De Novo Prediction of Drug Targets and Candidates by Chemical Similarity-Guided Network-Based Inference
Abstract
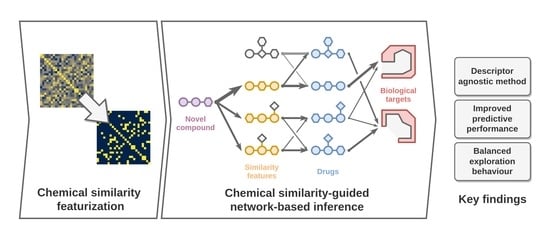
:1. Introduction
2. Results and Discusion
2.1. Selection of Optimal Method Parameters
2.2. Prediction Performance Compared to Other Methods
2.3. Prediction Performance Estimated by External Time-Split Validation
2.4. Analysis of the Exploration Behavior in Ligand and Target Space
3. Materials and Methods
3.1. Dataset Selection and Preparation
3.2. Molecular Descriptors and Similarity Measures
3.3. Chemical Similarity Feature Matrix Construction
3.4. De Novo Network-Based Inference Method
3.5. SimSpread Algorithm
3.6. Other Predictive Algorithms
3.7. Cross-Validation and Time-Split Validation
3.8. Evaluation Metrics
3.9. Exploration of Chemical and Biological Space
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, J.J.; Pan, W.; Hu, Y.J.; Wang, Y.T. Multi-Target Drugs: The Trend of Drug Research and Development. PLoS ONE 2012, 7, e40262. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Anighoro, A.; Bajorath, J.; Rastelli, G. Polypharmacology: Challenges and Opportunities in Drug Discovery: Miniperspective. J. Med. Chem. 2014, 57, 7874–7887. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Fridkin, M.; Youdim, M. From Single Target to Multitarget/Network Therapeutics in Alzheimer’s Therapy. Pharmaceuticals 2014, 7, 113–135. [Google Scholar] [CrossRef]
- Lin, H.H.; Zhang, L.L.; Yan, R.; Lu, J.J.; Hu, Y. Network Analysis of Drug–target Interactions: A Study on FDA-approved New Molecular Entities Between 2000 to 2015. Sci. Rep. 2017, 7, 12230. [Google Scholar] [CrossRef]
- Kola, I.; Landis, J. Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 2004, 3, 711–716. [Google Scholar] [CrossRef]
- Xie, L.; Xie, L.; Bourne, P.E. Structure-based systems biology for analyzing off-target binding. Curr. Opin. Struct. Biol. 2011, 21, 189–199. [Google Scholar] [CrossRef]
- Chaudhari, R.; Fong, L.W.; Tan, Z.; Huang, B.; Zhang, S. An up-to-date overview of computational polypharmacology in modern drug discovery. Expert Opin. Drug Discov. 2020, 15, 1025–1044. [Google Scholar] [CrossRef] [PubMed]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014, 42, W32–W38. [Google Scholar] [CrossRef]
- Huang, T.; Mi, H.; Yuan Lin, C.; Zhao, L.; Zhong, L.L.D.; Bin Liu, F.; Zhang, G.; Ping Lu, A.; Xiang Bian, Z. MOST: Most-similar ligand based approach to target prediction. BMC Bioinform. 2017, 18, 165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gfeller, D.; Michielin, O.; Zoete, V. Shaping the interaction landscape of bioactive molecules. Bioinformatics 2013, 29, 3073–3079. [Google Scholar] [CrossRef]
- Najmanovich, R.; Kurbatova, N.; Thornton, J. Detection of 3D atomic similarities and their use in the discrimination of small molecule protein-binding sites. Bioinformatics 2008, 24, i105–i111. [Google Scholar] [CrossRef]
- Zhou, H.; Skolnick, J. FINDSITE comb: A Threading/Structure-Based, Proteomic-Scale Virtual Ligand Screening Approach. J. Chem. Inf. Model. 2013, 53, 230–240. [Google Scholar] [CrossRef]
- Zhou, H.; Gao, M.; Skolnick, J. Comprehensive prediction of drug-protein interactions and side effects for the human proteome. Sci. Rep. 2015, 5, 11090. [Google Scholar] [CrossRef]
- Yabuuchi, H.; Niijima, S.; Takematsu, H.; Ida, T.; Hirokawa, T.; Hara, T.; Ogawa, T.; Minowa, Y.; Tsujimoto, G.; Okuno, Y. Analysis of multiple compound–protein interactions reveals novel bioactive molecules. Mol. Syst. Biol. 2011, 7, 472. [Google Scholar] [CrossRef]
- Jones, D.; Bopaiah, J.; Alghamedy, F.; Jacobs, N.; Weiss, H.L.; de Jong, W.A.; Ellingson, S.R. Polypharmacology Within the Full Kinome: A Machine Learning Approach. AMIA Jt. Summits Transl. Sci. Proc. AMIA Jt. Summits Transl. Sci. 2018, 2017, 98–107. [Google Scholar]
- van Laarhoven, T.; Nabuurs, S.B.; Marchiori, E. Gaussian interaction profile kernels for predicting drug–target interaction. Bioinformatics 2011, 27, 3036–3043. [Google Scholar] [CrossRef]
- van Laarhoven, T.; Marchiori, E. Predicting Drug-Target Interactions for New Drug Compounds Using a Weighted Nearest Neighbor Profile. PLoS ONE 2013, 8, e66952. [Google Scholar] [CrossRef]
- Nidhi; Glick, M.; Davies, J.W.; Jenkins, J.L. Prediction of Biological Targets for Compounds Using Multiple-Category Bayesian Models Trained on Chemogenomics Databases. J. Chem. Inf. Model. 2006, 46, 1124–1133. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, J. Predicting drug-target interactions using restricted Boltzmann machines. Bioinformatics 2013, 29, i126–i134. [Google Scholar] [CrossRef]
