Identification of the Gene Repertoire of the IMD Pathway and Expression of Antimicrobial Peptide Genes in Several Tissues and Hemolymph of the Cockroach Blattella germanica
Abstract
:1. Introduction
2. Results
2.1. Genes of the IMD Pathway in B. germanica
2.2. Expression of IMD Pathway Genes in Six Tissues and Hemolymph of B. germanica
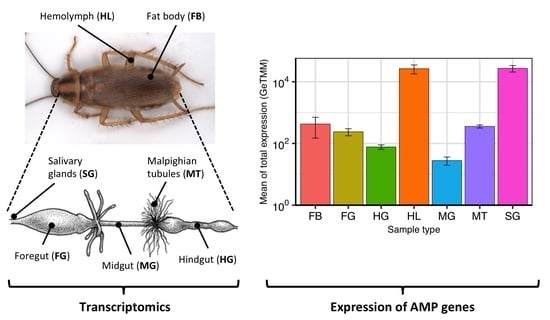
2.3. Expression of AMP Genes in B. germanica Tissues and Hemolymph
3. Discussion
4. Materials and Methods
4.1. IMD Signaling Pathway Characterization
4.2. Transcriptome Assembly
4.3. Review of Results and CDS Gathering
4.4. Insect Rearing
4.5. Insect Dissection and Collection of Samples
4.6. RNA Extraction and Sequencing
4.7. Generation of Normalized Expression Levels
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bell, W.J.; Roth, L.M.; Nalepa, C.A. Cockroaches: Ecology, Behavior, and Natural History; Johns Hopkins University Press: Baltimore, MD, USA, 2007. [Google Scholar]
- Li, S.; Zhu, S.; Jia, Q.; Yuan, D.; Ren, C.; Li, K.; Liu, S.; Cui, Y.; Zhao, H.; Cao, Y.; et al. The genomic and functional landscapes of developmental plasticity in the American cockroach. Nat. Commun. 2018, 9, 1008. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.C.; Jongepier, E.; Robertson, H.M.; Arning, N.; Bitard-Feildel, T.; Chao, H.; Childers, C.P.; Dinh, H.; Doddapaneni, H.; Dugan, S.; et al. Hemimetabolous genomes reveal molecular basis of termite eusociality. Nat. Ecol. Evol. 2018, 2, 557–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, F.J.; Muñoz-Benavent, M.; García-Ferris, C.; Latorre, A. Blattella germanica displays a large arsenal of antimicrobial peptide genes. Sci. Rep. 2020, 10, 21058. [Google Scholar] [CrossRef] [PubMed]
- Latorre, A.; Domínguez-Santos, R.; García-Ferris, C.; Gil, R. Of cockroaches and symbionts: Recent advances in the characterization of the relationship between Blattella germanica and its dual symbiotic system. Life 2022, 12, 290. [Google Scholar] [CrossRef] [PubMed]
- Bandi, C.; Sironi, M.; Damiani, G.; Magrassi, L.; Nalepa, C.; Laudani, U.; Sacchi, L. The establishment of intracellular symbiosis in an ancestor of cockroaches and termites. Proc. R. Soc. Lond. B 1995, 259, 293–299. [Google Scholar] [CrossRef]
- Evangelista, D.A.; Wipfler, B.; Béthoux, O.; Donath, A.; Fujita, M.; Kohli, M.K.; Legendre, F.; Liu, S.; Machida, R.; Misof, B.; et al. An integrative phylogenomic approach illuminates the evolutionary history of cockroaches and termites (Blattodea). Proc. R. Soc. B 2019, 286, 20182076. [Google Scholar] [CrossRef] [Green Version]
- López-Sánchez, M.J.; Neef, A.; Peretó, J.; Patiño-Navarrete, R.; Pignatelli, M.; Latorre, A.; Moya, A. Evolutionary convergence and nitrogen metabolism in Blattabacterium strain Bge, primary endosymbiont of the cockroach Blattella germanica. PLoS Genet. 2009, 5, e1000721. [Google Scholar] [CrossRef] [Green Version]
- Sabree, Z.L.; Kambhampati, S.; Moran, N.A. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc. Natl. Acad. Sci. USA 2009, 106, 19521–19526. [Google Scholar] [CrossRef] [Green Version]
- Neef, A.; Latorre, A.; Peretó, J.; Silva, F.J.; Pignatelli, M.; Moya, A. Genome economization in the endosymbiont of the wood roach Cryptocercus punctulatus due to drastic loss of amino acid synthesis capabilities. Genome Biol. Evol. 2011, 3, 1437–1448. [Google Scholar] [CrossRef] [Green Version]
