Multiple Parallel Fusion Network for Predicting Protein Subcellular Localization from Stimulated Raman Scattering (SRS) Microscopy Images in Living Cells
Abstract
1. Introduction
2. Results
2.1. Experimental Settings
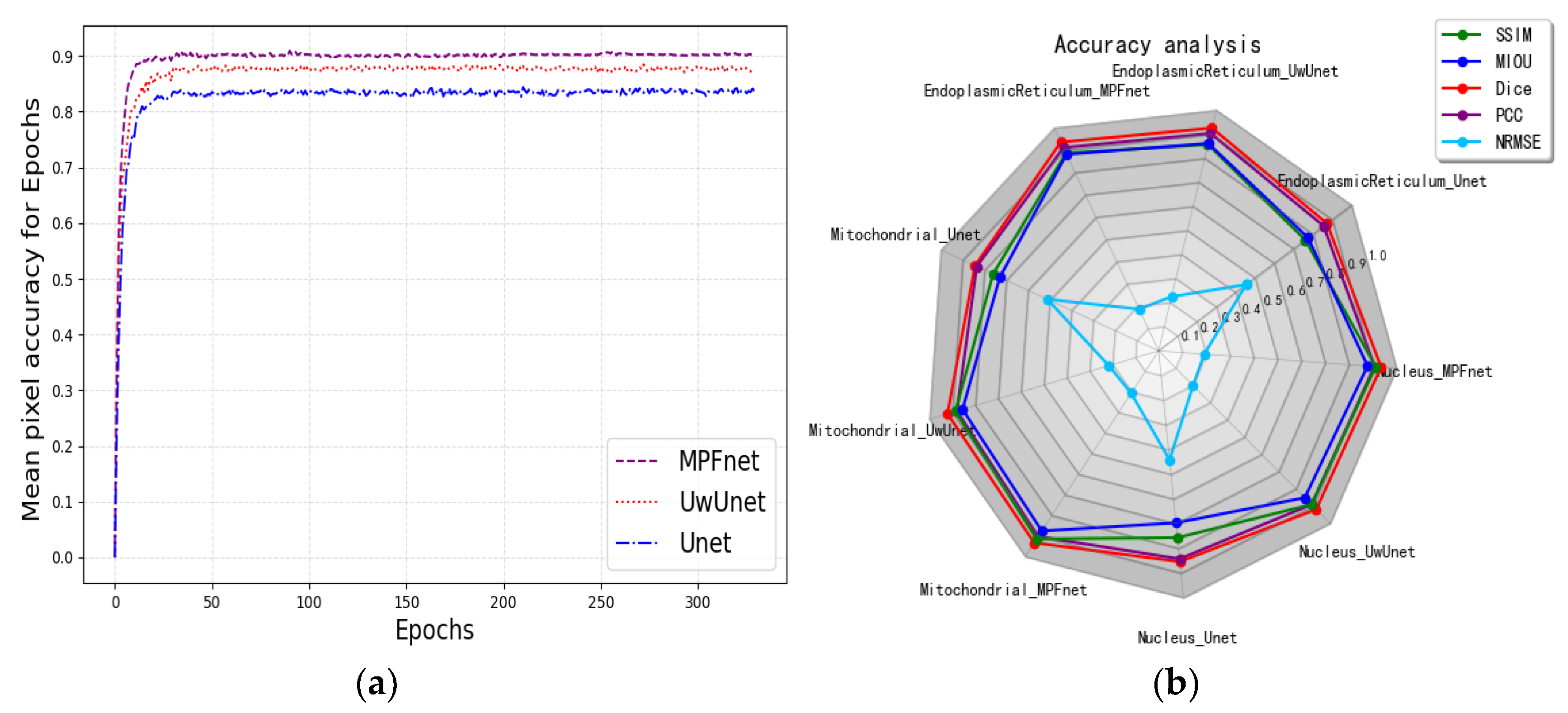
