Differentially CTCF-Binding Sites in Cattle Rumen Tissue during Weaning
Abstract
:1. Introduction
2. Results
2.1. Data Quality and CTCF Peaks
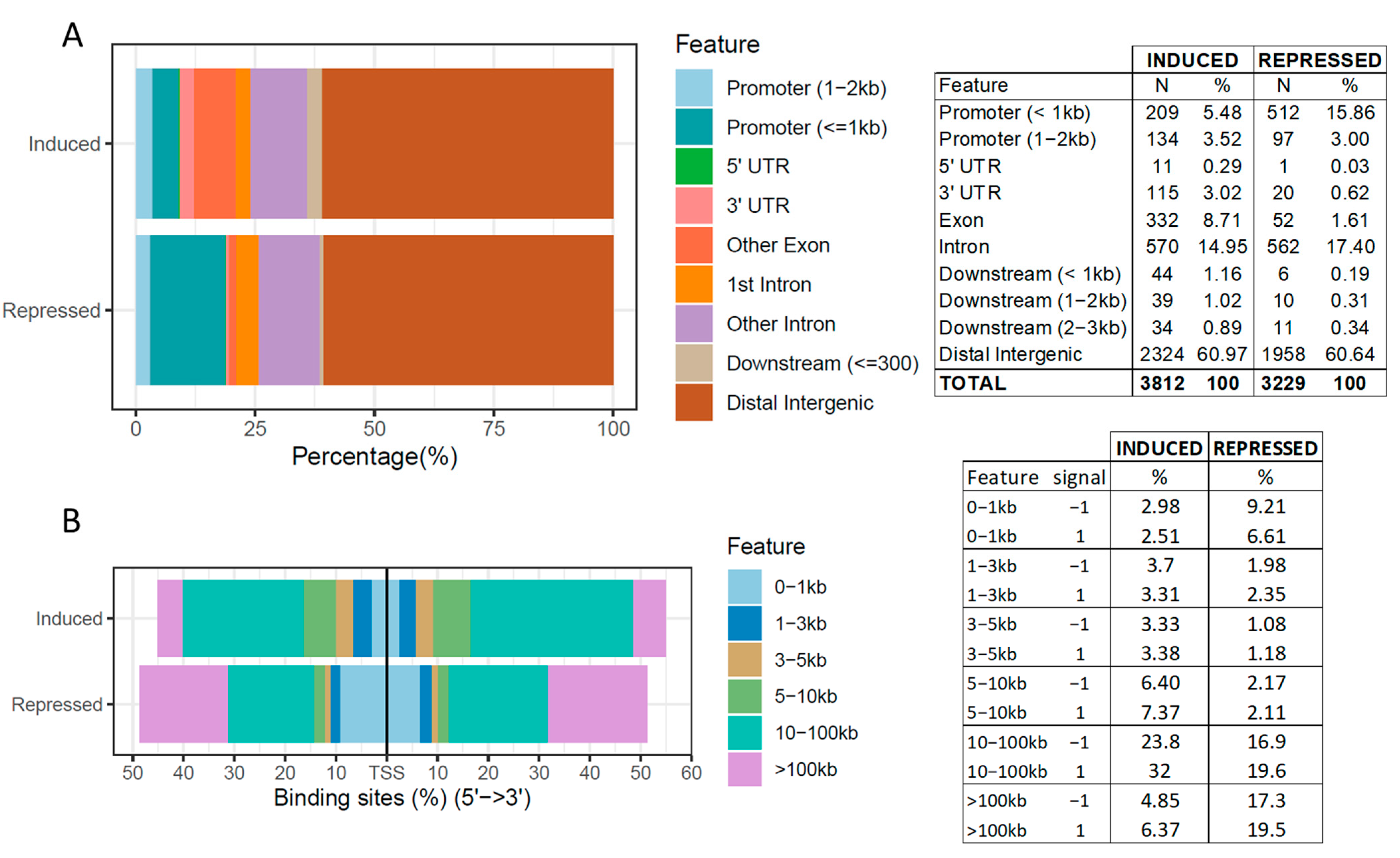
2.2. Differentially CTCF-Binding Sites
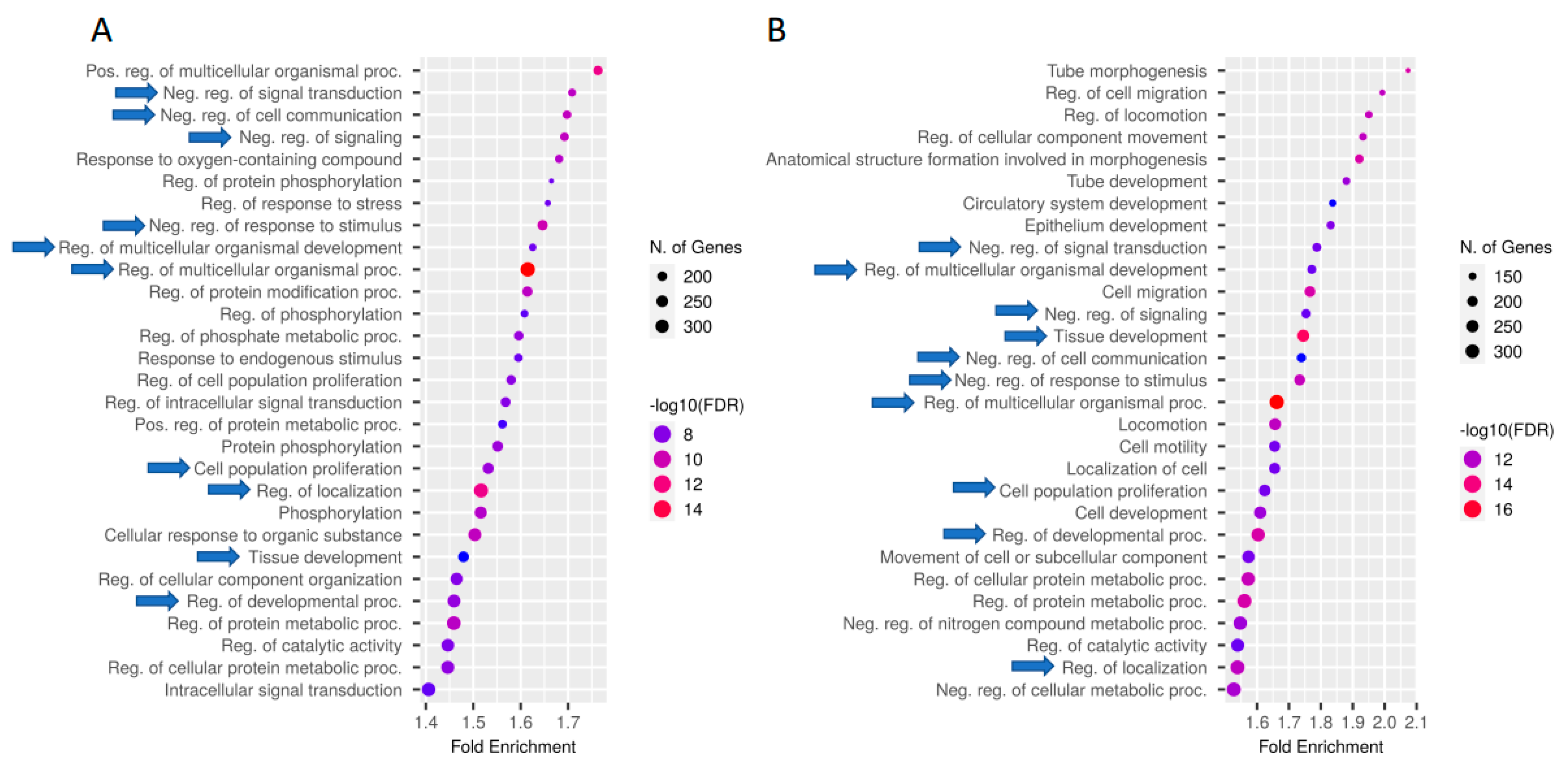
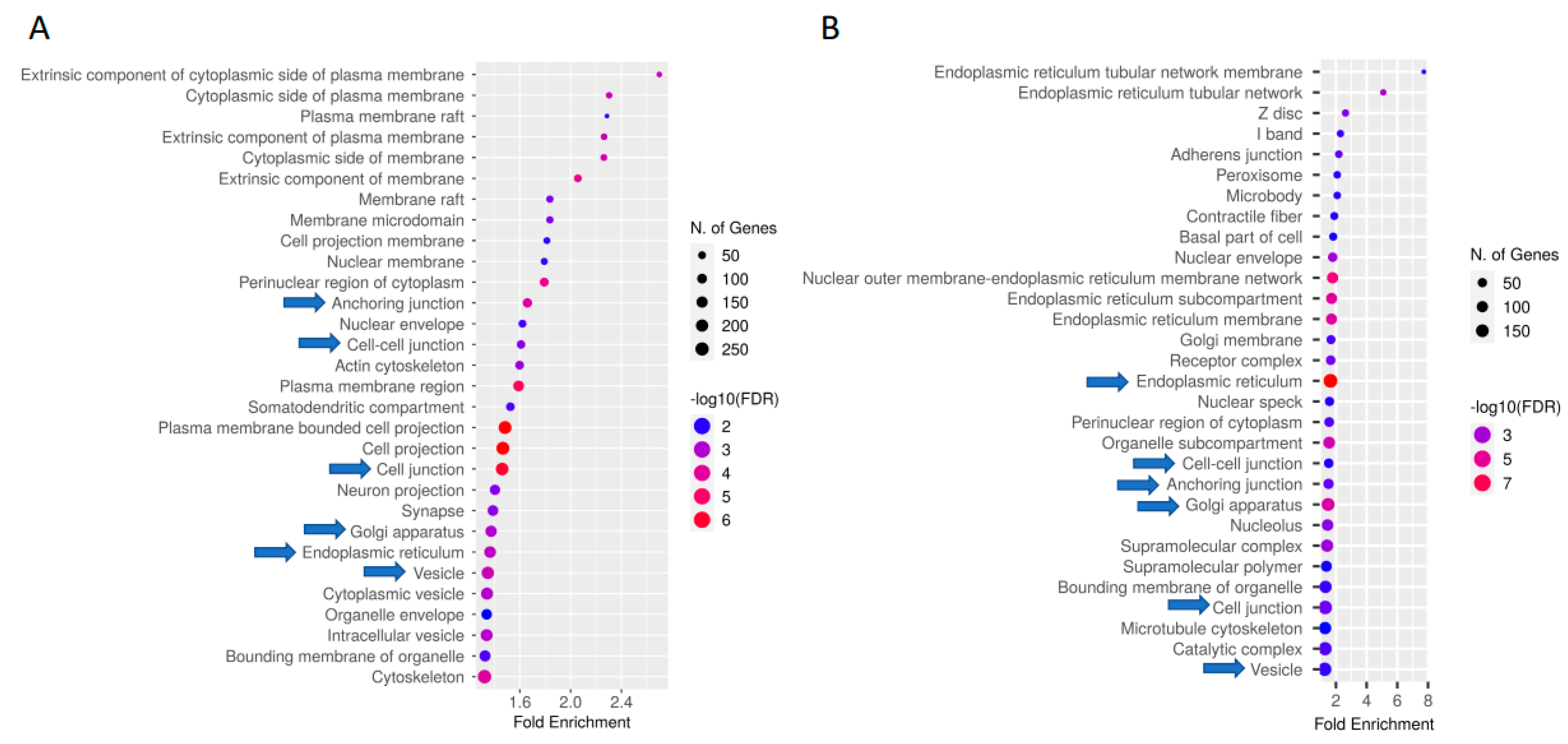
2.3. Functional Annotation of Differentially CTCF-Binding Sites
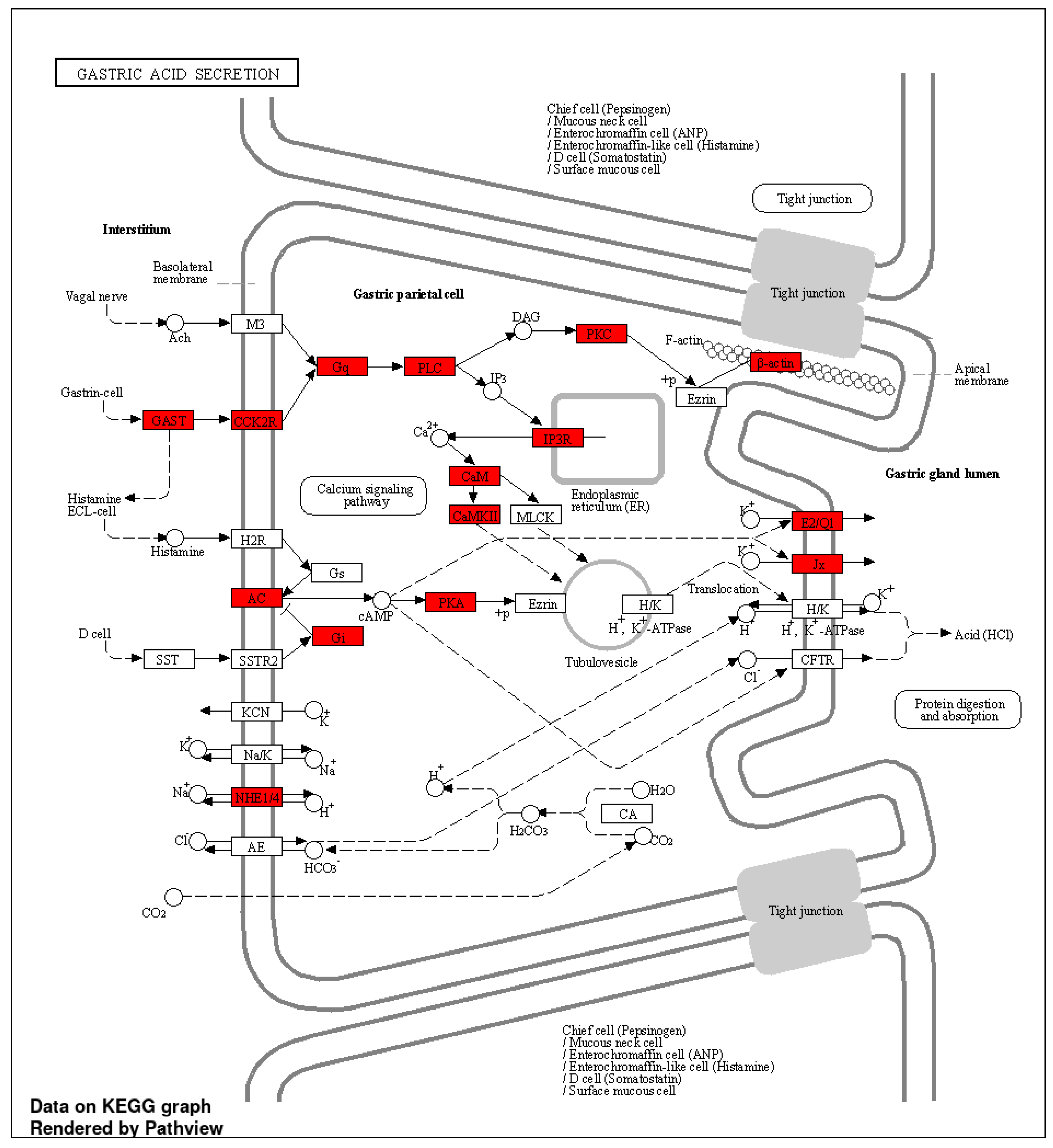
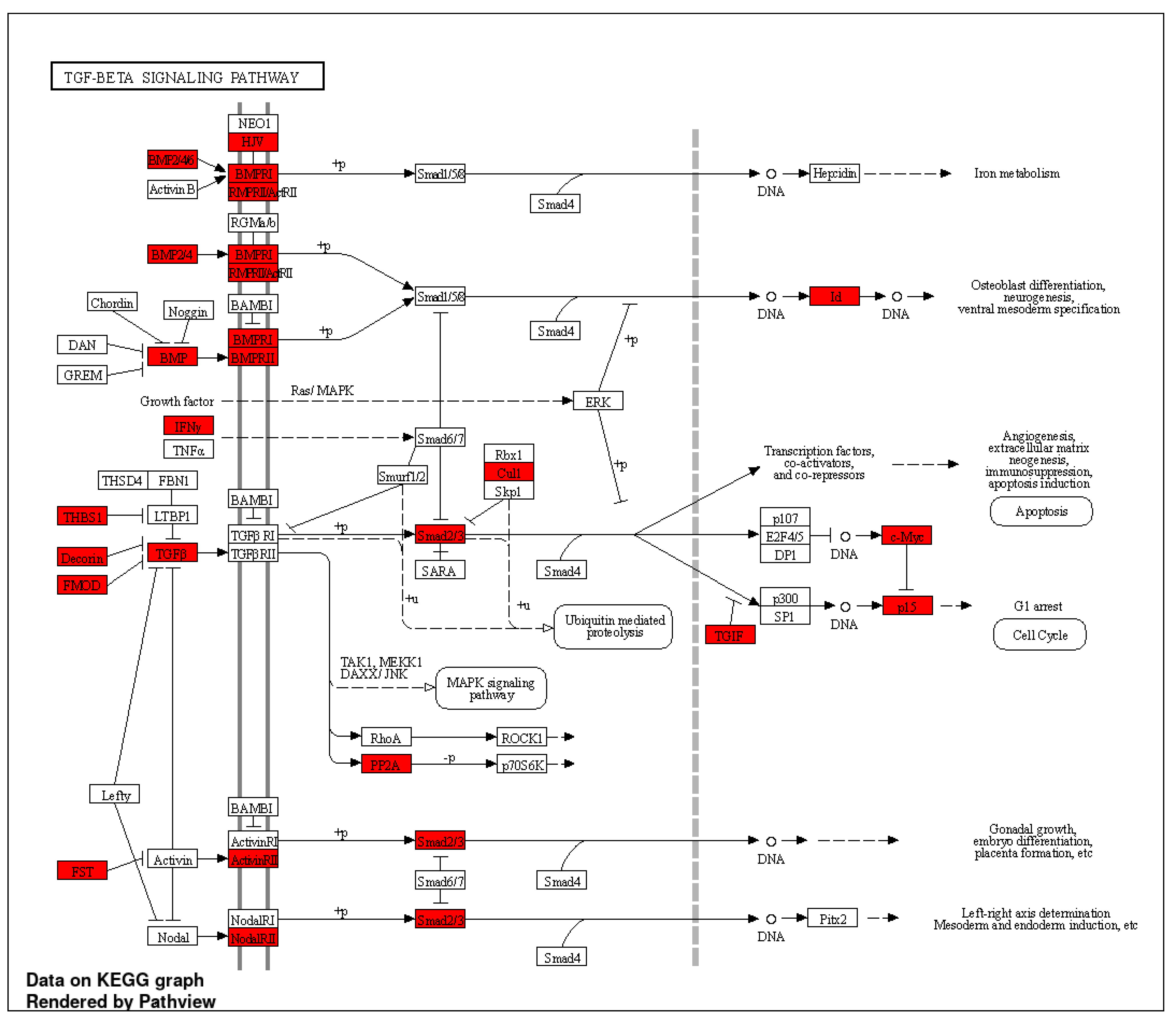
2.4. IPA Results
2.5. Identification of Putative Transcription Factor Binding Sites
2.6. RNA-Seq Integration with CTCF ChIP-Seq Data
2.7. Visualization of CTCF Signals in Selected Genes
3. Discussion
3.1. Identification of CTCF Peaks
3.2. Differentially CTCF-Binding Sites
3.3. Enrichment and Pathway Results of DCBS
3.4. Putative Transcription Factor Binding Sites
3.5. RNA-Seq Integration with CTCF-Binding Sites
3.6. CTCF Signals in Selected Genes
4. Materials and Methods
4.1. Rumen Epithelial Tissue Collection
4.2. CTCF ChIP-Sequencing
4.3. ChIP-Seq Data Processing and Mapping
4.4. ChIP-Seq Peak Calling and Quality Check
4.5. Identification of Differentially CTCF-Binding Sites
4.6. DCBS Annotation
4.7. Enrichment and Pathway Analysis
4.8. Motif Enrichment
4.9. RNA-Seq Integration with CTCF ChIP-Seq Data
4.10. Visualization of the CTCF Signals in Selected Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Diao, Q.; Zhang, R.; Fu, T. Review of Strategies to Promote Rumen Development in Calves. Animals 2019, 9, 490. [Google Scholar] [CrossRef] [PubMed]
- Warner, R.G.; Flatt, W.P.; Loosli, J.K. Ruminant Nutrition, Dietary Factors Influencing Development of Ruminant Stomach. J. Agric. Food Chem. 1956, 4, 788–792. [Google Scholar] [CrossRef]
- Steele, M.A.; Penner, G.B.; Chaucheyras-Durand, F.; Guan, L.L. Development and physiology of the rumen and the lower gut: Targets for improving gut health. J. Dairy Sci. 2016, 99, 4955–4966. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Fang, L.; Baldwin, R.L.T.; Connor, E.E.; Cole, J.B.; Van Tassell, C.P.; Ma, L.; Li, C.J.; Liu, G.E. Single-cell transcriptomic analyses of dairy cattle ruminal epithelial cells during weaning. Genomics 2021, 113, 2045–2055. [Google Scholar] [CrossRef]
- Connor, E.E.; Baldwin, R.L.; Li, C.-J.; Li, R.W.; Chung, H. Gene expression in bovine rumen epithelium during weaning identifies molecular regulators of rumen development and growth. Funct. Integr. Genom. 2013, 13, 133–142. [Google Scholar] [CrossRef]
- Connor, E.E.; Baldwin, R.L.T.; Walker, M.P.; Ellis, S.E.; Li, C.; Kahl, S.; Chung, H.; Li, R.W. Transcriptional regulators transforming growth factor-beta1 and estrogen- related receptor-alpha identified as putative mediators of calf rumen epithelial tissue development and function during weaning. J. Dairy Sci. 2014, 97, 4193–4207. [Google Scholar] [CrossRef]
