Antiviral Activity of N1,N3-Disubstituted Uracil Derivatives against SARS-CoV-2 Variants of Concern
Abstract
:1. Introduction
2. Results
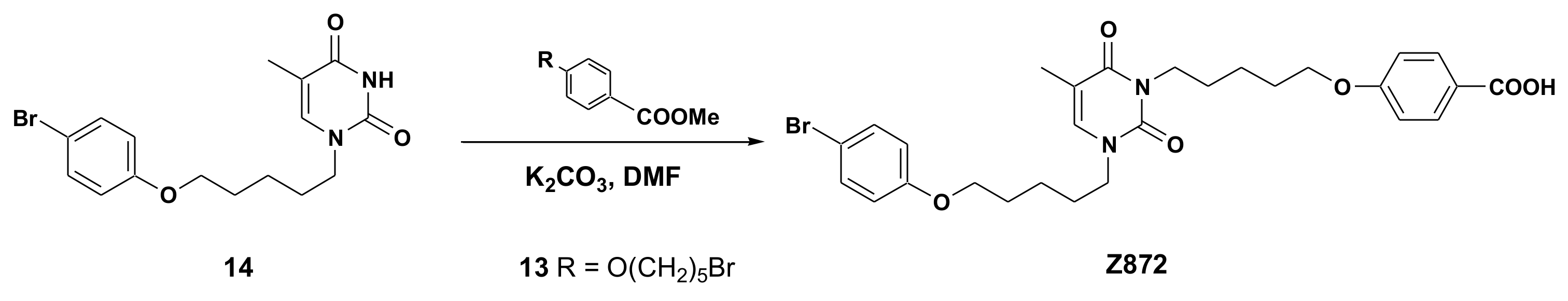
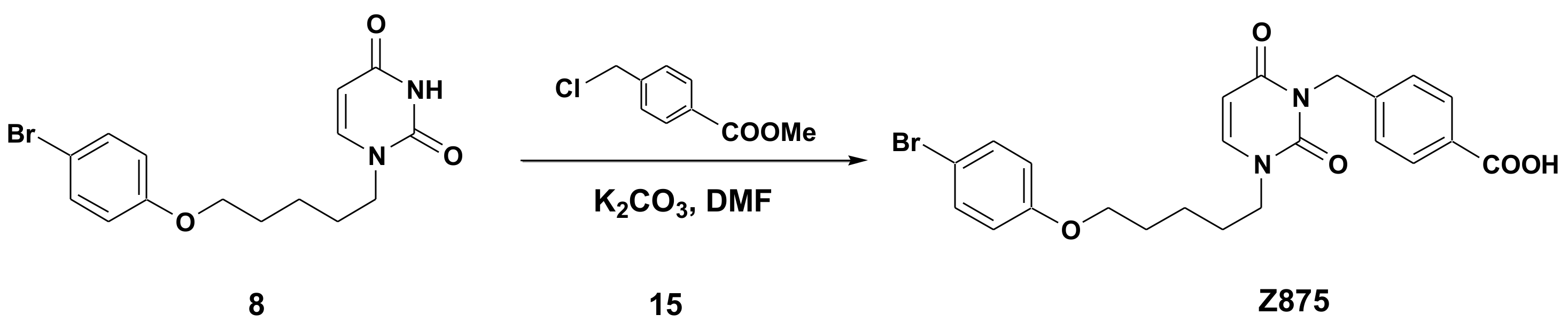
2.1. Chemical Synthesis of Panel Compounds
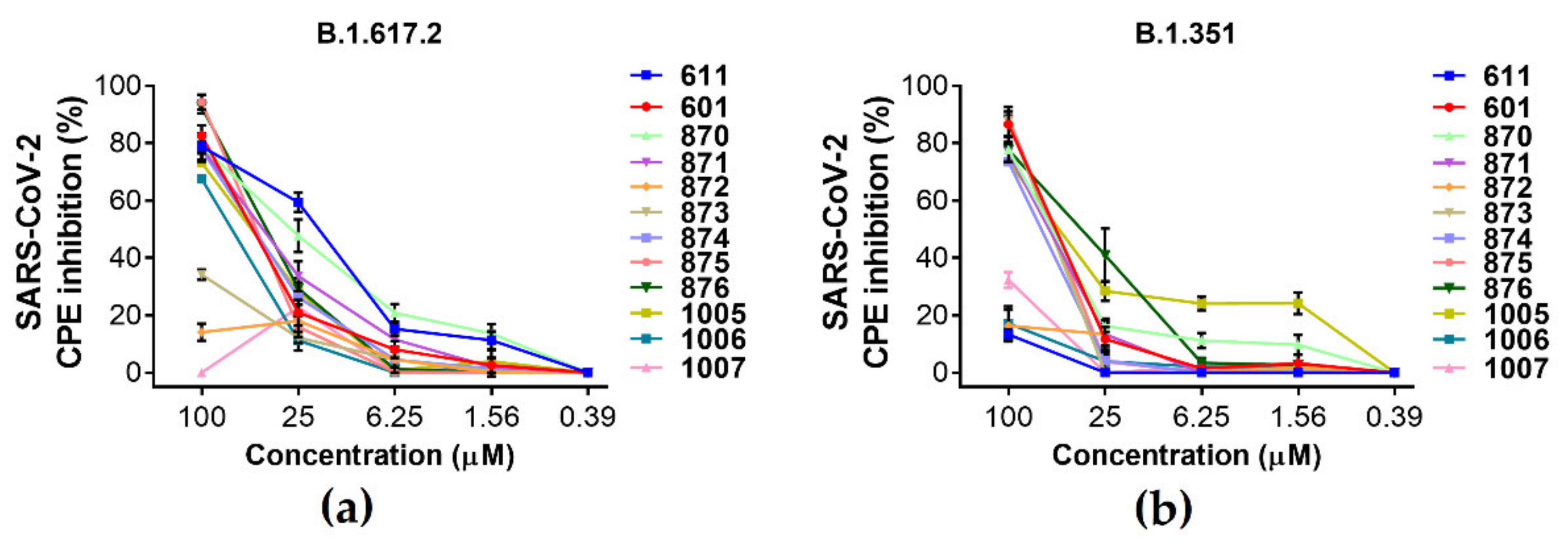
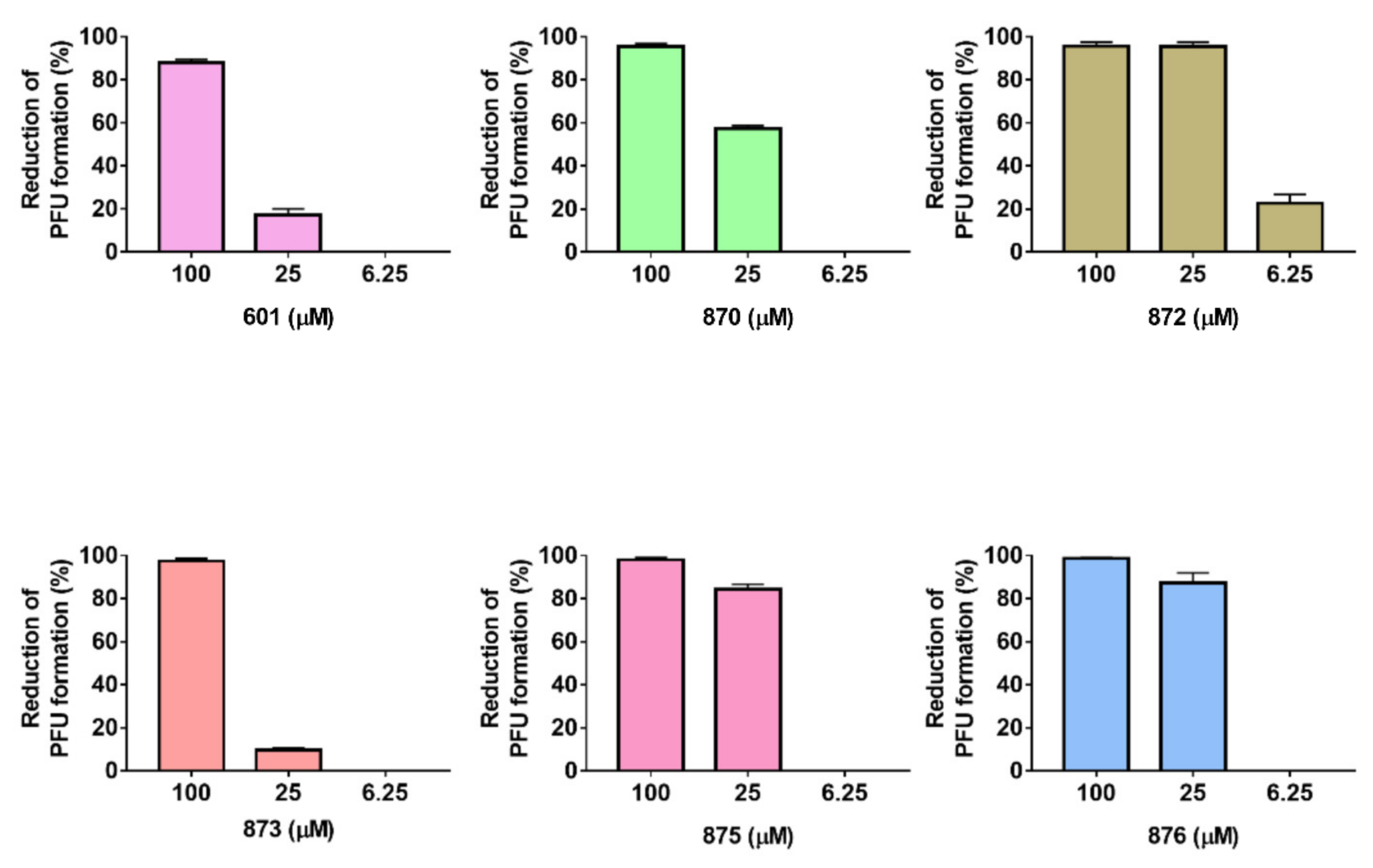
2.2. Antiviral Activity
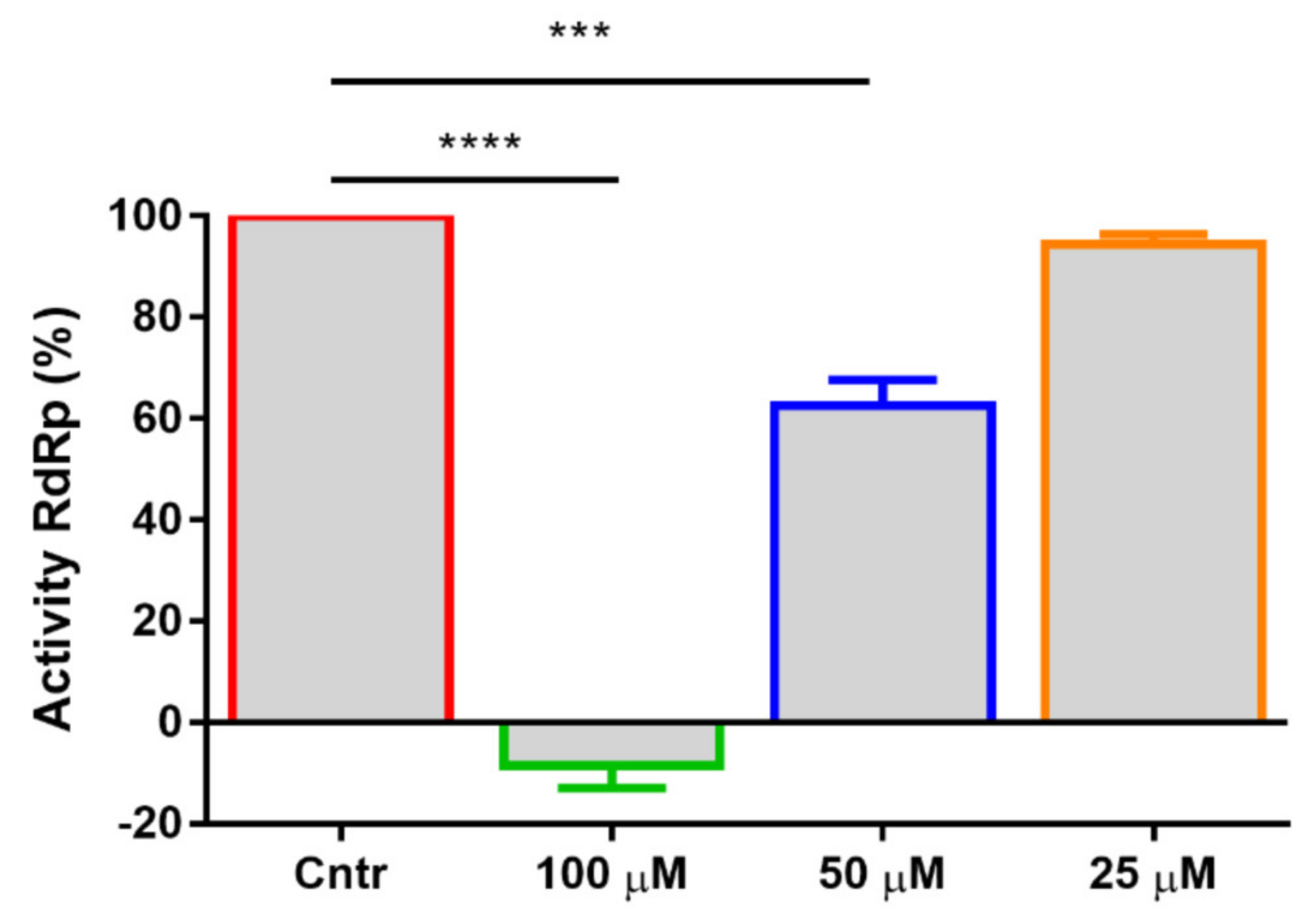
2.3. Mechanism of Action and Inhibition of SARS-CoV-2 RdRp
3. Discussion
4. Materials and Methods
4.1. General
4.2. General Procedure for the Synthesis of Acids 601, 870, 871, 872, 873 and 874
4.3. Cells and Viruses
4.4. Cytopathic Effect Inhibition Antiviral Assay
4.5. Plaque Reduction Assay
4.6. Time-of-Addition Experiments
4.7. Molecular Docking
4.8. Inhibition Activity of SARS-CoV-2 RNA-Dependent RNA Polymerase (RdRp)
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shankar, P.R.; Nadarajah, V.D.; Wilson, I.G. Implications of the ongoing coronavirus disease 2019 pandemic for primary care. Aust. J. Prim. Health 2022, 28, 200–203. [Google Scholar] [CrossRef] [PubMed]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Stadler, K.; Masignani, V.; Eickmann, M.; Becker, S.; Abrignani, S.; Klenk, H.-D.; Rappuoli, R. SARS—Beginning to understand a new virus. Nat. Rev. Genet. 2003, 1, 209–218. [Google Scholar] [CrossRef]
- Yadav, R.; Chaudhary, J.; Jain, N.; Chaudhary, P.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821. [Google Scholar] [CrossRef]
- Markov, P.V.; Katzourakis, A.; Stilianakis, N.I. Antigenic evolution will lead to new SARS-CoV-2 variants with unpredictable severity. Nat. Rev. Genet. 2022, 20, 251–252. [Google Scholar] [CrossRef] [PubMed]
