Ultrastructural Characterization of Human Bronchial Epithelial Cells during SARS-CoV-2 Infection: Morphological Comparison of Wild-Type and CFTR-Modified Cells
Abstract
:1. Introduction
2. Results
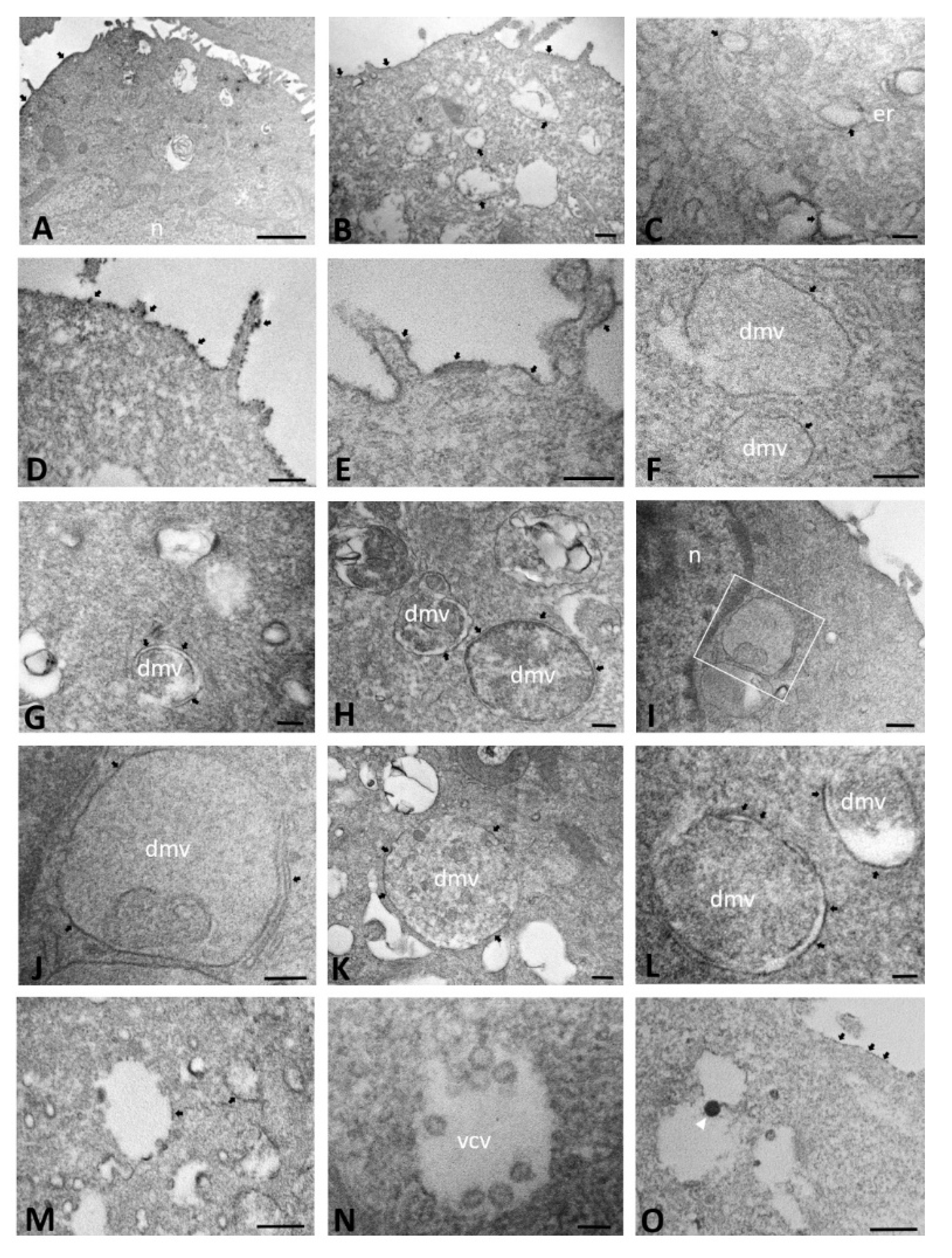
2.1. Ultrastructural Morphology of Mock-Infected and SARS-CoV-2-Infected CFBE41o- Wild-Type Cells
2.2. Ultrastructural Morphology of Mock-Infected and SARS-CoV-2-Infected CFBE41o- ΔF Cells
2.3. ACE2 Immunoelectron Microscopic Localization in Mock-Infected and SARS-CoV-2-Infected CFBE41o- Wild-Type Cells
2.4. ACE2 Immuno-Electron Microscopic Localization in Mock-Infected and SARS-CoV-2-Infected CFBE41o- ΔF Cells
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Virus Strain
4.2. Cell Culture and Infection
4.3. Transmission Electron Microscopy (TEM)
4.4. Immunoelectron Microscopy
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Burrell, C.J.; Howard, C.R.; Murphy, F.A. Coronaviruses. Fenner and White’s Medical Virology; Academic Press: Cambridge, MA, USA, 2017; pp. 437–446. [Google Scholar]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef] [PubMed]
- Stadler, K.; Masignani, V.; Eickmann, M.; Becker, S.; Abrignani, S.; Klenk, H.-D.; Rappuoli, R. SARS—Beginning to understand a new virus. Nat. Rev. Microbiol. 2003, 1, 209–218. [Google Scholar] [CrossRef] [PubMed]
- de Groot, R.J.; Baker, S.C.; Baric, R.S.; Brown, C.S.; Drosten, C.; Enjuanes, L.; Fouchier, R.A.; Galiano, M.; Gorbalenya, A.E.; Memish, Z.A.; et al. Middle East respiratory syndrome coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. J. Virol. 2013, 87, 7790–7792. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef]
- Künkel, F.; Herrler, G. Structural and Functional Analysis of the Surface Protein of Human Coronavirus OC43. Virology 1993, 195, 195–202. [Google Scholar] [CrossRef]
- Bradburne, A.F.; Bynoe, M.L.; Tyrrell, D.A. Effects of a "new" human respiratory virus in volunteers. BMJ 1967, 3, 767–769. [Google Scholar] [CrossRef]
- Hendley, J.; Fishburne, H.B.; Gwaltney, J.M., Jr. Coronavirus infections in working adults. Eight-year study with 229 E and OC 43. Am. Rev. Respir. Dis. 1972, 105, 805–811. [Google Scholar] [CrossRef]