- Sabree, Z.L.; Huang, C.Y.; Arakawa, G.; Tokuda, G.; Lo, N.; Watanabe, H.; Moran, N.A. Genome shrinkage and loss of nutrient-providing potential in the obligate symbiont of the primitive termite Mastotermes darwiniensis. Appl. Environ. Microbiol. 2012, 78, 204–210. [Google Scholar] [CrossRef] [Green Version]
- Patiño-Navarrete, R.; Moya, A.; Latorre, A.; Peretó, J. Comparative genomics of Blattabacterium cuenoti: The frozen legacy of an ancient endosymbiont genome. Genome Biol. Evol. 2013, 5, 351–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Domenech, C.M.; Belda, E.; Patiño-Navarrete, R.; Moya, A.; Peretó, J.; Latorre, A. Metabolic stasis in an ancient symbiosis: Genome-scale metabolic networks from two Blattabacterium cuenoti strains, primary endosymbionts of cockroaches. BMC Microbiol. 2012, 12, S5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patiño-Navarrete, R.; Piulachs, M.D.; Bellés, X.; Moya, A.; Latorre, A.; Peretó, J. The cockroach Blattella germanica obtains nitrogen from uric acid through a metabolic pathway shared with its bacterial endosymbiont. Biol. Lett. 2014, 10, 20140407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moran, N.A.; Ochman, H.; Hammer, T.J. Evolutionary and ecological consequences of gut microbial communities. Annu. Rev. Ecol. Evol. Syst. 2019, 50, 451–475. [Google Scholar] [CrossRef] [PubMed]
- Nishide, Y.; Kageyama, D.; Yokoi, K.; Jouraku, A.; Tanaka, H.; Futahashi, R.; Fukatsu, T. Functional crosstalk across IMD and Toll pathways: Insight into the evolution of incomplete immune cascades. Proc. R. Soc. B 2019, 286, 20182207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, M.A.; Lemaitre, B. New insights on Drosophila antimicrobial peptide function in host defense and beyond. Curr. Opin. Immunol. 2020, 62, 22–30. [Google Scholar] [CrossRef]
- Mergaert, P.; Kikuchi, Y.; Shigenobu, S.; Nowack, E.C.M. Metabolic integration of bacterial endosymbionts through antimicrobial peptides. Trends Microbiol. 2017, 25, 703–712. [Google Scholar] [CrossRef]
- Mergaert, P. Role of antimicrobial peptides in controlling symbiotic bacterial populations. Nat. Prod. Rep. 2018, 35, 336–356. [Google Scholar] [CrossRef] [PubMed]
- Marra, A.; Hanson, M.A.; Kondo, S.; Erkosar, B.; Lemaitre, B. Drosophila antimicrobial peptides and lysozymes regulate gut microbiota composition and abundance. mBio 2021, 12, e0082421. [Google Scholar] [CrossRef]
- Login, F.H.; Balmand, S.; Vallier, A.; Vincent-Monégat, C.; Vigneron, A.; Weiss-Gayet, M.; Rochat, D.; Heddi, A. Antimicrobial peptides keep insect endosymbionts under control. Science 2011, 334, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Oakeson, K.F.; Gil, R.; Clayton, A.L.; Dunn, D.M.; von Niederhausern, A.C.; Hamil, C.; Aoyagi, A.; Duval, B.; Baca, A.; Silva, F.J.; et al. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol. Evol. 2014, 6, 76–93. [Google Scholar] [CrossRef] [PubMed]
- Masson, F.; Zaidman-Rémy, A.; Heddi, A. Antimicrobial peptides and cell processes tracking endosymbiont dynamics. Phil. Trans. R. Soc. B 2016, 371, 20150298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondorosi, E.; Mergaert, P.; Kereszt, A. A paradigm for endosymbiotic life: Cell differentiation of Rhizobium bacteria provoked by host plant factors. Ann. Rev. Microbiol. 2013, 67, 611–628. [Google Scholar] [CrossRef] [PubMed]