- Mei, M.; Tan, X. Current Strategies of Antiviral Drug Discovery for COVID-19. Front. Mol. Biosci. 2021, 8, 671263. [Google Scholar] [CrossRef]
- Gottlieb, R.L.; Vaca, C.E.; Paredes, R.; Mera, J.; Webb, B.J.; Perez, G.; Oguchi, G.; Ryan, P.; Nielsen, B.U.; Brown, M.; et al. Early Remdesivir to Prevent Progression to Severe COVID-19 in Outpatients. N. Engl. J. Med. 2022, 386, 305–315. [Google Scholar] [CrossRef]
- Jayk Bernal, A.; Gomes da Silva, M.M.; Musungaie, D.B.; Kovalchuk, E.; Gonzalez, A.; Delos Reyes, V.; Martín-Quirós, A.; Caraco, Y.; Williams-Diaz, A.; Brown, M.L.; et al. Molnupiravir for Oral Treatment of COVID-19 in Nonhospitalized Patients. N. Engl. J. Med. 2022, 386, 509–520. [Google Scholar] [CrossRef]
- Hammond, J.; Leister-Tebbe, H.; Gardner, A.; Abreu, P.; Bao, W.; Wisemandle, W.; Baniecki, M.; Hendrick, V.M.; Damle, B.; Simón-Campos, A.; et al. Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with COVID-19. N. Engl. J. Med. 2022, 386, 1397–1408. [Google Scholar] [CrossRef]
- Usach, I.; Melis, V.; Peris, J.-E. Non-nucleoside reverse transcriptase inhibitors: A review on pharmacokinetics, pharmacodynamics, safety and tolerability. J. Int. AIDS Soc. 2013, 16, 18567. [Google Scholar] [CrossRef]
- Tian, L.; Qiang, T.; Liang, C.; Ren, X.; Jia, M.; Zhang, J.; Li, J.; Wan, M.; YuWen, X.; Li, H.; et al. RNA-dependent RNA polymerase (RdRp) inhibitors: The current landscape and repurposing for the COVID-19 pandemic. Eur. J. Med. Chem. 2021, 213, 113201. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Nandi, R.; Vishwakarma, P.; Prakash, A.; Kumar, D. Discovering Potential RNA Dependent RNA Polymerase Inhibitors as Prospective Drugs Against COVID-19: An in Silico Approach. Front. Pharmacol. 