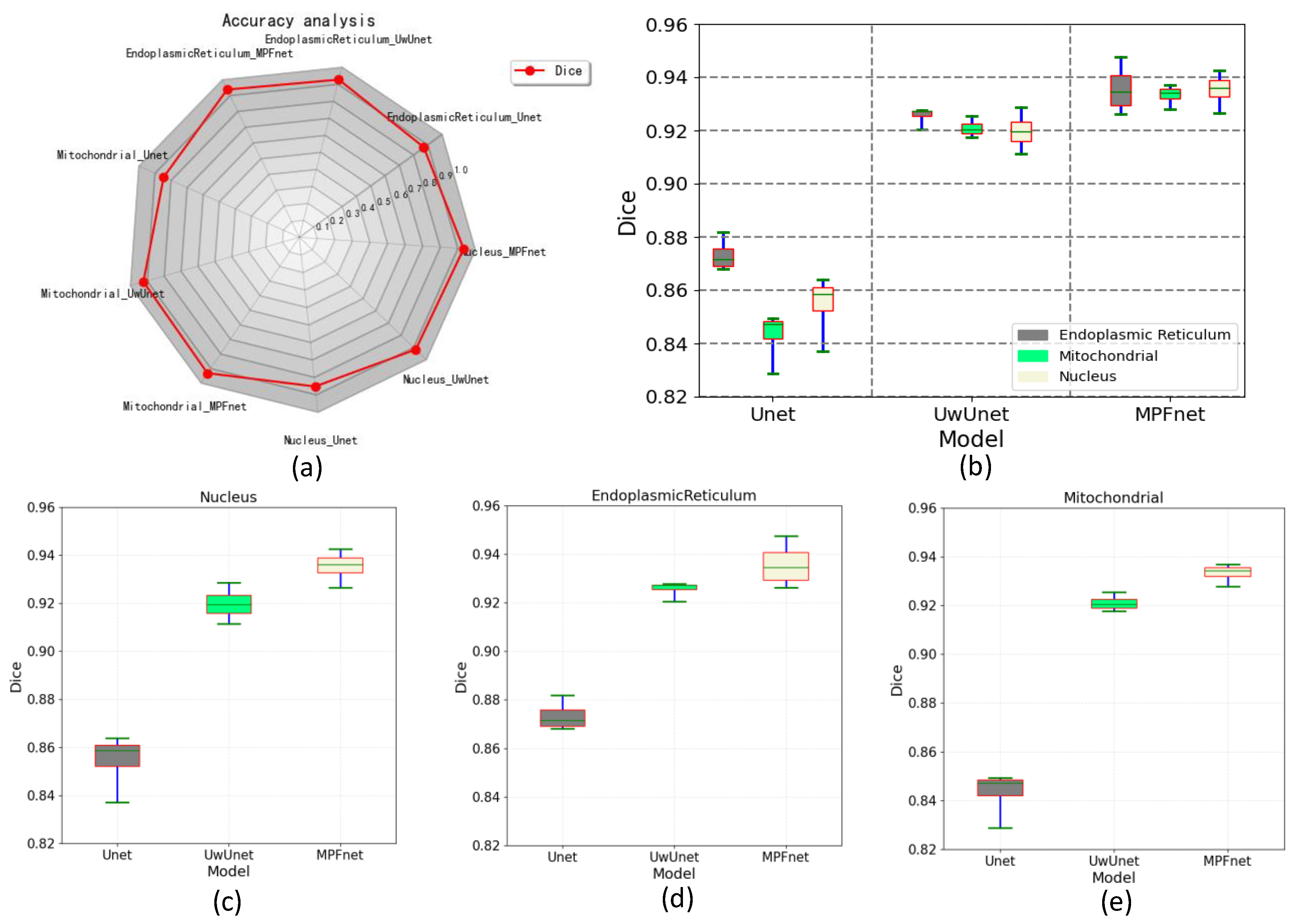
2.2. Metrics for Performance Evaluation
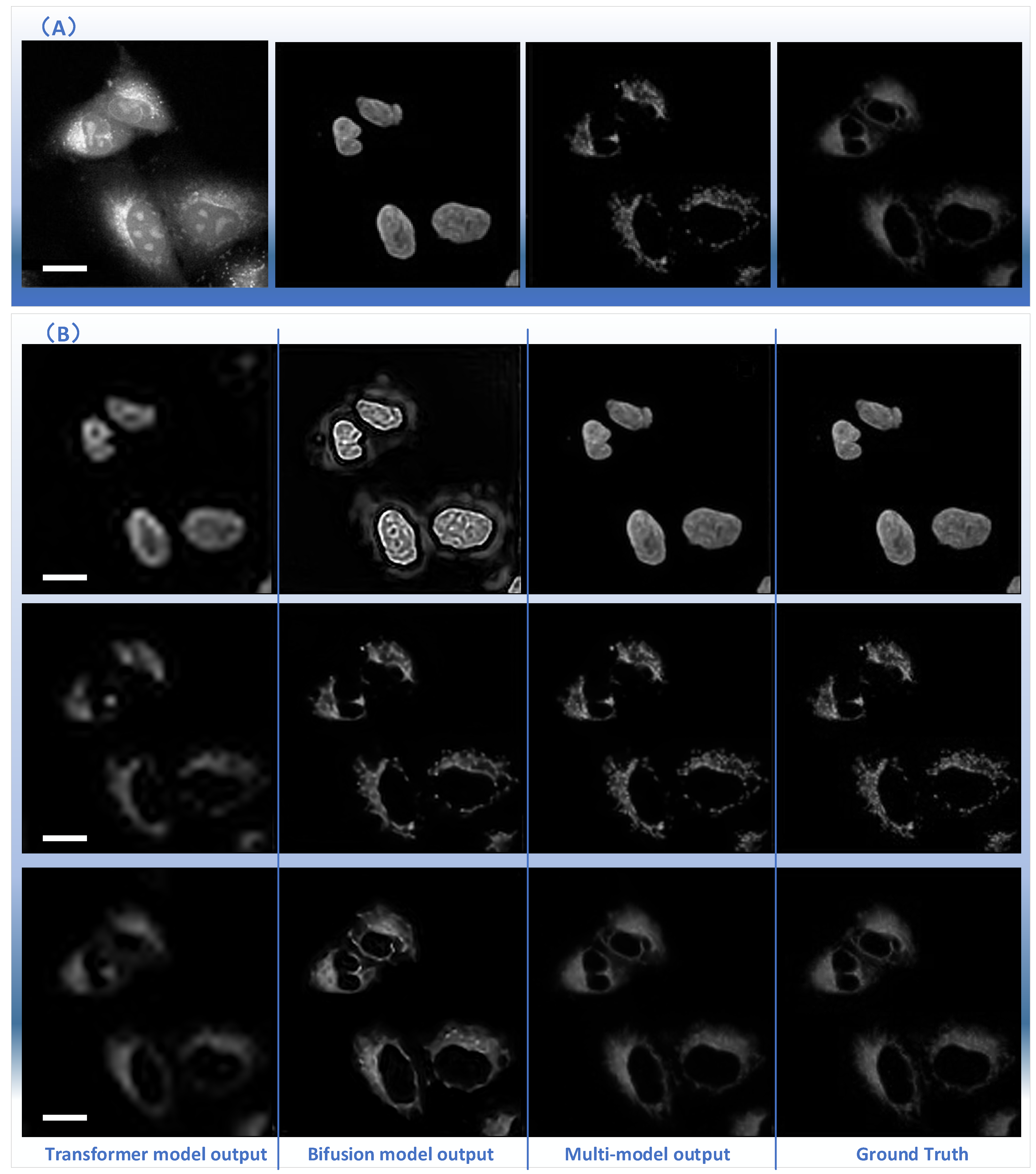
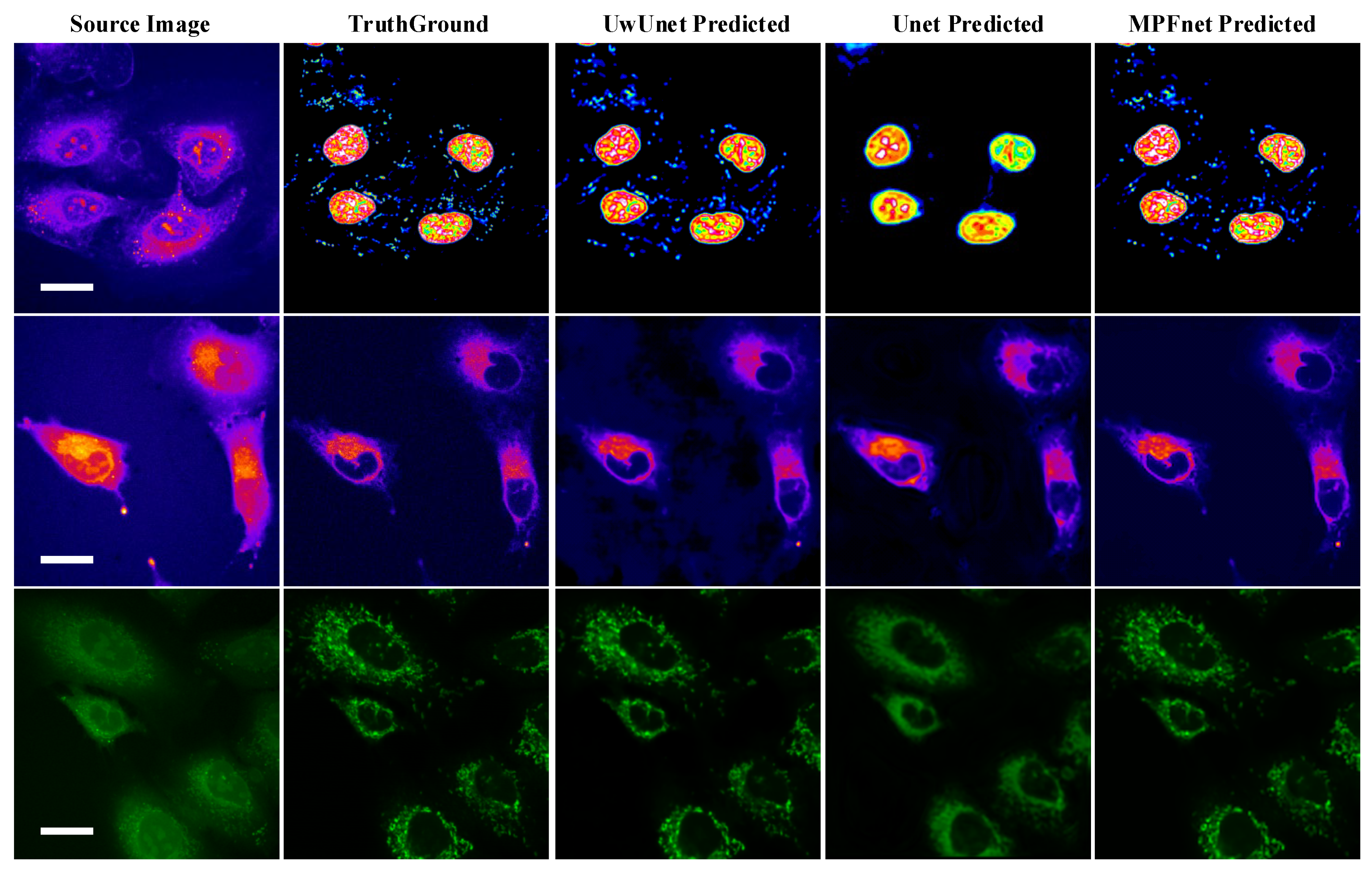