- van der Hoek, L.; Pyrc, K.; Jebbink, M.F.; Vermeulen-Oost, W.; Berkhout, R.J.; Wolthers, K.C.; Wertheim-van Dillen, P.M.; Kaandorp, J.; Spaargaren, J.; Berkhout, B. Identification of a new human coronavirus. Nat. Med. 2004, 10, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Chu, C.-M.; Chan, K.-H.; Tsoi, H.-W.; Huang, Y.; Wong, B.H.L.; Poon, R.W.S.; Cai, J.J.; Luk, W.-K.; et al. Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia. J. Virol. 2005, 79, 884–895. [Google Scholar] [CrossRef] [PubMed]
- Monto, A.S.; Lim, S.K. The Tecumseh study of respiratory illness. VI. Frequency of and relationship between outbreaks of coronavirus infection. J. Infect. Dis. 1974, 129, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Rahbar Saadat, Y.; Hosseiniyan Khatibi, S.M.; Zununi Vahed, S.; Ardalan, M. Host Serine Proteases: A Potential Targeted Therapy for COVID-19 and Influenza. Front. Mol. Biosci. 2021, 8, 725528. [Google Scholar] [CrossRef]
- Bradley, B.T.; Maioli, H.; Johnston, R.; Chaudhry, I.; Fink, S.L.; Xu, H.; Najafian, B.; Deutsch, G.; Lacy, J.M.; Williams, T.; et al. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: A case series. Lancet 2020, 396, 320–332. [Google Scholar] [CrossRef]
- Harak, C.; Lohmann, V. Ultrastructure of the replication sites of positive-strand RNA viruses. Virology 2015, 479–480, 418–433. [Google Scholar] [CrossRef] [Green Version]
- Ulasli, M.; Verheije, M.H.; de Haan, C.A.; Reggiori, F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 2010, 12, 844–861. [Google Scholar] [CrossRef]
- Knoops, K.; Kikkert, M.; Worm, S.H.E.V.D.; Zevenhoven-Dobbe, J.C.; Van Der Meer, Y.; Koster, A.J.; Mommaas, A.M.; Snijder, E.J. SARS-Coronavirus Replication Is Supported by a Reticulovesicular Network of Modified Endoplasmic Reticulum. PLoS Biol. 2008, 6, e226. [Google Scholar] [CrossRef] [PubMed]
- Wolff, G.; Melia, C.E.; Snijder, E.J.; Bárcena, M. Double-Membrane Vesicles as Platforms for Viral Replication. Trends Microbiol. 2020, 28, 1022–1033. [Google Scholar] [CrossRef]
- Snijder, E.J.; Limpens, R.W.A.L.; de Wilde, A.H.; de Jong, A.W.M.; Zevenhoven-Dobbe, J.C.; Maier, H.J.; Faas, F.F.G.A.; Koster, A.J.; Bárcena, M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020, 18, e3000715. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.L.; Tan, S.H.; See, E.E.; Ooi, E.E.; Ling, A.E. Early events of SARS coronavirus infection in vero cells. J. Med Virol. 2003, 71, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Qinfen, Z.; Jinming, C.; Xiaojun, H.; Huanying, Z.; Jicheng, H.; Ling, F.; Kunpeng, L.; Jingqiang, Z. The life cycle of SARS coronavirus in Vero E6 cells. J. Med Virol. 2004, 73, 332–337. [Google Scholar] [CrossRef]