2021, 12, 267. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Sheahan, T.P.; Sims, A.C.; Leist, S.R.; Schäfer, A.; Won, J.; Brown, A.J.; Montgomery, S.A.; Hogg, A.; Babusis, D.; Clarke, M.O.; et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020, 11, 222. [Google Scholar] [CrossRef]
- Menéndez-Arias, L. Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase. Virus Res. 2008, 134, 124–146. [Google Scholar] [CrossRef] [PubMed]
- Paton, N.I.; Kityo, C.; Thompson, J.; Nankya, I.; Bagenda, L.; Hoppe, A.; Hakim, J.; Kambugu, A.; van Oosterhout, J.J.; Kiconco, M.; et al. Nucleoside Reverse-Transcriptase Inhibitor Cross-Resistance and Outcomes from Second-Line Antiretroviral Therapy in the Public Health Approach: An Observational Analysis within the Randomised, Open-Label, EARNEST Trial. Lancet HIV 2017, 4, e341–e348. [Google Scholar] [CrossRef]
- Paramonova, M.P.; Snoeck, R.; Andrei, G.; Khandazhinskaya, A.L.; Novikov, M.S. New acetamide derivatives containing (ω-p-bromophenoxyalkyl)uracil moiety and their anticytomegalovirus activity. Mendeleev Commun. 2020, 30, 602–603. [Google Scholar] [CrossRef]
- Babkov, D.A.; Khandazhinskaya, A.L.; Chizhov, A.O.; Andrei, G.; Snoeck, R.; Seley-Radtke, K.L.; Novikov, M.S. Toward the discovery of dual HCMV–VZV inhibitors: Synthesis, structure activity relationship analysis, and cytotoxicity studies of long chained 2-uracil-3-yl-N-(4-phenoxyphenyl)acetamides. Bioorg. Med. Chem. 2015, 23, 7035–7044. [Google Scholar] [CrossRef]
