [Google Scholar]
- Wu, P.; Liu, A.; Zhang, Y.; Feng, K.; Zhao, S.; Li, L. NnABI4-Mediated ABA Regulation of Starch Biosynthesis in Lotus (Nelumbo nucifera Gaertn). Int. J. Mol. Sci. 2021, 22, 13506. [Google Scholar] [CrossRef]
- Han, T.; Li, W.; Wu, W.; Shi, C.; Xiong, Z. Quality analysis and metabolomic profiling of the effects of exogenous abscisic acid on rabbiteye blueberry. BMC Plant Biol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Jian-Lan, X.U.; Rui-Juan, M.A.; Zhang, B.B.; Lin-Jian, N.I. Effects of Different Concentration of ABA on Fruit Quality of Peach Cultivar ‘Meixiang’. Southwest China J. Agric. Sci. 2012, 25, 1027–1030. [Google Scholar]
- Emes, M.J.; Bowsher, C.G.; Hedley, C.; Burrell, M.M.; Tetlow, I.J. Starch synthesis and carbon partitioning in developing endosperm. J. Exp. Bot. 2003, 54, 569–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khlestkin, V.K.; Peltek, S.E.; Kolchanov, N.A. Review of direct chemical and biochemical transformations of starch. Carbohydr. Polym. 2018, 181, 460–476. [Google Scholar] [CrossRef]
- Vamadevan, V.; Bertoft, E. Structure-function relationships of starch components. Starch-Stärke 2015, 67, 55–68. [Google Scholar] [CrossRef]
- Li, J.; Guan, Y.L.; Yuan, L.Y.; Hou, J.F.; Wang, C.G.; Liu, F.F.; Yang, Y.; Lu, Z.Y.; Chen, G.H.; Zhu, S.D. Effects of exogenous IAA in regulating photosynthetic capacity, carbohydrate metabolism and yield of Zizania latifolia. Sci. Hortic. 2019, 253, 276–285. [Google Scholar] [CrossRef]
- Zhou, Y.-F.; Yan, T.; Zhang, J.; Wang, Y.-T.; Wang, N.; Wu, Q.; Sun, F.; Xu, W.-J.; Huang, R.-D. Effects of exogenous IAA application on endogenous hormone contents and tillering in sorghum. Shengtaixue Zazhi. 2017, 36, 2191–2197. [Google Scholar]
- Chi, C.D.; Li, X.X.; Zhang, Y.P.; Chen, L.; Li, L.; Wang, Z.J. Digestibility and supramolecular structural changes of maize starch by non-covalent interactions with gallic acid. Food Function. 2017, 8, 720–730. [Google Scholar] [CrossRef]
- Wang, X.H.; Yin, W.; Wu, J.X.; Chai, L.J.; Yi, H.L. Effects of exogenous abscisic acid on the expression of citrus fruit ripening-related genes and fruit ripening. Sci. Hortic. 2016, 201, 175–183. [Google Scholar] [CrossRef]
- Wang, X.; Xu, C.; Cang, J.; Zeng, Y.; Yu, J.; Liu, L.; Zhang, D.; Wang, J. Effects of Exogenous GA (3) on Wheat Cold Tolerance. J. Agric. Sci. Technol. 2015, 17, 921–934. [Google Scholar]
- Pan, T.; Lin, L.; Wang, J.; Liu, Q.; Wei, C. Long branch-chains of amylopectin with B-type crystallinity in rice seed with inhibition of starch branching enzyme I and IIb resist in situ degradation and inhibit plant growth during seedling development. BMC Plant Biol. 2018, 18, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Zhang, J.; Wang, Z.; Xu, G.; Zhu, Q. Activities of Key Enzymes in Sucrose-to-Starch Conversion in Wheat Grains Subjected to Water Deficit during Grain Filling. Plant Physiol. 2004, 135, 1621–1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, D.; Zhen, Y.U.; Yong, L.I.; Song, Y.U. Dynamic Changes of Enzyme Activities Involving in Starch Synthesis in Superior and Inferior Grains of High-yield Winter Wheat. Sci. Agric. Sin. 2002, 35, 378–383. [Google Scholar]
- Keeling, P.L.; Wood, J.R.; Tyson, R.H.; Bridges, I.G. Starch biosynthesis in developing wheat grain. Plant Physiol. 1988, 87, 311–319. [Google Scholar] [CrossRef] [Green Version]