- Kim, S.-H.; Chang, S.Y.; Sung, M.; Park, J.H.; BIN Kim, H.; Lee, H.; Choi, J.-P.; Choi, W.S.; Min, J.-Y. Extensive Viable Middle East Respiratory Syndrome (MERS) Coronavirus Contamination in Air and Surrounding Environment in MERS Isolation Wards. Clin. Infect. Dis. 2016, 63, 363–369. [Google Scholar] [CrossRef]
- Park, W.B.; Kwon, N.-J.; Choe, P.G.; Choi, S.-J.; Oh, H.S.; Lee, S.M.; Chong, H.; Kim, J.-I.; Song, K.-H.; Bang, J.H.; et al. Isolation of Middle East Respiratory Syndrome Coronavirus from a Patient of the 2015 Korean Outbreak. J. Korean Med. Sci. 2016, 31, 315–320. [Google Scholar] [CrossRef]
- Alsaad, K.O.; Hajeer, A.H.; Al Balwi, M.; Al Moaiqel, M.; Al Oudah, N.; Al Ajlan, A.; AlJohani, S.; Alsolamy, S.; Gmati, G.E.; Balkhy, H.; et al. Histopathology of Middle East respiratory syndrome coronovirus (MERS-CoV) infection—Clinicopathological and ultrastructural study. Histopathology 2018, 72, 516–524. [Google Scholar] [CrossRef]
- Caly, L.; Druce, J.; Roberts, J.; Bond, K.; Tran, T.; Kostecki, R.; Yoga, Y.; Naughton, W.; Taiaroa, G.; Seemann, T.; et al. Isolation and rapid sharing of the 2019 novel coronavirus (SARS-CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med. J. Aust. 2020, 212, 459–462. [Google Scholar] [CrossRef] [Green Version]
- Colson, P.; Lagier, J.C.; Baudoin, J.P.; Bou Khalil, J.; La Scola, B.; Raoult, D. Ultrarapid diagnosis, microscope imaging, genome sequencing, and culture isolation of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1601–1603. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-M.; Chung, Y.-S.; Jo, H.J.; Lee, N.-J.; Kim, M.S.; Woo, S.H.; Park, S.; Kim, J.W.; Kim, H.M.; Han, M.-G. Identification of Coronavirus Isolated from a Patient in Korea with COVID-19. Osong Public Health Res. Perspect. 2020, 11, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Lotti, V.; Merigo, F.; Lagni, A.; Di Clemente, A.; Ligozzi, M.; Bernardi, P.; Rossini, G.; Concia, E.; Plebani, R.; Romano, M.; et al. CFTR Modulation Reduces SARS-CoV-2 Infection in Human Bronchial Epithelial Cells. Cells 2022, 11, 1347. [Google Scholar] [CrossRef] [PubMed]
- Collins, F. Cystic fibrosis: Molecular biology and therapeutic implications. Science 1992, 256, 774–779. [Google Scholar] [CrossRef]