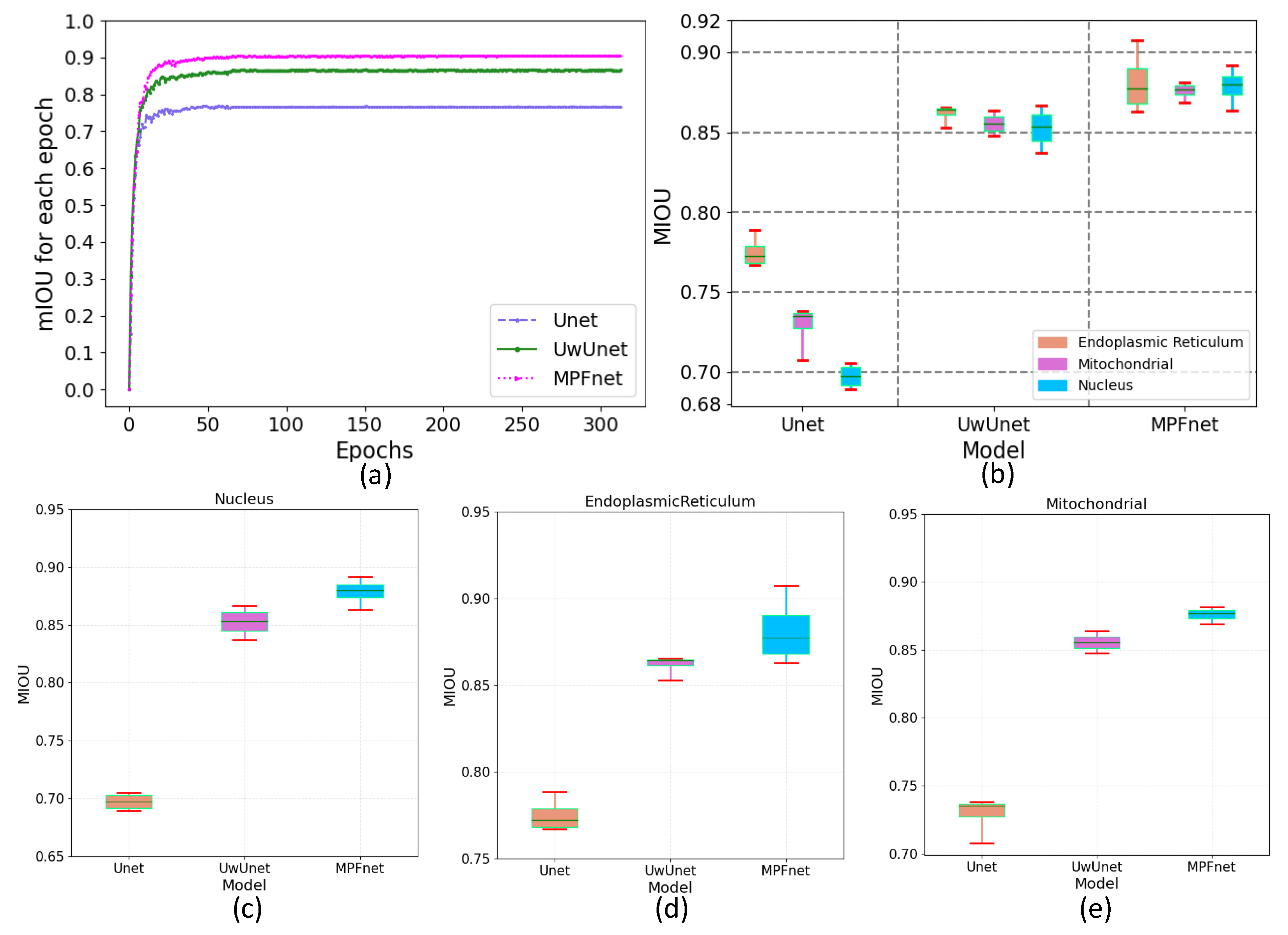
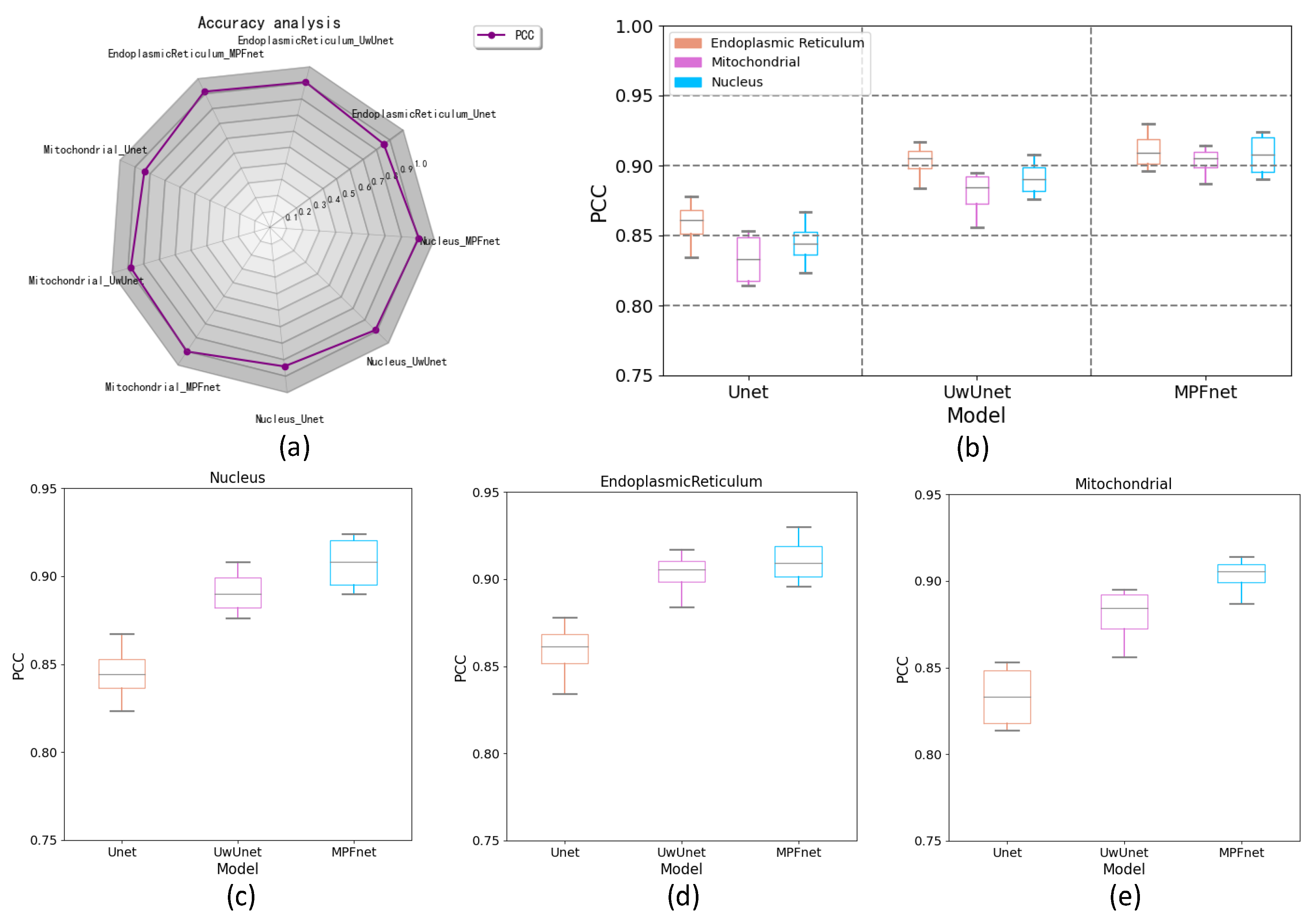
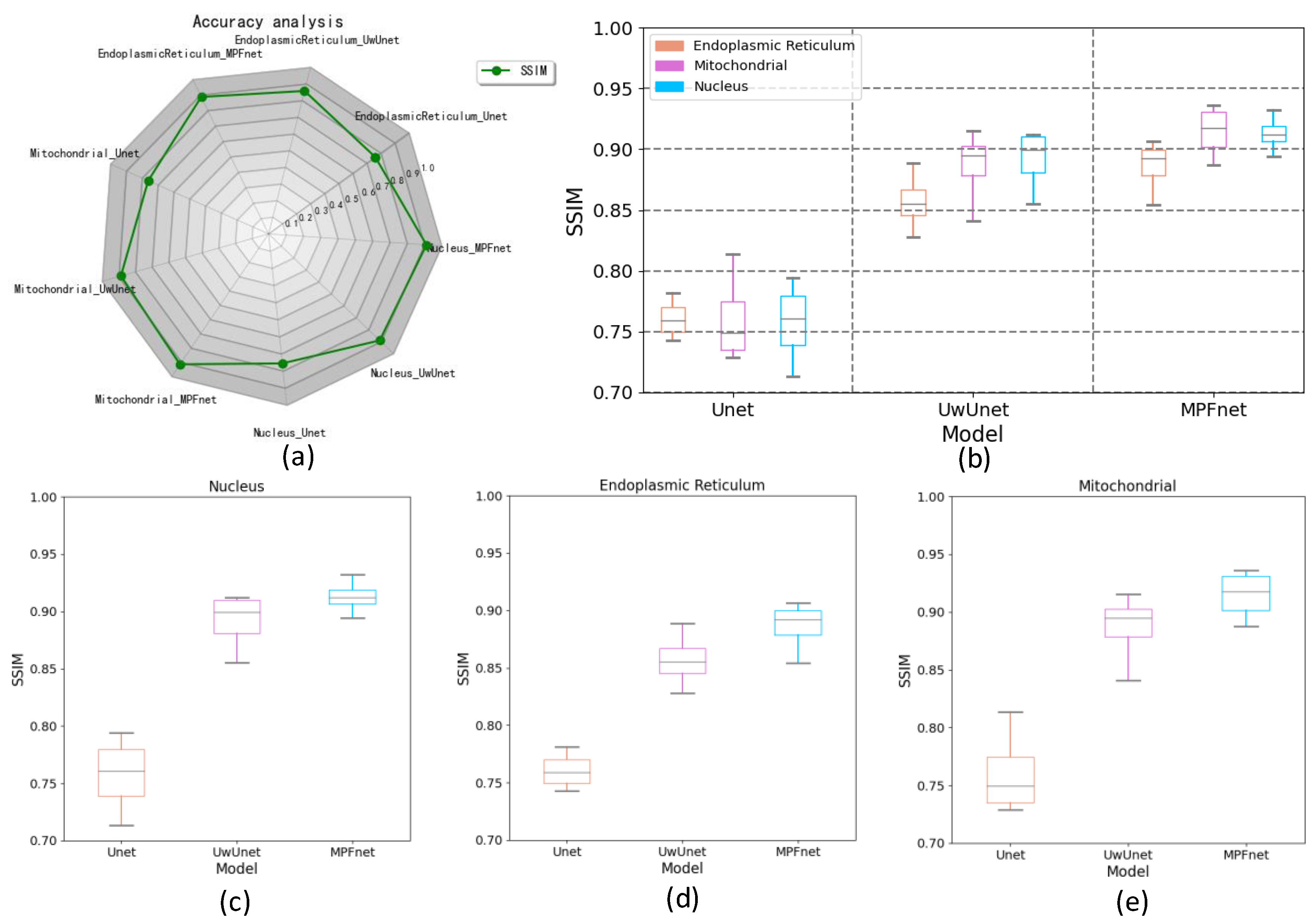