- Li, T.; Zhao, X.; Li, L. Co-VAE: Drug-target binding affinity prediction by co-regularized variational autoencoders. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 1. [Google Scholar] [CrossRef]
- Karimi, M.; Wu, D.; Wang, Z.; Shen, Y. DeepAffinity: Interpretable deep learning of compound–protein affinity through unified recurrent and convolutional neural networks. Bioinformatics 2019, 35, 3329–3338. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, E.N.; Sur, D.; Wu, Z.; Husic, B.E.; Mai, H.; Li, Y.; Sun, S.; Yang, J.; Ramsundar, B.; Pande, V.S. PotentialNet for Molecular Property Prediction. ACS Cent. Sci. 2018, 4, 1520–1530. [Google Scholar] [CrossRef] [PubMed]
- Maggiora, G.; Vogt, M.; Stumpfe, D.; Bajorath, J. Molecular Similarity in Medicinal Chemistry: Miniperspective. J. Med. Chem. 2014, 57, 3186–3204. [Google Scholar] [CrossRef] [PubMed]
- Yamanishi, Y.; Araki, M.; Gutteridge, A.; Honda, W.; Kanehisa, M. Prediction of drug-target interaction networks from the integration of chemical and genomic spaces. Bioinformatics 2008, 24, i232–i240. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.C.; Senese, S.; Li, C.M.; Hu, Q.; Huang, Y.; Torres, R.D.n.J.Z. Large-Scale Chemical Similarity Networks for Target Profiling of Compounds Identified in Cell-Based Chemical Screens. PLoS Comput. Biol. 2015, 11, 1–23. [Google Scholar] [CrossRef]
- Lo, Y.C.; Senese, S.; Damoiseaux, R.; Torres, J.Z. 3D Chemical Similarity Networks for Structure-Based Target Prediction and Scaffold Hopping. ACS Chem. Biol. 2016, 11, 2244–2253. [Google Scholar] [CrossRef]
- Martínez-Jiménez, F.; Marti-Renom, M.A. Ligand-Target Prediction by Structural Network Biology Using nAnnoLyze. PLoS Comput. Biol. 2015, 11, e1004157. [Google Scholar] [CrossRef]
- Ba-alawi, W.; Soufan, O.; Essack, M.; Kalnis, P.; Bajic, V.B. DASPfind: New efficient method to predict drug–target interactions. J. Cheminform. 2016, 8, 15. [Google Scholar] [CrossRef]
- Zhou, T.; Ren, J.; Medo, M.; Zhang, Y.C. Bipartite network projection and personal recommendation. Phys. Rev. E 2007, 76, 046115. [Google Scholar] [CrossRef]
- Cheng, F.; Liu, C.; Jiang, J.; Lu, W.; Li, W.; Liu, G.; Zhou, W.; Huang, J.; Tang, Y. Prediction of Drug-Target Interactions and Drug Repositioning via Network-Based Inference. PLoS Comput. Biol. 2012, 8, e1002503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Cheng, F.; Li, J.; Li, W.; Liu, G.; Tang, Y. SDTNBI: An integrated network and chemoinformatics tool for systematic prediction of drug–target interactions and drug repositioning. Briefings Bioinform. 2016, 18, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Hattori, M.; Tanaka, N.; Kanehisa, M.; Goto, S. SIMCOMP/SUBCOMP: Chemical structure search servers for network analyses. Nucleic Acids Res. 2010, 38, W652–W656. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Xie, Q.; Ge, W.; Qian, F.; Fang, H.; Shi, L.; Su, Z.; Perkins, R.; Tong, W. Mold2, Molecular Descriptors from 2D Structures for Chemoinformatics and Toxicoinformatics. J. Chem. Inf. Model. 2008, 48, 1337–1344. [Google Scholar] [CrossRef]
- Taminau, J.; Thijs, G.; Winter, H.D. Pharao: Pharmacophore alignment and optimization. J. Mol. Graph. Model. 2008, 27, 161–169. [Google Scholar] [CrossRef]
- Willett, P.; Barnard, J.M.; Downs, G.M. Chemical Similarity Searching. J. Chem. Inf. Comput. Sci. 1998, 38, 983–996. [Google Scholar] [CrossRef]
- Sheridan, R.P. Time-Split Cross-Validation as a Method for Estimating the Goodness of Prospective Prediction. J. Chem. Inf. Model. 2013, 53, 783–790. [Google Scholar] [CrossRef]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; Veij, M.D.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2018, 47, D930–D940. [Google Scholar] [CrossRef]
- Bemis, G.W.; Murcko, M.A. The Properties of Known Drugs. 1. Molecular Frameworks. J. Med. Chem. 1996, 39, 2887–2893. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Landrum, G. RDKit: Open-Source Cheminformatics Software; Zenodo: Genève, Switzerland, 2016. [Google Scholar]
- Yap, C.W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2010, 32, 1466–1474. [Google Scholar] [CrossRef]
- Bajusz, D.; Rácz, A.; Héberger, K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminform. 2015, 7, 20. [Google Scholar] [CrossRef]
- Mold2 | FDA. Available online: https://www.fda.gov/science-research/bioinformatics-tools/mold2 (accessed on 27 July 2022).