- Rommens, J.M.; Iannuzzi, M.C.; Kerem, B.; Drumm, M.L.; Melmer, G.; Dean, M.; Rozmahel, R.; Cole, J.L.; Kennedy, D.; Hidaka, N.; et al. Identification of the cystic fibrosis gene: Chromosome walking and jumping. Science 1989, 245, 1059–1065. [Google Scholar] [CrossRef]
- Anderson, M.P.; Gregory, R.J.; Thompson, S.; Souza, D.W.; Paul, S.; Mulligan, R.C.; Smith, A.E.; Welsh, M.J. Demonstration That CFTR Is a Chloride Channel by Alteration of Its Anion Selectivity. Science 1991, 253, 202–205. [Google Scholar] [CrossRef]
- Collawn, J.F.; Matalon, S. CFTR and lung homeostasis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014, 307, L917–L923. [Google Scholar] [CrossRef]
- Rowe, S.M.; Miller, S.; Sorscher, E.J. Cystic fibrosis. N. Engl. J. Med. 2005, 352, 1992–2001. [Google Scholar] [CrossRef]
- Elborn, J.S. Cystic fibrosis. Lancet 2016, 388, 2519–2531. [Google Scholar] [CrossRef]
- O’Sullivan, B.P.; Freedman, S.D. Cystic fibrosis. Lancet 2009, 373, 1891–1904. [Google Scholar] [CrossRef]
- Fainardi, V.; Longo, F.; Chetta, A.; Esposito, S.; Pisi, G. SARS-CoV-2 infection in patients with cystic fibrosis. An overwiew. Acta Biomed. Atenei Parm. 2020, 91, e2020035. [Google Scholar] [CrossRef]
- Bezzerri, V.; Lucca, F.; Volpi, S.; Cipolli, M. Does cystic fibrosis constitute an advantage in COVID-19 infection? Ital. J. Pediatr. 2020, 46, 143. [Google Scholar] [CrossRef] [PubMed]
- Cosgriff, R.; Ahern, S.; Bell, S.C.; Brownlee, K.; Burgel, P.R.; Byrnes, C.; Corvol, H.; Cheng, S.Y.; Elbert, A.; Faro, A.; et al. A multinational report to characterise SARS-CoV-2 infection in people with cystic fibrosis. J. Cyst. Fibros. 2020, 19, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Brahim Belhaouari, D.; Fontanini, A.; Baudoin, J.-P.; Haddad, G.; Le Bideau, M.; Bou Khalil, J.Y.; Raoult, D.; La Scola, B. The Strengths of Scanning Electron Microscopy in Deciphering SARS-CoV-2 Infectious Cycle. Front. Microbiol. 2020, 11, 2014. [Google Scholar] [CrossRef]
- Stertz, S.; Reichelt, M.; Spiegel, M.; Kuri, T.; Martínez-Sobrido, L.; García-Sastre, A.; Weber, F.; Kochs, G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology 2007, 361, 304–315. [Google Scholar] [CrossRef]
- Knoops, K.; Bárcena, M.; Limpens, R.W.A.L.; Koster, A.J.; Mommaas, A.M.; Snijder, E.J. Ultrastructural Characterization of Arterivirus Replication Structures: Reshaping the Endoplasmic Reticulum To Accommodate Viral RNA Synthesis. J. Virol. 2012, 86, 2474–2487. [Google Scholar] [CrossRef]