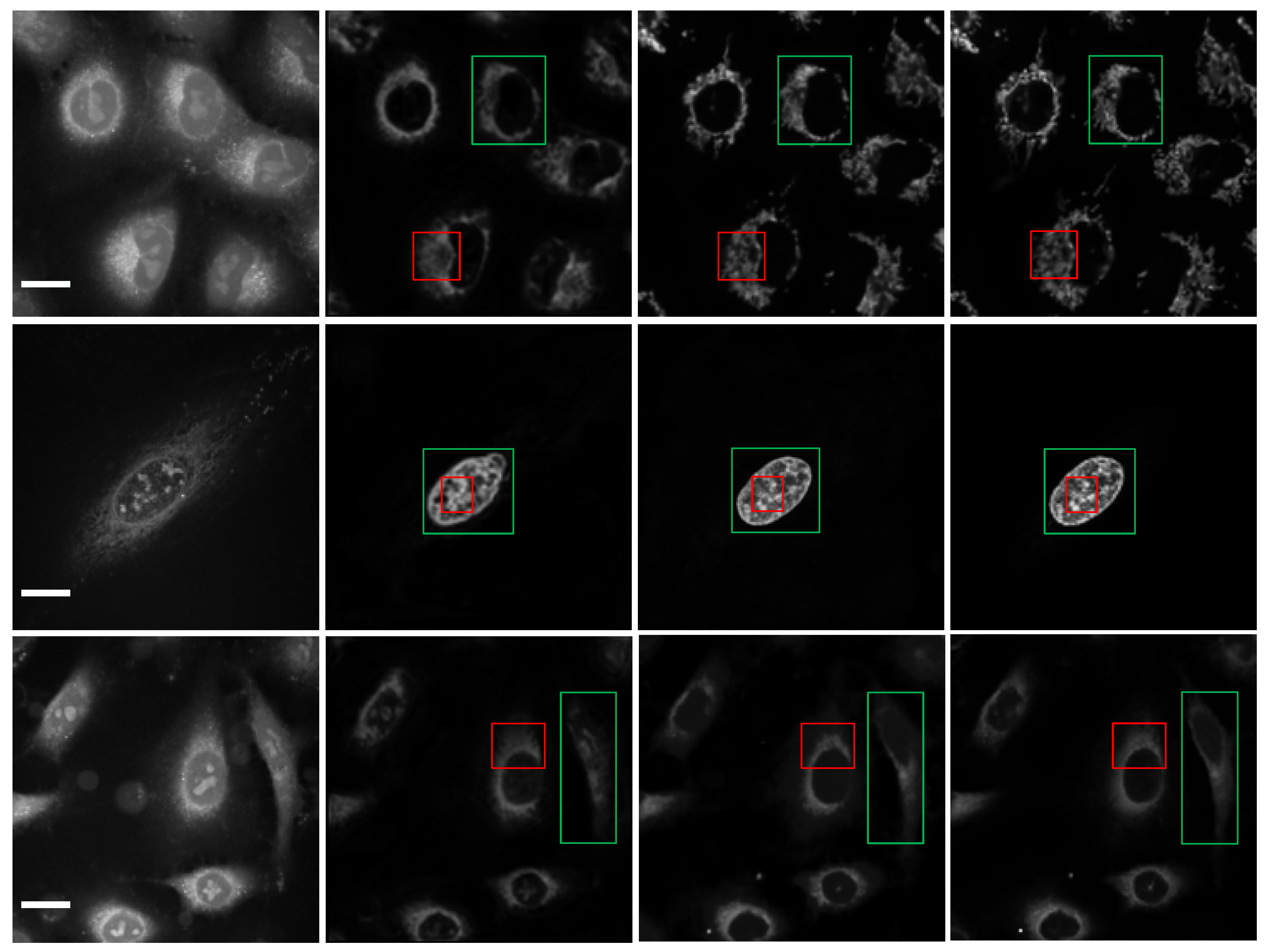
2.3. The Analysis of the Performance of Multiple Parallel Fusion Deep Networks