- Borzenkova, R.A.; Borovkova, M.P. Developmental patterns of phytohormone content in the cortex and pith of potato tubers as related to their growth and starch content. Russ. J. Plant Physiol. 2003, 50, 119–124. [Google Scholar] [CrossRef]
- Kato, T.; Sakurai, N.; Kuraishi, S. The Changes of Endogenous Abscisic Acid in Developing Grain of Two Rice Cultivars with Different Grain Size. Jpn. J. Crop Sci. 1993, 62, 456–461. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.F.; Li, Y.P.; Zhang, J.; Liu, H.; Tian, M.; Huang, Y. Binding of ABI4 to a CACCG motif mediates the ABA-induced expression of the ZmSSI gene in maize (Zea mays L.) endosperm. J. Exp. Bot. 2012, 63, 5979–5989. [Google Scholar] [CrossRef] [Green Version]
- Rook, F.; Corke, F.; Baier, M.; Holman, R.; May, A.G.; Bevan, M.W. Impaired sucrose induction1 encodes a conserved plant-specific protein that couples carbohydrate availability to gene expression and plant growth. Plant J. 2006, 46, 1045–1058. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Gampala, S.S.L.; Rock, C.D. Abscisic Acid Signaling in Seeds and Seedlings. Plant Cell. 2002, 14, S15–S45. [Google Scholar] [CrossRef] [Green Version]
- Arenas-Huertero, F.; Arroyo, A.; Li, Z.; Sheen, J.; León, P. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 2000, 14, 2085–2096. [Google Scholar] [CrossRef]
- Huijser, C.; Kortstee, A.; Pego, J.; Weisbeek, P.; Wisman, E.; Smeekens, S. The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: Involvement of abscisic acid in sugar responses. Plant J. 2010, 23, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yu, X.; Song, L.; An, C. ABI4 Activates DGAT1 Expression in Arabidopsis Seedlings during Nitrogen Deficiency. Plant Physiol. 2011, 156, 873–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramon, M.; Rolland, F.; Thevelein, J.M.; Van Dijck, P.; Leyman, B. ABI4 mediates the effects of exogenous trehalose on Arabidopsis growth and starch breakdown. Plant Mol. Biol. 2007, 63, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Peng, C. The Molecular Regulation Mechanism of Starch Synthesis by ABI4 in Maize Endosperm. Master’s Thesis, Sichuan Agricultural University, Yaan, China, 2016. [Google Scholar]
- Zhao, H.; Zhao, S.; Guillaume, D.; Sun, C. New cerebrosides from Euryale ferox. J. Nat. Prod. 1994, 57, 138–141. [Google Scholar] [CrossRef]
- Zhao, H.R.; Zhao, S.X.; Sun, C.Q.; Guillaume, D. Glucosylsterols in extracts of Euryale ferox identified by high resolution NMR and mass spectrometry. J. Lipid Res. 1989, 30, 1633–1637. [Google Scholar] [CrossRef]
- Lin, L.; Guo, K.; Zhang, L.; Zhang, C.; Liu, Q.; Wei, C. Effects of molecular compositions on crystalline structure and functional properties of rice starches with different amylopectin extra-long chains. Food Hydrocoll. 2019, 88, 137–145. [Google Scholar] [CrossRef]
- Liu, X.; He, Z.; Yin, Y.; Xu, X.; Wu, W.; Li, L. Transcriptome sequencing and analysis during seed growth and development in Euryale ferox Salisb. BMC Genom. 2018, 19, 343. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Yan, Z.J. Study on Physico-chemical Properties of Euryale ferox Salisb Starch. Food Sci. 2006, 27, 119–122. [Google Scholar]
- Xu, X. The Study of Starch Formation and the EfSBEl Expression in the Development of Gordon Euryale Seed (Euryale ferox Salisb). Master’s Thesis, Yangzhou University, Yangzhou, China, 2019. [Google Scholar]
- Zhao, L.X.; Huang, J.; Man, J.M.; Huai, H.Y.; Chen, Y.F.; Wei, C.X. Physicochemical Properties of Euryale ferox Kernel Starches from Two Different Regions. Int. J. Food Prop. 2015, 19, 289–299. [Google Scholar] [CrossRef]