- Novikov, M.S.; Ivanova, O.N.; Ivanov, A.V.; Ozerov, A.A.; Valuev-Elliston, V.T.; Temburnikar, K.; Gurskaya, G.V.; Kochetkov, S.N.; Pannecouque, C.; Balzarini, J.; et al. 1-[2-(2-Benzoyl- and 2-benzylphenoxy)ethyl]uracils as potent anti-HIV-1 agents. Bioorg. Med. Chem. 2011, 19, 5794–5802. [Google Scholar] [CrossRef]
- Magri, A.; Ozerov, A.; Tunitskaya, V.L.; Valuev-Elliston, V.T.; Wahid, A.; Pirisi, M.; Simmonds, P.; Ivanov, A.V.; Novikov, M.S.; Patel, A.H. Exploration of acetanilide derivatives of 1-(ω-phenoxyalkyl)uracils as novel inhibitors of Hepatitis C Virus replication. Sci. Rep. 2016, 6, 29487. [Google Scholar] [CrossRef] [Green Version]
- Sapozhnikova, K.A.; Slesarchuk, N.A.; Orlov, A.A.; Khvatov, E.V.; Radchenko, E.V.; Chistov, A.A.; Ustinov, A.V.; Palyulin, V.A.; Kozlovskaya, L.I.; Osolodkin, D.I.; et al. Ramified derivatives of 5-(perylen-3-ylethynyl)uracil-1-acetic acid and their antiviral properties. RSC Adv. 2019, 9, 26014–26023. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.R.; Kelley, J.L. Irreversible Enzyme Inhibitors. CLXXI. Inhibition of FUDR Phosphorylase from Walker 256 Rat Tumor by 5-Substituted Uracils. J. Med. Chem. 1970, 13, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Malik, V.; Singh, P.; Kumar, S. Regioselective synthesis of 1-allyl- and 1-arylmethyl uracil and thymine derivatives. Tetrahedron 2005, 61, 4009–4014. [Google Scholar] [CrossRef]
- Novikov, M.S.; Babkov, D.A.; Paramonova, M.P.; Khandazhinskaya, A.L.; Ozerov, A.A.; Chizhov, A.O.; Andrei, G.; Snoeck, R.; Balzarini, J.; Seley-Radtke, K.L. Synthesis and anti-HCMV activity of 1-[ω-(phenoxy)alkyl]uracil derivatives and analogues thereof. Bioorg. Med. Chem. 2013, 21, 4151–4157. [Google Scholar] [CrossRef] [PubMed]