- Baldwin Vi, R.L.; Liu, M.; Connor, E.E.; Ramsay, T.G.; Liu, G.E.; Li, C.J. Transcriptional Reprogramming in Rumen Epithelium during the Developmental Transition of Pre-Ruminant to the Ruminant in Cattle. Animals 2021, 11, 2870. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, S.; Baldwin VI, R.L.; Connor, E.E.; Cole, J.B.; Ma, L.; Fang, L.; Li, C.-J.; Liu, G.E. Functional annotation of regulatory elements in cattle genome reveals the roles of extracellular interaction and dynamic change of chromatin states in rumen development during weaning. BMC Genom. 2022, 114, 110296. [Google Scholar] [CrossRef]
- Consortium, E.P. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004, 306, 636–640. [Google Scholar] [CrossRef]
- Yue, F.; Cheng, Y.; Breschi, A.; Vierstra, J.; Wu, W.; Ryba, T.; Sandstrom, R.; Ma, Z.; Davis, C.; Pope, B.D.; et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 2014, 515, 355–364. [Google Scholar] [CrossRef]
- Gerstein Mark, B.; Lu Zhi, J.; Van Nostrand Eric, L.; Cheng, C.; Bradley, I.A.; Liu, T.; Kevin, Y.Y.; Robilotto, R.; Rechtsteiner, A.; Ikegami, K.; et al. Integrative Analysis of the Caenorhabditis elegans Genome by the modENCODE Project. Science 2010, 330, 1775–1787. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Ernst, J.; Kharchenko, P.V.; Kheradpour, P.; Negre, N.; Eaton, M.L.; Landolin, J.M.; Bristow, C.A.; Ma, L.; Lin, M.F.; et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 2010, 330, 1787–1797. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, P.A.; Naval-Sanchez, M.; Menzies, M.; Nguyen, L.T.; Porto-Neto, L.R.; Fortes, M.R.S.; Reverter, A. Chromatin accessibility and regulatory vocabulary across indicine cattle tissues. Genome Biol. 2021, 22, 273. [Google Scholar] [CrossRef]
- Cao, X.; Cheng, J.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. Comparative Enhancer Map of Cattle Muscle Genome Annotated by ATAC-Seq. Front. Vet. Sci. 2021, 8, 1467. [Google Scholar] [CrossRef]
- Fang, L.; Liu, S.; Liu, M.; Kang, X.; Lin, S.; Li, B.; Connor, E.E.; Baldwin, R.L.T.; Tenesa, A.; Ma, L.; et al. Functional annotation of the cattle genome through systematic discovery and characterization of chromatin states and butyrate-induced variations. BMC Biol. 2019, 17, 68. [Google Scholar] [CrossRef]
- Halstead, M.M.; Kern, C.; Saelao, P.; Wang, Y.; Chanthavixay, G.; Medrano, J.F.; Van Eenennaam, A.L.; Korf, I.; Tuggle, C.K.; Ernst, C.W.; et al. A comparative analysis of chromatin accessibility in cattle, pig, and mouse tissues. BMC Genom. 2020, 21, 698. [Google Scholar] [CrossRef]