- Carro, L.; Pujic, P.; Alloisio, N.; Fournier, P.; Boubakri, H.; Hay, A.E.; Poly, F.; François, P.; Hocher, V.; Mergaert, P.; et al. Alnus peptides modify membrane porosity and induce the release of nitrogen-rich metabolites from nitrogen-fixing Frankia. ISME J. 2015, 9, 1723–1733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleino, A.; Silverman, N. The Drosophila IMD pathway in the activation of the humoral immune response. Dev. Comp. Immunol. 2014, 42, 25–35. [Google Scholar] [CrossRef] [Green Version]
- Ryu, J.-H.; Kim, S.-H.; Lee, H.-Y.; Bai, J.Y.; Nam, Y.-D.; Bae, J.-W.; Lee, D.G.; Shin, S.C.; Ha, E.-M.; Lee, W.-J. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 2008, 319, 777–782. [Google Scholar] [CrossRef] [Green Version]
- Park, M.S.; Park, P.; Takeda, M. Starvation induces apoptosis in the midgut nidi of Periplaneta americana: A histochemical and ultrastructural study. Cell Tissue Res. 2009, 335, 631–638. [Google Scholar] [CrossRef]
- Huang, X.; Warren, J.T.; Buchanan, J.; Gilbert, L.I.; Scott, M.P. Drosophila Niemann-Pick Type C-2 genes control sterol homeostasis and steroid biosynthesis: A model of human neurodegenerative disease. Development 2007, 134, 3733–3742. [Google Scholar] [CrossRef] [Green Version]
- Erkosar, B.; Defaye, A.; Bozonnet, N.; Puthier, D.; Royet, J.; Leulier, F. Drosophila microbiota modulates host metabolic gene expression via IMD/NF-κb signaling. PLoS ONE 2014, 9, e94729. [Google Scholar] [CrossRef]
- Buonocore, F.; Fausto, A.M.; Della Pelle, G.; Roncevic, T.; Gerdol, M.; Picchietti, S. Attacins: A promising class of insect antimicrobial peptides. Antibiotics 2021, 10, 212. [Google Scholar] [CrossRef]
- Tanji, T.; Hu, X.; Weber, A.N.; Ip, Y.T. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol. Cell. Biol. 2007, 27, 4578–4588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamberty, M.; Zachary, D.; Lanot, R.; Bordereau, C.; Robert, A.; Hoffmann, J.A.; Bulet, P. Insect immunity. Constitutive expression of a cysteine-rich antifungal and a linear antibacterial peptide in a termite insect. J. Biol. Chem. 2001, 276, 4085–4092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulet, P.; Stöcklin, R. Insect antimicrobial peptides: Structures, properties and gene regulation. Protein Pept. Lett. 2005, 12, 3–11. [Google Scholar] [CrossRef]
- Harrison, M.C.; Arning, N.; Kremer, L.P.M.; Ylla, G.; Belles, X.; Bornberg-Bauer, E.; Huylmans, A.K.; Jongepier, E.; Piulachs, M.D.; Richards, S.; et al. Expansions of key protein families in the German cockroach highlight the molecular basis of its remarkable success as a global indoor pest. J. Exp. Zool. B Mol. Dev. Evol. 2018, 330, 254–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sackton, T.B.; Werren, J.H.; Clark, A.G. Characterizing the infection-induced transcriptome of Nasonia vitripennis reveals a preponderance of taxonomically-restricted immune genes. PLoS ONE 2013, 8, e83984. [Google Scholar] [CrossRef] [Green Version]
- Sackton, T.B.; Lazzaro, B.P.; Clark, A.G. Rapid expansion of immune-related gene families in the house fly, Musca domestica. Mol. Biol. Evol. 2017, 34, 857–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sackton, T. Comparative genomics and transcriptomics of host–pathogen interactions in insects: Evolutionary insights and future directions. Curr. Opin. Insect Sci. 2019, 31, 106–113. [Google Scholar] [CrossRef] [Green Version]