- Wang, Q.; Yan, H.Y.S.; Jing, Z.; Liu, L.; Bao, K.; Wu, Q. Cloning and bioinformatics analysis of SBE3 gene from Euryale ferox. Mol. Plant Breed. 2021, 18, 1–13. [Google Scholar]
- Oh, H.D.; Yu, D.J.; Chung, S.W.; Chea, S.; Lee, H.J. Abscisic acid stimulates anthocyanin accumulation in ‘Jersey’ highbush blueberry fruits during ripening. Food Chem. 2018, 244, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Pei, C.; Duan, C.; Pang, T. Transcriptional Regulation of Genes Encoding Key Enzymes of Abscisic Acid Metabolism During Melon (Cucumis melo L.) Fruit Development and Ripening. J. Plant Growth Regul. 2013, 32, 233–244. [Google Scholar] [CrossRef]
- Jia, H.F.; Chai, Y.M.; Li, C.L.; Lu, D.; Shen, Y.Y. Abscisic Acid Plays an Important Role in the Regulation of Strawberry Fruit Ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afoufa-Bastien, D.; Medici, A.; Jeauffre, J.; Coutos-Thevenot, P.; Lemoine, R.; Atanassova, R.; Laloi, M. The Vitis vinifera sugar transporter gene family phylogenetic overview and macroarray expression profiling. BMC Plant Biol. 2010, 12, 245. [Google Scholar] [CrossRef] [Green Version]
- Murcia, G.N.; Pontin, M.; Piccoli, P. Role of ABA and Gibberellin A3 on gene expression pattern of sugar transporters and invertases in Vitis vinifera cv. Malbec during berry ripening. Plant Growth Regul. 2018, 84, 275–283. [Google Scholar] [CrossRef]
- Xu, Y.J.; Zhang, W.Y.; Ju, C.X.; Li, Y.Y.; Yang, J.C.; Zhang, J.H. Involvement of abscisic acid in fructan hydrolysis and starch biosynthesis in wheat under soil drying. Plant Growth Regul. 2016, 80, 265–279. [Google Scholar] [CrossRef]
- Tripathi, P.; Rabara, R.C.; Rushton, P.J. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 2014, 239, 255–266. [Google Scholar] [CrossRef]
- Eiji, N. Abscisic acid and the control of seed dormancy and germination. Seed Sci. Res. 2010, 20, 55–67. [Google Scholar]
- Akihiro, T.; Mizuno, K.; Fujimura, T. Gene expression of ADP-glucose pyrophosphorylase and starch contents in rice cultured cells are cooperatively regulated by sucrose and ABA. Plant Cell Physiol. 2005, 46, 937–946. [Google Scholar] [CrossRef]
- Bossi, F.; Cordoba, E.; Dupre, P.; Mendoza, M.S.; Roman, C.S.; Leon, P. The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J. 2009, 59, 359–374. [Google Scholar] [CrossRef]
- Wu, P.; Zhang, L.; Zhang, K.; Yin, Y.; Liu, A.; Zhu, Y.; Fu, Y.; Sun, F.; Zhao, S.; Feng, K.; et al. The adaptive evolution of Euryale ferox to the aquatic environment through paleo-hexaploidization. Plant J. 2022, 110, 627–645. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, P.; Zhu, Y.; Liu, A.; Wang, Y.; Zhao, S.; Feng, K.; Li, L. EfABI4 Transcription Factor Is Involved in the Regulation of Starch Biosynthesis in Euryale ferox Salisb Seeds. Int. J. Mol. Sci. 2022, 23, 7598. https://doi.org/10.3390/ijms23147598
Wu P, Zhu Y, Liu A, Wang Y, Zhao S, Feng K, Li L. EfABI4 Transcription Factor Is Involved in the Regulation of Starch Biosynthesis in Euryale ferox Salisb Seeds. International Journal of Molecular Sciences. 2022; 23(14):7598. https://doi.org/10.3390/ijms23147598
Chicago/Turabian StyleWu, Peng, Yue Zhu, Ailian Liu, Yuhao Wang, Shuping Zhao, Kai Feng, and Liangjun Li. 2022. "EfABI4 Transcription Factor Is Involved in the Regulation of Starch Biosynthesis in Euryale ferox Salisb Seeds" International Journal of Molecular Sciences 23, no. 14: 7598. https://doi.org/10.3390/ijms23147598