- Johnston, D.; Kim, J.; Taylor, J.F.; Earley, B.; McCabe, M.S.; Lemon, K.; Duffy, C.; McMenamy, M.; Cosby, S.L.; Waters, S.M. ATAC-Seq identifies regions of open chromatin in the bronchial lymph nodes of dairy calves experimentally challenged with bovine respiratory syncytial virus. BMC Genom. 2021, 22, 14. [Google Scholar] [CrossRef]
- Kern, C.; Wang, Y.; Xu, X.; Pan, Z.; Halstead, M.; Chanthavixay, G.; Saelao, P.; Waters, S.; Xiang, R.; Chamberlain, A.; et al. Functional annotations of three domestic animal genomes provide vital resources for comparative and agricultural research. Nat. Commun. 2021, 12, 1821. [Google Scholar] [CrossRef]
- Boschiero, C.; Gao, Y.; Baldwin, R.L.T.; Ma, L.; Liu, G.E.; Li, C.-J. Characterization of Accessible Chromatin Regions in Cattle Rumen Epithelial Tissue during Weaning. Genes 2022, 13, 535. [Google Scholar] [CrossRef]
- Zhou, V.W.; Goren, A.; Bernstein, B.E. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011, 12, 7–18. [Google Scholar] [CrossRef]
- Kim, S.; Yu, N.-K.; Kaang, B.-K. CTCF as a multifunctional protein in genome regulation and gene expression. Exp. Mol. Med. 2015, 47, e166. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wu, D.; Wang, X. Roles of CTCF in conformation and functions of chromosome. Semin. Cell Dev. Biol. 2019, 90, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Chao, W.; Huynh, K.D.; Spencer, R.J.; Davidow, L.S.; Lee, J.T. CTCF, a candidate trans-acting factor for X-inactivation choice. Science 2002, 295, 345–347. [Google Scholar] [CrossRef]
- Kang, M.A.; Lee, J.-S. A Newly Assigned Role of CTCF in Cellular Response to Broken DNAs. Biomolecules 2021, 11, 363. [Google Scholar] [CrossRef]
- Burgess-Beusse, B.; Farrell, C.; Gaszner, M.; Litt, M.; Mutskov, V.; Recillas-Targa, F.; Simpson, M.; West, A.; Felsenfeld, G. The insulation of genes from external enhancers and silencing chromatin. Proc. Natl. Acad. Sci. USA 2002, 99, 16433–16437. [Google Scholar] [CrossRef]
- Cuddapah, S.; Jothi, R.; Schones, D.E.; Roh, T.-Y.; Cui, K.; Zhao, K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009, 19, 24–32. [Google Scholar] [CrossRef]
- Kim, T.H.; Abdullaev, Z.K.; Smith, A.D.; Ching, K.A.; Loukinov, D.I.; Green, R.D.; Zhang, M.Q.; Lobanenkov, V.V.; Ren, B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 2007, 128, 1231–1245. [Google Scholar] [CrossRef]
- Phillips, J.E.; Corces, V.G. CTCF: Master weaver of the genome. Cell 2009, 137, 1194–1211. [Google Scholar] [CrossRef]
- Matthews, B.J.; Waxman, D.J. Impact of 3D genome organization, guided by cohesin and CTCF looping, on sex-biased chromatin interactions and gene expression in mouse liver. Epigenetics Chromatin 2020, 13, 30. [Google Scholar] [CrossRef]