- Wolff, G.; Limpens, R.W.A.L.; Zevenhoven-Dobbe, J.C.; Laugks, U.; Zheng, S.; De Jong, A.W.M.; Koning, R.I.; Agard, D.A.; Grünewald, K.; Koster, A.J.; et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 2020, 369, 1395–1398. [Google Scholar] [CrossRef]
- Akilesh, S.; Nicosia, R.F.; Alpers, C.E.; Tretiakova, M.; Hsiang, T.Y.; Gale, M., Jr.; Smith, K.D. Characterizing Viral Infection by Electron Microscopy: Lessons from the Coronavirus Disease 2019 Pandemic. Am. J. Pathol. 2021, 191, 222–227. [Google Scholar] [CrossRef]
- Weber, F.; Wagner, V.; Rasmussen, S.B.; Hartmann, R.; Paludan, S.R. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006, 80, 5059–5064. [Google Scholar] [CrossRef]
- Harcourt, B.H.; Jukneliene, D.; Kanjanahaluethai, A.; Bechill, J.; Severson, K.M.; Smith, C.M.; Rota, P.A.; Baker, S.C. Identification of Severe Acute Respiratory Syndrome Coronavirus Replicase Products and Characterization of Papain-Like Protease Activity. J. Virol. 2004, 78, 13600–13612. [Google Scholar] [CrossRef] [Green Version]
- Oudshoorn, D.; Rijs, K.; Limpens, R.; Groen, K.; Koster, A.J.; Snijder, E.J.; Kikkert, M.; Bárcena, M. Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication. mBio 2017, 8, e01658-17. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, C.S.; Tatti, K.M.; Ksiazek, T.G.; Rollin, P.; Comer, J.A.; Lee, W.W.; Rota, P.A.; Bankamp, B.; Bellini, W.J.; Zaki, S.R. Ultrastructural Characterization of SARS Coronavirus. Emerg. Infect. Dis. 2004, 10, 320–326. [Google Scholar] [CrossRef]
- Mao, J.; Lin, E.; He, L.; Yu, J.; Tan, P.; Zhou, Y. Autophagy and Viral Infection. Adv. Exp. Med. Biol. 2019, 1209, 55–78. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Bowman, J.W.; Jung, J.U. Autophagy during viral infection—A double-edged sword. Nat. Rev. Microbiol. 2018, 16, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Luo, H. Interplay between the cellular autophagy machinery and positive-stranded RNA viruses. Acta Biochim. Biophys. Sin. 2012, 44, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Czaja, M.J. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013, 20, 3–11. [Google Scholar] [CrossRef]
- Gassen, N.C.; Niemeyer, D.; Muth, D.; Corman, V.M.; Martinelli, S.; Gassen, A.; Hafner, K.; Papies, J.; Mösbauer, K.; Zellner, A.; et al. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 2019, 10, 5770. [Google Scholar] [CrossRef]