- Paramonova, M.P.; Ozerov, A.A.; Chizhov, A.O.; Snoeck, R.; Andrei, G.; Khandazhinskaya, A.L.; Novikov, M.S. Synthesis of uracil–coumarin conjugates as potential inhibitors of virus replication. Mendeleev Commun. 2019, 29, 638–639. [Google Scholar] [CrossRef]
- Paramonova, M.P.; Babkov, D.A.; Valuev-Elliston, V.T.; Ivanov, A.V.; Kochetkov, S.N.; Pannecouque, C.; Ozerov, A.A.; Balzarini, J.; Novikov, M.S. Synthesis and Anti-HIV-1 Activity of 1-[ω-(Phenoxy)Alkyl and -Alkenyl]Uracil Derivatives. Pharm. Chem. J. 2013, 47, 459–463. [Google Scholar] [CrossRef]
- Shi, Y.; Zhang, X.; Mu, K.; Peng, C.; Zhu, Z.; Wang, X.; Yang, Y.; Xu, Z.; Zhu, W. D3Targets-2019-nCoV: A webserver for predicting drug targets and for multi-target and multi-site based virtual screening against COVID-19. Acta Pharm. Sin. B 2020, 10, 1239–1248. [Google Scholar] [CrossRef]
- Grein, J.; Ohmagari, N.; Shin, D.; Diaz, G.; Asperges, E.; Castagna, A.; Feldt, T.; Green, G.; Green, M.L.; Lescure, F.-X.; et al. Compassionate Use of Remdesivir for Patients with Severe COVID-19. N. Engl. J. Med. 2020, 382, 2327–2336. [Google Scholar] [CrossRef]
- Painter, W.P.; Holman, W.; Bush, J.A.; Almazedi, F.; Malik, H.; Eraut, N.C.J.E.; Morin, M.J.; Szewczyk, L.J.; Painter, G.R. Human Safety, Tolerability, and Pharmacokinetics of Molnupiravir, a Novel Broad-Spectrum Oral Antiviral Agent with Activity against SARS-CoV-2. Antimicrob. Agents Chemother. 2021, 65, e02428-20. [Google Scholar] [CrossRef]
- Owen, D.R.; Allerton, C.M.N.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J.; et al. An Oral SARS-CoV-2 Mpro Inhibitor Clinical Candidate for the Treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef]