- Tian, G.G.; Zhao, X.; Hou, C.; Xie, W.; Li, X.; Wang, Y.; Wang, L.; Li, H.; Zhao, X.; Li, J.; et al. Integrative analysis of the 3D genome structure reveals that CTCF maintains the properties of mouse female germline stem cells. Cell. Mol. Life Sci. 2022, 79, 22. [Google Scholar] [CrossRef]
- Wen, Z.; Zhang, L.; Ruan, H.; Li, G. Histone variant H2A.Z regulates nucleosome unwrapping and CTCF binding in mouse ES cells. Nucleic Acids Res. 2020, 48, 5939–5952. [Google Scholar] [CrossRef] [PubMed]
- Massa, A.T.; Mousel, M.R.; Herndon, M.K.; Herndon, D.R.; Murdoch, B.M.; White, S.N. Genome-Wide Histone Modifications and CTCF Enrichment Predict Gene Expression in Sheep Macrophages. Front. Genet. 2021, 11, 612031. [Google Scholar] [CrossRef] [PubMed]
- Davenport, K.M.; Massa, A.T.; Bhattarai, S.; McKay, S.D.; Mousel, M.R.; Herndon, M.K.; White, S.N.; Cockett, N.E.; Smith, T.P.L.; Murdoch, B.M.; et al. Characterizing Genetic Regulatory Elements in Ovine Tissues. Front. Genet. 2021, 12, 628849. [Google Scholar] [CrossRef] [PubMed]
- Prowse-Wilkins, C.P.; Wang, J.; Xiang, R.; Garner, J.B.; Goddard, M.E.; Chamberlain, A.J. Putative Causal Variants Are Enriched in Annotated Functional Regions From Six Bovine Tissues. Front. Genet. 2021, 12, 664379. [Google Scholar] [CrossRef]
- Rosen, B.D.; Bickhart, D.M.; Schnabel, R.D.; Koren, S.; Elsik, C.G.; Tseng, E.; Rowan, T.N.; Low, W.Y.; Zimin, A.; Couldrey, C.; et al. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience 2020, 9, giaa021. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Shen, L.; Shao, N.Y.; Liu, X.; Maze, I.; Feng, J.; Nestler, E.J. diffReps: Detecting differential chromatin modification sites from ChIP- seq data with biological replicates. PLoS ONE 2013, 8, e65598. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef]
- Wang, S.; Sun, H.; Ma, J.; Zang, C.; Wang, C.; Wang, J.; Tang, Q.; Meyer, C.A.; Zhang, Y.; Liu, X.S. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc. 2013, 8, 2502–2515. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- de Wit, E.; Vos, E.S.; Holwerda, S.J.; Valdes-Quezada, C.; Verstegen, M.J.; Teunissen, H.; Splinter, E.; Wijchers, P.J.; Krijger, P.H.; de Laat, W. CTCF Binding Polarity Determines Chromatin Looping. Mol. Cell 2015, 60, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.S.; Pustova, I.; Cattoglio, C.; Tjian, R.; Darzacq, X. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife 2017, 6, e25776. [Google Scholar] [CrossRef]