- SimSpread GitHub Repository. Available online: https://github.com/cvigilv/simspread (accessed on 21 August 2022).
- Chen, X.; Liu, M.X.; Yan, G.Y. Drug–target interaction prediction by random walk on the heterogeneous network. Mol. Biosyst. 2012, 8, 1970. [Google Scholar] [CrossRef]
- Zhao, W.; Hevener, K.E.; White, S.W.; Lee, R.E.; Boyett, J.M. A statistical framework to evaluate virtual screening. BMC Bioinform. 2009, 10, 225. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Lamb, Y.N. Nintedanib: A Review in Fibrotic Interstitial Lung Diseases. Drugs 2021, 81, 575–586. [Google Scholar] [CrossRef]
- Aimo, A.; Spitaleri, G.; Nieri, D.; Tavanti, L.M.; Meschi, C.; Panichella, G.; Lupón, J.; Pistelli, F.; Carrozzi, L.; Bayes-Genis, A.; et al. Pirfenidone for Idiopathic Pulmonary Fibrosis and Beyond. Card. Fail. Rev. 2022, 8, 1–11. [Google Scholar] [CrossRef]





| Method | AuROC | AuPRC | AuBEDROC | P@20 | R@20 |
|---|---|---|---|---|---|
| 1-NN | 0.942 (0.699, 0.997) | 0.137 (0.014, 0.500) | 0.527 (0.016, 0.951) | 0.050 (0.000, 0.100) | 0.667 (0.000, 1.000) |
| SDTNBI | 0.873 (0.623, 0.990) | 0.067 (0.009, 0.417) | 0.305 (0.002, 0.840) | 0.050 (0.000, 0.050) | 0.333 (0.000, 1.000) |
| SimSpread | 0.912 (0.500, 0.994) | 0.090 (0.009, 0.500) | 0.434 (0.004, 0.885) | 0.050 (0.000, 0.100) | 0.500 (0.000, 1.000) |
| SimSpread | 0.932 (0.500, 0.995) | 0.111 (0.010, 0.539) | 0.477 (0.006, 0.909) | 0.050 (0.000, 0.100) | 0.500 (0.000, 1.000) |
| Statistics | |||||||
|---|---|---|---|---|---|---|---|
| Dataset | Density | ||||||
| Enzyme | 445 | 664 | 2.926 | 6.57 ± 12.74 | 4.41 ± 6.33 | [7, 101] | 0.99 |
| Ion Channel | 223 | 95 | 635 | 7.03 ± 12.14 | 7.24 ± 6.63 | [7, 95] | 3.45 |
| GPCR | 210 | 205 | 1.476 | 2.85 ± 3.35 | 6.68 ± 8.43 | [7, 94] | 3.00 |
| Nuclear Receptor | 54 | 26 | 90 | 1.67 ± 1.60 | 3.46 ± 3.39 | [14, 59] | 6.41 |
| Global | 1844 | 1032 | 10,185 | 5.52 ± 15.79 | 9.87 ± 14.95 | [5, 118] | 0.54 |
| ChEMBL24→28 | 1513 | 395 | 4863 | 3.21 ± 5.66 | 12.31 ± 12.24 | [7, 78] | 0.81 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vigil-Vásquez, C.; Schüller, A. De Novo Prediction of Drug Targets and Candidates by Chemical Similarity-Guided Network-Based Inference. Int. J. Mol. Sci. 2022, 23, 9666. https://doi.org/10.3390/ijms23179666
Vigil-Vásquez C, Schüller A. De Novo Prediction of Drug Targets and Candidates by Chemical Similarity-Guided Network-Based Inference. International Journal of Molecular Sciences. 2022; 23(17):9666. https://doi.org/10.3390/ijms23179666
Chicago/Turabian StyleVigil-Vásquez, Carlos, and Andreas Schüller. 2022. "De Novo Prediction of Drug Targets and Candidates by Chemical Similarity-Guided Network-Based Inference" International Journal of Molecular Sciences 23, no. 17: 9666. https://doi.org/10.3390/ijms23179666