- Fakher, S.; Peymani, P.; Ghavami, S.; Mokarram, P. The Role of Autophagy in Respiratory Complications of COVID-19. Shiraz E-Med. J. 2020, 21, e102967. [Google Scholar] [CrossRef]
- Sargazi, S.; Sheervalilou, R.; Rokni, M.; Shirvaliloo, M.; Shahraki, O.; Rezaei, N. The role of autophagy in controlling SARS-CoV-2 infection: An overview on virophagy-mediated molecular drug targets. Cell Biol. Int. 2021, 45, 1599–1612. [Google Scholar] [CrossRef]
- Gies, V.; Bekaddour, N.; Dieudonné, Y.; Guffroy, A.; Frenger, Q.; Gros, F.; Rodero, M.P.; Herbeuval, J.-P.; Korganow, A.-S. Beyond Anti-viral Effects of Chloroquine/Hydroxychloroquine. Front. Immunol. 2020, 11, 1409. [Google Scholar] [CrossRef]
- Liu, J.; Cao, R.; Xu, M.; Wang, X.; Zhang, H.; Hu, H.; Li, Y.; Hu, Z.; Zhong, W.; Wang, M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Luciani, A.; Villella, V.R.; Esposito, S.; Brunetti-Pierri, N.; Medina, D.L.; Settembre, C.; Gavina, M.; Raia, V.; Ballabio, A.; Maiuri, L. Cystic fibrosis: A disorder with defective autophagy. Autophagy 2011, 7, 104–106. [Google Scholar] [CrossRef] [PubMed]
- Luciani, A.; Villella, V.R.; Esposito, S.; Brunetti-Pierri, N.; Medina, D.L.; Settembre, C.; Gavina, M.; Pulze, L.; Giardino, I.; Mantovani, M.P.; et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat. Cell Biol. 2010, 12, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Villella, V.R.; Esposito, S.; Bruscia, E.M.; Maiuri, M.C.; Raia, V.; Kroemer, G.; Maiuri, L. Targeting the Intracellular Environment in Cystic Fibrosis: Restoring Autophagy as a Novel Strategy to Circumvent the CFTR Defect. Front. Pharmacol. 2013, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Villella, V.R.; Esposito, S.; Bruscia, E.M.; Vicinanza, M.; Cenci, S.; Guido, S.; Pettoello-Mantovani, M.; Carnuccio, R.; A De Matteis, M.; Luini, A.; et al. Disease-relevant proteostasis regulation of cystic fibrosis transmembrane conductance regulator. Cell Death Differ. 2013, 20, 1101–1115. [Google Scholar] [CrossRef]
- Villella, V.R.; Esposito, S.; Ferrari, E.; Monzani, R.; Tosco, A.; Rossin, F.; Castaldo, A.; Silano, M.; Marseglia, G.L.; Romani, L.; et al. Autophagy suppresses the pathogenic immune response to dietary antigens in cystic fibrosis. Cell Death Dis. 2019, 10, 258. [Google Scholar] [CrossRef]
- Komatsu, M.; Waguri, S.; Koike, M.; Sou, Y.-S.; Ueno, T.; Hara, T.; Mizushima, N.; Iwata, J.-I.; Ezaki, J.; Murata, S.; et al. Homeostatic Levels of p62 Control Cytoplasmic Inclusion Body Formation in Autophagy-Deficient Mice. Cell 2007, 131, 1149–1163. [Google Scholar] [CrossRef]