- Landt, S.G.; Marinov, G.K.; Kundaje, A.; Kheradpour, P.; Pauli, F.; Batzoglou, S.; Bernstein, B.E.; Bickel, P.; Brown, J.B.; Cayting, P.; et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012, 22, 1813–1831. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Tian, Y.; Shu, W.; Bo, X.; Wang, S. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS ONE 2012, 7, e41374. [Google Scholar] [CrossRef] [PubMed]
- Ricci, S.; Rivera-Chacon, R.; Petri, R.M.; Sener-Aydemir, A.; Sharma, S.; Reisinger, N.; Zebeli, Q.; Castillo-Lopez, E. Supplementation With Phytogenic Compounds Modulates Salivation and Salivary Physico-Chemical Composition in Cattle Fed a High-Concentrate Diet. Front. Physiol. 2021, 12, 645529. [Google Scholar] [CrossRef]
- Allen, M.S. Relationship between fermentation acid production in the rumen and the requirement for physically effective fiber. J. Dairy Sci. 1997, 80, 1447–1462. [Google Scholar] [CrossRef]
- Toullec, R.; Chayvialle, J.A.; Guilloteau, P.; Bernard, C. Early-life patterns of plasma gut regulatory peptide levels in calves. Effects of age, weaning and feeding. Comp. Biochem. Physiol. Comp. Physiol. 1992, 102, 203–209. [Google Scholar] [CrossRef]
- Bergstrom, K.S.; Xia, L. Mucin-type O-glycans and their roles in intestinal homeostasis. Glycobiology 2013, 23, 1026–1037. [Google Scholar] [CrossRef]
- Hoorens, P.R.; Rinaldi, M.; Li, R.W.; Goddeeris, B.; Claerebout, E.; Vercruysse, J.; Geldhof, P. Genome wide analysis of the bovine mucin genes and their gastrointestinal transcription profile. BMC Genom. 2011, 12, 140. [Google Scholar] [CrossRef]
- Deplancke, B.; Gaskins, H.R. Microbial modulation of innate defense: Goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 2001, 73, 1131s–1141s. [Google Scholar] [CrossRef]
- Hu, G.; Wang, Z.; Zhang, R.; Sun, W.; Chen, X. The Role of Apelin/Apelin Receptor in Energy Metabolism and Water Homeostasis: A Comprehensive Narrative Review. Front. Physiol. 2021, 12, 632886. [Google Scholar] [CrossRef]
- Wang, G.; Kundu, R.; Han, S.; Qi, X.; Englander, E.W.; Quertermous, T.; Greeley, G.H. Ontogeny of apelin and its receptor in the rodent gastrointestinal tract. Regul. Pept. 2009, 158, 32–39. [Google Scholar] [CrossRef]
- Mesmin, C.; Fenaille, F.; Becher, F.; Tabet, J.C.; Ezan, E. Identification and characterization of apelin peptides in bovine colostrum and milk by liquid chromatography-mass spectrometry. J. Proteome Res. 2011, 10, 5222–5231. [Google Scholar] [CrossRef] [PubMed]
- Gorka, P.; Kowalski, Z.M.; Zabielski, R.; Guilloteau, P. Invited review: Use of butyrate to promote gastrointestinal tract development in calves. J. Dairy Sci. 2018, 101, 4785–4800. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.C.; Humphries, J.D.; Byron, A.; Millon-Frémillon, A.; Robertson, J.; Paul, N.R.; Ng, D.H.J.; Askari, J.A.; Humphries, M.J. Isolation of integrin-based adhesion complexes. Curr. Protoc. Cell Biol. 2015, 66, 9.8.1–9.8.15. [Google Scholar] [CrossRef] [PubMed]