- Engel, P.; Moran, N.A. The gut microbiota of insects–Diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Lee, Y.K.; Mazmanian, S.K. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 2010, 330, 1768–1773. [Google Scholar] [CrossRef] [Green Version]
- Fraune, S.; Augustin, R.; Anton-Erxleben, F.; Wittlieb, J.; Gelhaus, C.; Klimovich, V.B.; Samoilovich, M.P.; Bosch, T.C. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2010, 107, 18067–18072. [Google Scholar] [CrossRef] [Green Version]
- Bosch, T.C.G.; Zasloff, M. Antimicrobial peptides—Or how our ancestors learned to control the microbiome. mBio 2021, 12, e01847-21. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-Z.; Shi, M.; Ye, X.-Q.; Chen, M.-Y.; Chen, X.-X. Identification, characterization and expression of a defensin-like antifungal peptide from the whitefly Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae). Insect Mol. Biol. 2013, 22, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, B.A.; Kneubehl, A.R.; Mitchell, R.D.; Krishnavajhala, A.; Teel, P.D.; Pérez de León, A.A.; Lopez, J.E. Differential expression of putative Ornithodoros turicata defensins mediated by tick feeding. Front. Cell. Infect. Microbiol. 2020, 10, 152. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, J.A. Innate immunity of insects. Curr. Opin. Immunol. 1995, 7, 4–10. [Google Scholar] [CrossRef]
- Buchon, N.; Silverman, N.; Cherry, S. Immunity in Drosophila melanogaster—From microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 2014, 14, 796–810. [Google Scholar] [CrossRef] [PubMed]
- Shigenobu, S.; Stern, D.L. Aphids evolved novel secreted proteins for symbiosis with bacterial endosymbiont. Proc. R. Soc. B 2013, 280, 20121952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchi, N.; Fukudome, M.; Nozaki, N.; Suzuki, M.; Osuki, K.I.; Shigenobu, S.; Uchiumi, T. Antimicrobial activities of cysteine-rich peptides specific to bacteriocytes of the pea aphid Acyrthosiphon pisum. Microbes Environ. 2019, 34, 155–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Hillyer, J. The immune and circulatory systems are functionally integrated across insect evolution. Sci. Adv. 2020, 6, eabb3164. [Google Scholar] [CrossRef]
- Turner, M.; Pietri, J.E. Antimicrobial peptide expression in the cockroach gut during enterobacterial infection is specific and influenced by type III secretion. Biol. Open. 2022, 11, bio059414. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Eddy, S.R. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009, 23, 205–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, L.; Florea, L. Rcorrector: Efficient and accurate error correction for Illumina RNA-seq reads. GigaScience 2015, 4, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef] [PubMed]
- Kapustin, Y.; Souvorov, A.; Tatusova, T.; Lipman, D. Splign: Algorithms for computing spliced alignments with identification of paralogs. Biol. Direct 2008, 3, 20. [Google Scholar] [CrossRef] [Green Version]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; UGENE team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [Green Version]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Smid, M.; Coebergh van den Braak, R.R.J.; van de Werken, H.J.G.; van Riet, J.; van Galen, A.; de Weerd, V.; van der Vlugt-Daane, M.; Bril, S.I.; Lalmahomed, Z.S.; Kloosterman, W.P.; et al. Gene length corrected trimmed mean of M-values (GeTMM) processing of RNA-seq data performs similarly in intersample analyses while improving intrasample comparisons. BMC Bioinform. 