- Siniavin, A.E.; Streltsova, M.A.; Nikiforova, M.A.; Kudryavtsev, D.S.; Grinkina, S.D.; Gushchin, V.A.; Mozhaeva, V.A.; Starkov, V.G.; Osipov, A.V.; Lummis, S.C.R.; et al. Snake venom phospholipase A2s exhibit strong virucidal activity against SARS-CoV-2 and inhibit the viral spike glycoprotein interaction with ACE2. Experientia 2021, 78, 7777–7794. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Wang, R.; Chan, J.F.-W.; Zhang, A.J.; Cheng, T.; Chik, K.K.-H.; Ye, Z.-W.; Wang, S.; Lee, A.C.-Y.; Jin, L.; et al. Metallodrug ranitidine bismuth citrate suppresses SARS-CoV-2 replication and relieves virus-associated pneumonia in Syrian hamsters. Nat. Microbiol. 2020, 5, 1439–1448. [Google Scholar] [CrossRef] [PubMed]
- Shidlovskaya, E.V.; Kuznetsova, N.A.; Divisenko, E.V.; Nikiforova, M.A.; Siniavin, A.E.; Ogarkova, D.A.; Shagaev, A.V.; Semashko, M.A.; Tkachuk, A.P.; Burgasova, O.A.; et al. The Value of Rapid Antigen Tests for Identifying Carriers of Viable SARS-CoV-2. Viruses 2021, 13, 2012. [Google Scholar] [CrossRef] [PubMed]
- Koes, D.R.; Baumgartner, M.; Camacho, C.J. Lessons Learned in Empirical Scoring with smina from the CSAR 2011 Benchmarking Exercise. J. Chem. Inf. Model. 2013, 53, 1893–1904. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Yan, L.; Yang, Y.; Li, M.; Zhang, Y.; Zheng, L.; Ge, J.; Huang, Y.C.; Liu, Z.; Wang, T.; Gao, S.; et al. Coupling of N7-methyltransferase and 3′-5′ exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading. Cell 2021, 184, 3474–3485.e11. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007, 35, W522–W525. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- PyMOL. Available online: https://www.pymol.org/pymol.html? (accessed on 18 June 2022).





| N | IC50 (µM) | CC50 (µM) | ||