- Nezis, I.P.; Simonsen, A.; Sagona, A.P.; Finley, K.; Gaumer, S.; Contamine, D.; Rusten, T.E.; Stenmark, H.; Brech, A. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J. Cell Biol. 2008, 180, 1065–1071. [Google Scholar] [CrossRef]
- Gilardini Montani, M.S.; Santarelli, R.; Granato, M.; Gonnella, R.; Torrisi, M.R.; Faggioni, A.; Cirone, M. EBV reduces autophagy, intracellular ROS and mitochondria to impair monocyte survival and differentiation. Autophagy 2019, 15, 652–667. [Google Scholar] [CrossRef]
- Assani, K.; Tazi, M.F.; Amer, A.O.; Kopp, B.T. IFN-γ Stimulates Autophagy-Mediated Clearance of Burkholderia cenocepacia in Human Cystic Fibrosis Macrophages. PLoS ONE 2014, 9, e96681. [Google Scholar] [CrossRef] [Green Version]
- Del Porto, P.; Cifani, N.; Guarnieri, S.; Di Domenico, E.G.; Mariggiò, M.A.; Spadaro, F.; Guglietta, S.; Anile, M.; Venuta, F.; Quattrucci, S.; et al. Dysfunctional CFTR alters the bactericidal activity of human macrophages against Pseudomonas aeruginosa. PLoS ONE 2011, 6, e19970. [Google Scholar] [CrossRef]
- Singh, R.; Kaushik, S.; Wang, Y.; Xiang, Y.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Czaja, M.J. Regulation of lipid droplets by autophagy. Trends Endocrinol. Metab. 2011, 22, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Heaton, N.S.; Randall, G. Dengue Virus-Induced Autophagy Regulates Lipid Metabolism. Cell Host Microbe 2010, 8, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Illek, B.; Maurisse, R.; Wahler, L.; Kunzelmann, K.; Fischer, H.; Gruenert, D.C. Cl Transport in Complemented CF Bronchial Epithelial Cells Correlates with CFTR mRNA Expression Levels. Cell. Physiol. Biochem. 2008, 22, 057–068. [Google Scholar] [CrossRef] [PubMed]
- Bruscia, E.; Sangiuolo, F.; Sinibaldi, P.; Goncz, K.K.; Novelli, G.; Gruenert, D.C. Isolation of CF cell lines corrected at DeltaF508-CFTR locus by SFHR-mediated targeting. Gene Ther. 2002, 9, 683–685. [Google Scholar] [CrossRef]
- Bebok, Z.; Collawn, J.F.; Wakefield, J.; Parker, W.; Li, Y.; Varga, K.; Sorscher, E.J.; Clancy, J.P. Failure of cAMP agonists to activate rescued deltaF508 CFTR in CFBE41o- airway epithelial monolayers. J. Physiol. 2005, 569, 601–615. [Google Scholar] [CrossRef]