- Yohe, T.T.; Tucker, H.L.M.; Parsons, C.L.M.; Geiger, A.J.; Akers, R.M.; Daniels, K.M. Short communication: Initial evidence supporting existence of potential rumen epidermal stem and progenitor cells. J. Dairy Sci. 2016, 99, 7654–7660. [Google Scholar] [CrossRef]
- Rosa-Garrido, M.; Ceballos, L.; Alonso-Lecue, P.; Abraira, C.; Delgado, M.D.; Gandarillas, A. A cell cycle role for the epigenetic factor CTCF-L/BORIS. PLoS ONE 2012, 7, e39371. [Google Scholar] [CrossRef]
- Slattery, C.; Ryan, M.P.; McMorrow, T. E2A proteins: Regulators of cell phenotype in normal physiology and disease. Int. J. Biochem. Cell Biol. 2008, 40, 1431–1436. [Google Scholar] [CrossRef]
- Goding, C.R.; Arnheiter, H. MITF-the first 25 years. Genes Dev. 2019, 33, 983–1007. [Google Scholar] [CrossRef]
- Schuijers, J.; Junker, J.P.; Mokry, M.; Hatzis, P.; Koo, B.-K.; Sasselli, V.; van der Flier, L.G.; Cuppen, E.; van Oudenaarden, A.; Clevers, H. Ascl2 Acts as an R-spondin/Wnt-Responsive Switch to Control Stemness in Intestinal Crypts. Cell Stem Cell 2015, 16, 158–170. [Google Scholar] [CrossRef]
- Fayard, E.; Auwerx, J.; Schoonjans, K. LRH-1: An orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004, 14, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Ogawa, K.; Nagarajan, R.P.; Zhang, M.; Kuang, C.; Chen, Y. Regulation of TG-interacting factor by transforming growth factor-beta. Biochem. J. 2003, 371, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.A.; Pietenpol, J.A.; Moses, H.L. A tale of two proteins: Differential roles and regulation of Smad2 and Smad3 in TGF-beta signaling. J. Cell. Biochem. 2007, 101, 9–33. [Google Scholar] [CrossRef] [PubMed]
- Boros, J.; Donaldson, I.J.; O’Donnell, A.; Odrowaz, Z.A.; Zeef, L.; Lupien, M.; Meyer, C.A.; Liu, X.S.; Brown, M.; Sharrocks, A.D. Elucidation of the ELK1 target gene network reveals a role in the coordinate regulation of core components of the gene regulation machinery. Genome Res. 2009, 19, 1963–1973. [Google Scholar] [CrossRef]
- de Los Fayos Alonso, I.G.; Liang, H.C.; Turner, S.D.; Lagger, S.; Merkel, O.; Kenner, L. The Role of Activator Protein-1 (AP-1) Family Members in CD30-Positive Lymphomas. Cancers 2018, 10, 93. [Google Scholar] [CrossRef]
- Wasylyk, B.; Hahn, S.L.; Giovane, A. The Ets family of transcription factors. Eur. J. Biochem. 1993, 211, 7–18. [Google Scholar] [CrossRef]
- Oh, S.; Shin, S.; Janknecht, R. ETV1, 4 and 5: An oncogenic subfamily of ETS transcription factors. Biochim. Biophys. Acta 2012, 1826, 1–12. [Google Scholar] [CrossRef]