|---|---|---|---|---|
| B.1.617.2 | B.1.351 | B.1.1.529 | ||
| 601 | 32.72 | 32.68 | 27.1 | >50 |
| 611 | 13.3 | >50 | >50 | >50 |
| 870 | 14.6 | 34.14 | 24.42 | >50 |
| 871 | 26.57 | 27.33 | >50 | >25 |
| 872 | >50 | >50 | 7.92 | >50 |
| 873 | >50 | 49.97 | 28.21 | >50 |
| 874 | 30.24 | 29.06 | >50 | >25 |
| 875 | 30.66 | 33.06 | 22.55 | >50 |
| 876 | 29.33 | 23.82 | 22.39 | >50 |
| 1005 | 27.83 | 15.45 | >50 | >50 |
| 1006 | 27.44 | >50 | >50 | >50 |
| 1007 | >50 | >50 | >50 | >25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siniavin, A.E.; Novikov, M.S.; Gushchin, V.A.; Terechov, A.A.; Ivanov, I.A.; Paramonova, M.P.; Gureeva, E.S.; Russu, L.I.; Kuznetsova, N.A.; Shidlovskaya, E.V.; et al. Antiviral Activity of N1,N3-Disubstituted Uracil Derivatives against SARS-CoV-2 Variants of Concern. Int. J. Mol. Sci. 2022, 23, 10171. https://doi.org/10.3390/ijms231710171
Siniavin AE, Novikov MS, Gushchin VA, Terechov AA, Ivanov IA, Paramonova MP, Gureeva ES, Russu LI, Kuznetsova NA, Shidlovskaya EV, et al. Antiviral Activity of N1,N3-Disubstituted Uracil Derivatives against SARS-CoV-2 Variants of Concern. International Journal of Molecular Sciences. 2022; 23(17):10171. https://doi.org/10.3390/ijms231710171
Chicago/Turabian StyleSiniavin, Andrei E., Mikhail S. Novikov, Vladimir A. Gushchin, Alexander A. Terechov, Igor A. Ivanov, Maria P. Paramonova, Elena S. Gureeva, Leonid I. Russu, Nadezhda A. Kuznetsova, Elena V. Shidlovskaya, and et al. 2022. "Antiviral Activity of N1,N3-Disubstituted Uracil Derivatives against SARS-CoV-2 Variants of Concern" International Journal of Molecular Sciences 23, no. 17: 10171. https://doi.org/10.3390/ijms231710171