- Ogando, N.S.; Dalebout, T.J.; Zevenhoven-Dobbe, J.C.; Limpens, R.W.; van der Meer, Y.; Caly, L.; Druce, J.; de Vries, J.J.C.; Kikkert, M.; Bárcena, M.; et al. SARS-coronavirus-2 replication in Vero E6 cells: Replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020, 101, 925–940. [Google Scholar] [CrossRef]
- Valletta, E.A.; Bertini, M.; Sbarbati, A. Tannic Acid Supplemented Fixation Improves Ultrastructural Evaluation of Respiratory Epithelium in Children with Recurrent Respiratory Tract Infections. Biotech. Histochem. 1996, 71, 245–250. [Google Scholar] [CrossRef]

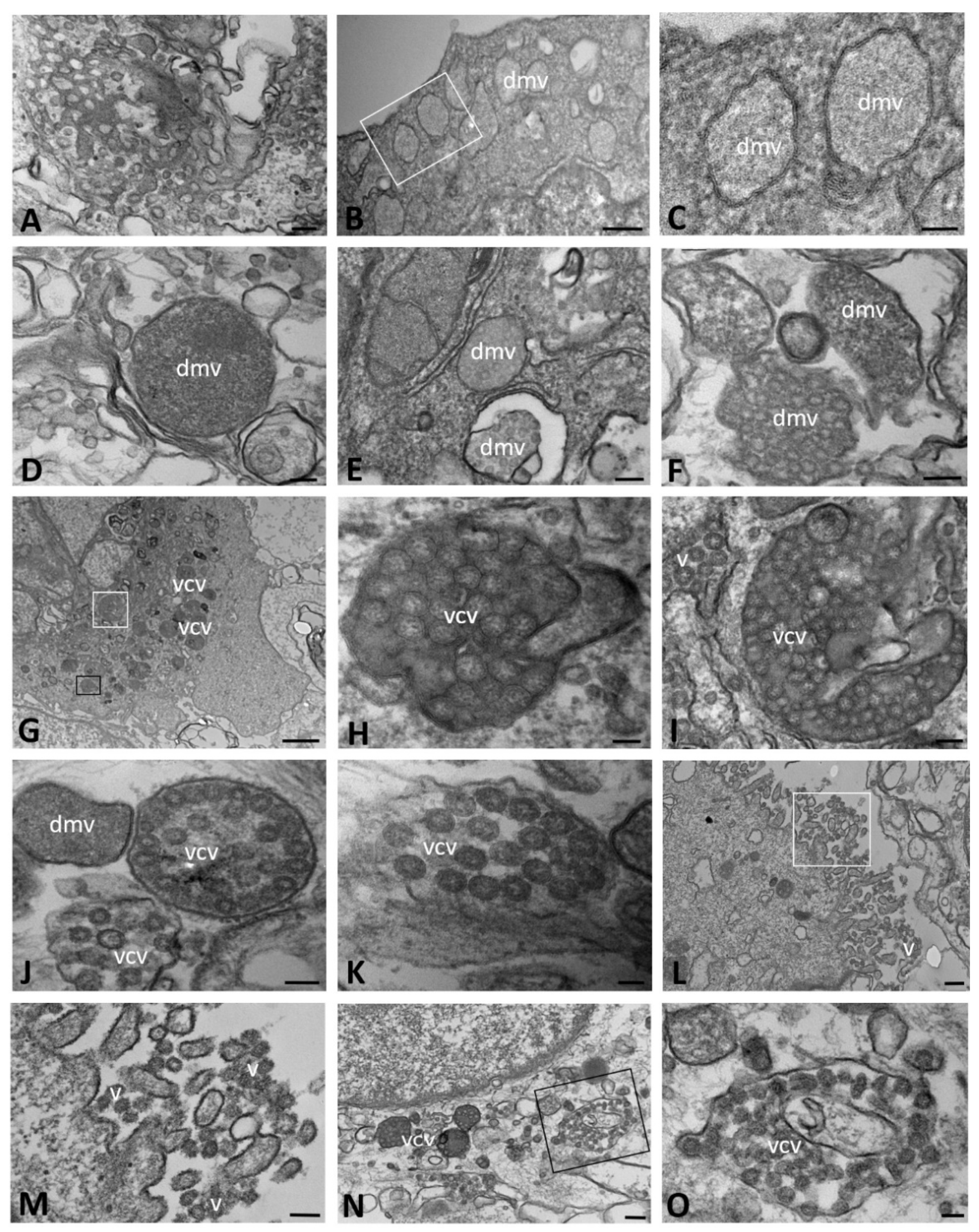
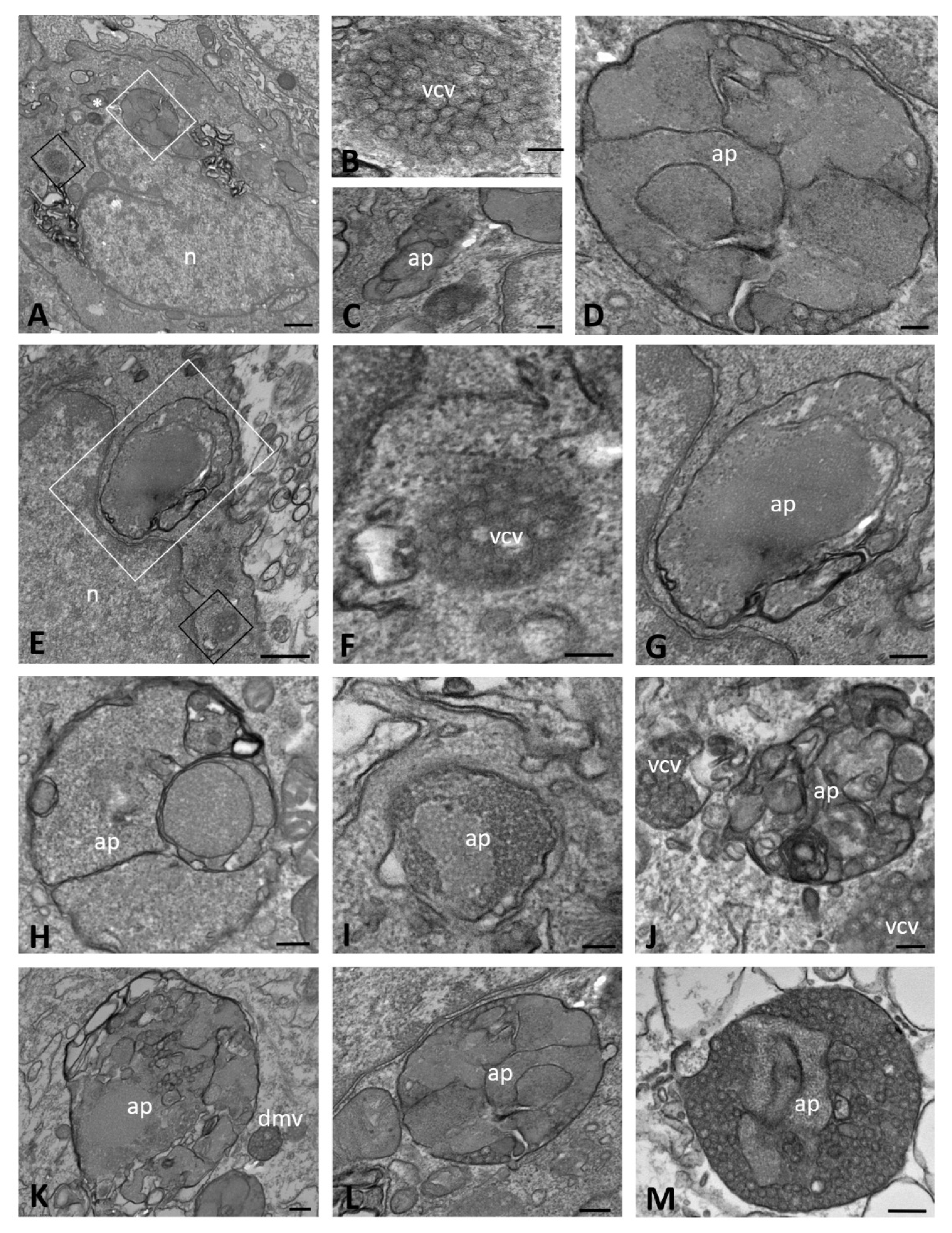

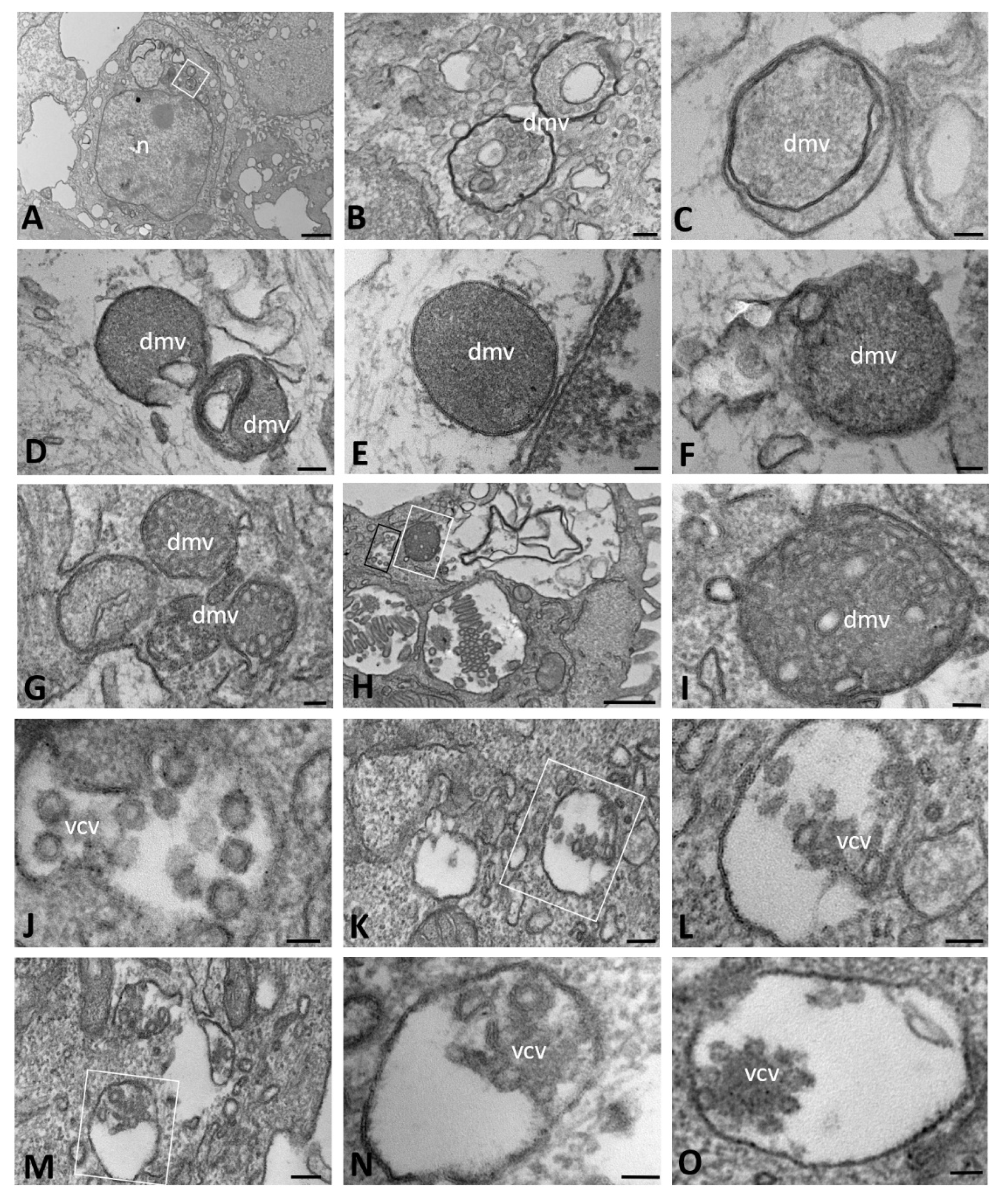
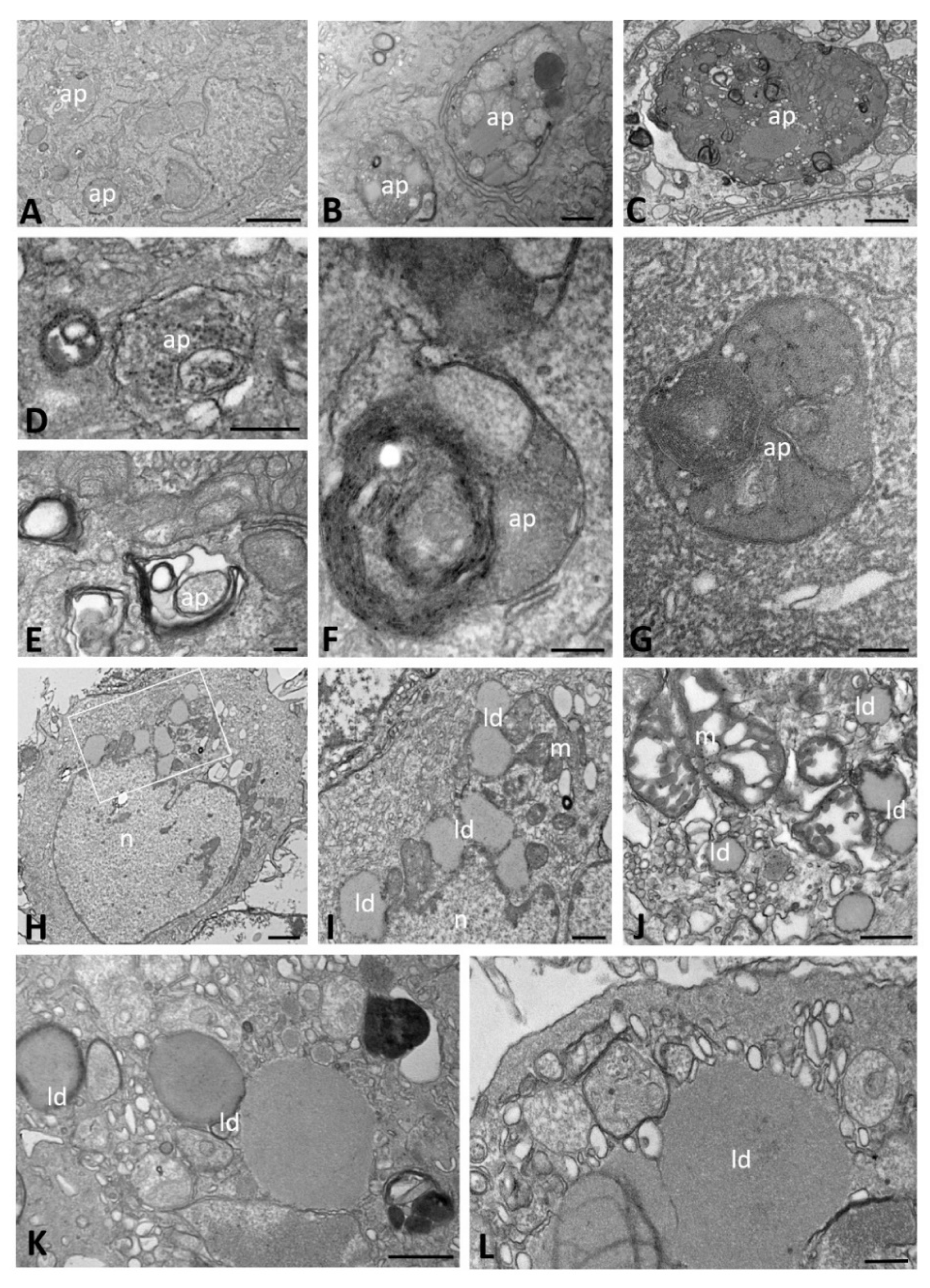

| Time of Infection | CFBE41o- Cells | DMVs | Autophagosomes | Lipid Droplets | Proteinaceous Material | Virions | ACE2 Expression | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Granular Content | Vesicle Structures | Cellular Material | Replicative Structure | Cytoplasmic Vesicles | Cell Membrane | Cytoplasmic Membrane | Cell Membrane | ||||
| Early | WT | ● | ● | - | - | - | ● | - | - | ● | ● |
| ΔF | ● | - | ● | - | ● | ● | - | - | ● | ● | |
| Late | WT | ● | ● | - | ● | - | ● | ● | ● | - | ● |
| ΔF | ● | ● | ● | - | ● | - | ● | - | ● | ● | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merigo, F.; Lotti, V.; Bernardi, P.; Conti, A.; Clemente, A.D.; Ligozzi, M.; Lagni, A.; Sorio, C.; Sbarbati, A.; Gibellini, D. Ultrastructural Characterization of Human Bronchial Epithelial Cells during SARS-CoV-2 Infection: Morphological Comparison of Wild-Type and CFTR-Modified Cells. Int. J. Mol. Sci. 2022, 23, 9724. https://doi.org/10.3390/ijms23179724
Merigo F, Lotti V, Bernardi P, Conti A, Clemente AD, Ligozzi M, Lagni A, Sorio C, Sbarbati A, Gibellini D. Ultrastructural Characterization of Human Bronchial Epithelial Cells during SARS-CoV-2 Infection: Morphological Comparison of Wild-Type and CFTR-Modified Cells. International Journal of Molecular Sciences. 2022; 23(17):9724. https://doi.org/10.3390/ijms23179724
Chicago/Turabian StyleMerigo, Flavia, Virginia Lotti, Paolo Bernardi, Anita Conti, Andrea Di Clemente, Marco Ligozzi, Anna Lagni, Claudio Sorio, Andrea Sbarbati, and Davide Gibellini. 2022. "Ultrastructural Characterization of Human Bronchial Epithelial Cells during SARS-CoV-2 Infection: Morphological Comparison of Wild-Type and CFTR-Modified Cells" International Journal of Molecular Sciences 23, no. 17: 9724. https://doi.org/10.3390/ijms23179724