- Scocco, P.; Mercati, F.; Brusaferro, A.; Ceccarelli, P.; Belardinelli, C.; Malfatti, A. Keratinisation degree of rumen epithelium and body condition score in sheep grazing on Brachypodium rupestre. Vet. Ital. 2013, 49, 211–217. [Google Scholar]
- Baldwin, R.L.; McLeod, K.R.; Klotz, J.L.; Heitmann, R.N. Rumen Development, Intestinal Growth and Hepatic Metabolism In The Pre- and Postweaning Ruminant. J. Dairy Sci. 2004, 87, E55–E65. [Google Scholar] [CrossRef]
- Ramos, S.C.; Jeong, C.D.; Mamuad, L.L.; Kim, S.H.; Kang, S.H.; Kim, E.T.; Cho, Y.I.; Lee, S.S.; Lee, S.S. Diet Transition from High-Forage to High-Concentrate Alters Rumen Bacterial Community Composition, Epithelial Transcriptomes and Ruminal Fermentation Parameters in Dairy Cows. Animals 2021, 11, 838. [Google Scholar] [CrossRef] [PubMed]
- Xiang, R.; Oddy, V.H.; Archibald, A.L.; Vercoe, P.E.; Dalrymple, B.P. Epithelial, metabolic and innate immunity transcriptomic signatures differentiating the rumen from other sheep and mammalian gastrointestinal tract tissues. PeerJ 2016, 4, e1762. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Atoji, Y.; Oohira, A.; Suzuki, Y. Immunohistochemical localization of chondroitin sulfate in the forestomach of the sheep. Eur. J. Histochem. 1995, 39, 265–272. [Google Scholar] [PubMed]
- Klatt, A.R.; Becker, A.-K.A.; Neacsu, C.D.; Paulsson, M.; Wagener, R. The matrilins: Modulators of extracellular matrix assembly. Int. J. Biochem. Cell Biol. 2011, 43, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.-H.; Lee, H.-J.; Cho, S.; Kim, H.J.; Hwang, J.Y.; Lee, C.-K.; Jeong, J.; Yoon, D.; Kim, H. Deleted copy number variation of Hanwoo and Holstein using next generation sequencing at the population level. BMC Genom. 2014, 15, 240. [Google Scholar] [CrossRef]
- Garcia-Mayea, Y.; Mir, C.; Carballo, L.; Sánchez-García, A.; Bataller, M.; Lleonart, M.E. TSPAN1, a novel tetraspanin member highly involved in carcinogenesis and chemoresistance. Biochim. Biophys. Acta BBA—Rev. Cancer 2022, 1877, 188674. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, C.A.; Lynch, A.M.; Hardin, J. Dynamic analysis identifies novel roles for DLG-1 subdomains in AJM-1 recruitment and LET-413-dependent apical focusing. J. Cell Sci. 2008, 121, 1477–1487. [Google Scholar] [CrossRef]
- Shafaq-Zadah, M.; Gomes-Santos, C.S.; Bardin, S.; Maiuri, P.; Maurin, M.; Iranzo, J.; Gautreau, A.; Lamaze, C.; Caswell, P.; Goud, B.; et al. Persistent cell migration and adhesion rely on retrograde transport of β1 integrin. Nat. Cell Biol. 2016, 18, 54–64. [Google Scholar] [CrossRef]