2018, 19, 236. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Sarkar, D. Lattice: Multivariate Data Visualization with R; Springer: New York, NY, USA, 2008. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Gene Name (Based on 4IN Database) | Presence in B. germanica | B. germanica Product Name | Protein Length | Located at Locus_Tag † |
|---|---|---|---|---|
| akirin | YES | Akirin | 180 | C0J52_14274 |
| ben | YES | Bendless | 151 | C0J52_14002 |
| Bruce | YES | BIR repeat containing Ubiquitin-conjugating enzyme | 4175 ‡ | C0J52_00682 |
| cad | YES | Caudal | 281 § | |
| casp | YES | Caspar | 669 | C0J52_10382 |
| CYLD | YES | Cylindromatosis | 1147 | C0J52_05308 |
| CASP | YES | Caspase | 307 | C0J52_04212 |
| Diap2 | YES | Death-associated inhibitor of apoptosis 2 | 578 | C0J52_01770 |
| dnr1 | YES | Defense repressor 1 | 544 | C0J52_09439 |
| Dredd_1 | YES | Death related ced-3/Nedd2-like caspase | 592 | C0J52 14092 |
| Dredd_2 | YES | Death related ced-3/Nedd2-like caspase | 572 § | C0J52 22862 |
| dsp1 | YES | Dorsal switch protein 1 | 205 | C0J52_00951 |
| Duox | Dual oxidase | 1544 | C0J52 05752 | |
| eff | YES | Effete | 147 | |
| FADD | YES | FADD | 245 | |
| imd | YES | Immune deficiency | 252 | C0J52_19439 |
| nemo | YES | NF-kappa-B essential modulator | 435 | |
| Npc2 | YES | Niemann-Pick type C-2 | 148 | C0J52_22057 |
| Ntf2 | YES | Nuclear transport factor 2 | 130 | |
| PGRP-LC | YES | Peptidoglycan recognition protein | 365 | C0J52_21009 |
| rel | YES | Relish | 1010 | C0J52_13050 |
| RYBP | YES | Ring and YY1 Binding Protein | 182 | C0J52_24359 # C0J52_26497 # |
| scny | YES | Scrawny | 740 | C0J52_00345 |
| Skp2 | YES | S-phase kinase-associated protein 2 Skp2 | 466 | C0J52_01896 |
| SkpA | YES | S-phase kinase-associated protein 1-related A | 162 | C0J52_18710 |
| Tab2 | YES | TAK1-associated Binding Protein 2 | 456 | C0J52_07566 |
| Tak1 | YES | TGF-beta activated kinase 1 | 471 | C0J52_05441 |
| Uev1A | YES | Ubiquitin-conjugating enzyme variant 1A | 144 | C0J52_05949 |
| IKKbeta | NOT | I-kappaB kinase beta | ||
| IKKg | NOT | IKK gamma | ||
| key | NOT | Kenny |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuber, L.; Domínguez-Santos, R.; García-Ferris, C.; Silva, F.J. Identification of the Gene Repertoire of the IMD Pathway and Expression of Antimicrobial Peptide Genes in Several Tissues and Hemolymph of the Cockroach Blattella germanica. Int. J. Mol. Sci. 2022, 23, 8444. https://doi.org/10.3390/ijms23158444
Zuber L, Domínguez-Santos R, García-Ferris C, Silva FJ. Identification of the Gene Repertoire of the IMD Pathway and Expression of Antimicrobial Peptide Genes in Several Tissues and Hemolymph of the Cockroach Blattella germanica. International Journal of Molecular Sciences. 2022; 23(15):8444. https://doi.org/10.3390/ijms23158444
Chicago/Turabian StyleZuber, Leo, Rebeca Domínguez-Santos, Carlos García-Ferris, and Francisco J. Silva. 2022. "Identification of the Gene Repertoire of the IMD Pathway and Expression of Antimicrobial Peptide Genes in Several Tissues and Hemolymph of the Cockroach Blattella germanica" International Journal of Molecular Sciences 23, no. 15: 8444. https://doi.org/10.3390/ijms23158444