- Pang, Y.; Zhang, Z.; Wang, Z.; Wang, Y.; Yan, Y.; Li, S.; Tong, H. Platelet endothelial aggregation receptor-1 regulates bovine muscle satellite cell migration and differentiation via integrin beta-1 and focal adhesion kinase. Cell Adhes. Migr. 2019, 13, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Kent, W.J.; Zweig, A.S.; Barber, G.; Hinrichs, A.S.; Karolchik, D. BigWig and BigBed: Enabling browsing of large distributed datasets. Bioinformatics 2010, 26, 2204–2207. [Google Scholar] [CrossRef]
| Condition | N of Reads | Mapped Reads (bosTau9) | Mapped Reads% | MT Reads | MT Reads% 1 | Duplicate Reads | Duplicate Reads% 1 | MAPQ < 10 Reads | MAPQ < 10 Reads% 1 | Clean Reads 2 |
|---|---|---|---|---|---|---|---|---|---|---|
| BW | 42,921,125 | 41,327,290 | 96.29 | 38,772 | 0.09 | 6,695,091 | 16.20 | 6,316,659 | 15.28 | 27,814,748 |
| AW | 44,702,240 | 42,063,278 | 94.10 | 72,080 | 0.16 | 4,140,259 | 9.84 | 5,655,767 | 13.45 | 31,627,368 |
| Total | 87,623,365 | 83,390,568 | - | 110,852 | - | 10,835,350 | - | 11,972,426 | - | 59,442,116 |
| Average | 43,811,683 | 41,695,284 | 95.19 | 55,426 | 0.13 | 5,417,675 | 13.02 | 5,986,213 | 14.37 | 29,721,058 |
| Condition | Clean Reads 1 | Clean Reads Used for FRiP 2 | CTCF Peaks 2 | N of Assigned Reads in Peaks 2 | FRiP 3 | Average Peak Length | Peaks Near TSS (±3 kb, %) |
|---|---|---|---|---|---|---|---|
| BW | 27,814,748 | 27,066,621 | 67,280 | 6,193,989 | 0.22 | 393 | 17.56 |
| AW | 31,627,368 | 30,864,883 | 39,891 | 4,401,267 | 0.14 | 498 | 20.61 |
| Total | 59,442,116 | 57,931,504 | 107,171 | 10,595,256 | - | - | - |
| Average | 29,721,058 | 28,965,752 | 53,586 | 5,297,628 | 0.18 | 446 | 19.09 |
| Filtration Steps of the After-Weaning DCBS | N | % |
|---|---|---|
| Initially detected DCBS (p < 0.05) | 26,871 | 100 |
| Filtered DCBS (FDR < 0.01 and |log2FC| > 1) | 8249 | 30.70 |
| Filtered DCBS that overlapped with peaks | 7041 | 26.20 |
| Total of filtered DCBS | 7041 | 100 |
| Induced sites with log2FC ≥ 1 | 3812 | 54.14 |
| Repressed sites with log2FC ≤ −1 | 3229 | 45.86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boschiero, C.; Gao, Y.; Baldwin, R.L., VI; Ma, L.; Li, C.-j.; Liu, G.E. Differentially CTCF-Binding Sites in Cattle Rumen Tissue during Weaning. Int. J. Mol. Sci. 2022, 23, 9070. https://doi.org/10.3390/ijms23169070
Boschiero C, Gao Y, Baldwin RL VI, Ma L, Li C-j, Liu GE. Differentially CTCF-Binding Sites in Cattle Rumen Tissue during Weaning. International Journal of Molecular Sciences. 2022; 23(16):9070. https://doi.org/10.3390/ijms23169070
Chicago/Turabian StyleBoschiero, Clarissa, Yahui Gao, Ransom L. Baldwin, VI, Li Ma, Cong-jun Li, and George E. Liu. 2022. "Differentially CTCF-Binding Sites in Cattle Rumen Tissue during Weaning" International Journal of Molecular Sciences 23, no. 16: 9070. https://doi.org/10.3390/ijms23169070