2.4. Comparison of the Performance with Various Prediction Models
3. Discussion
4. Methods and Materials
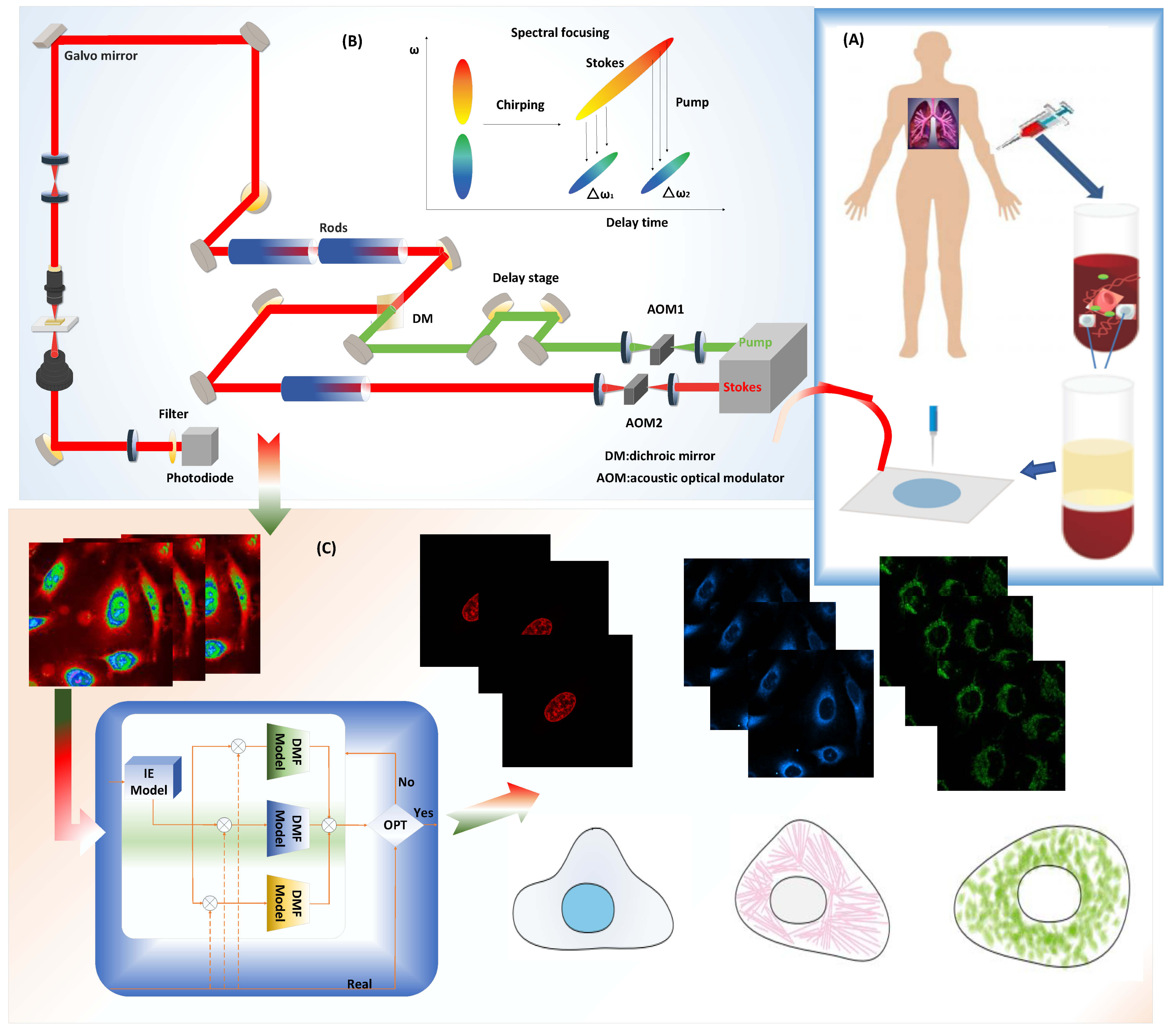
4.1. Experiment of the Simultaneous SRS and Fluorescence Microscopy
4.2. Protein Subcellular Localization Based on Multiple Parallel Fusion Deep Networks
4.3. Multiple Parallel Fusion Neural Network Architecture and Implementation
4.3.1. CNN Branch
4.3.2. Transformer Branch
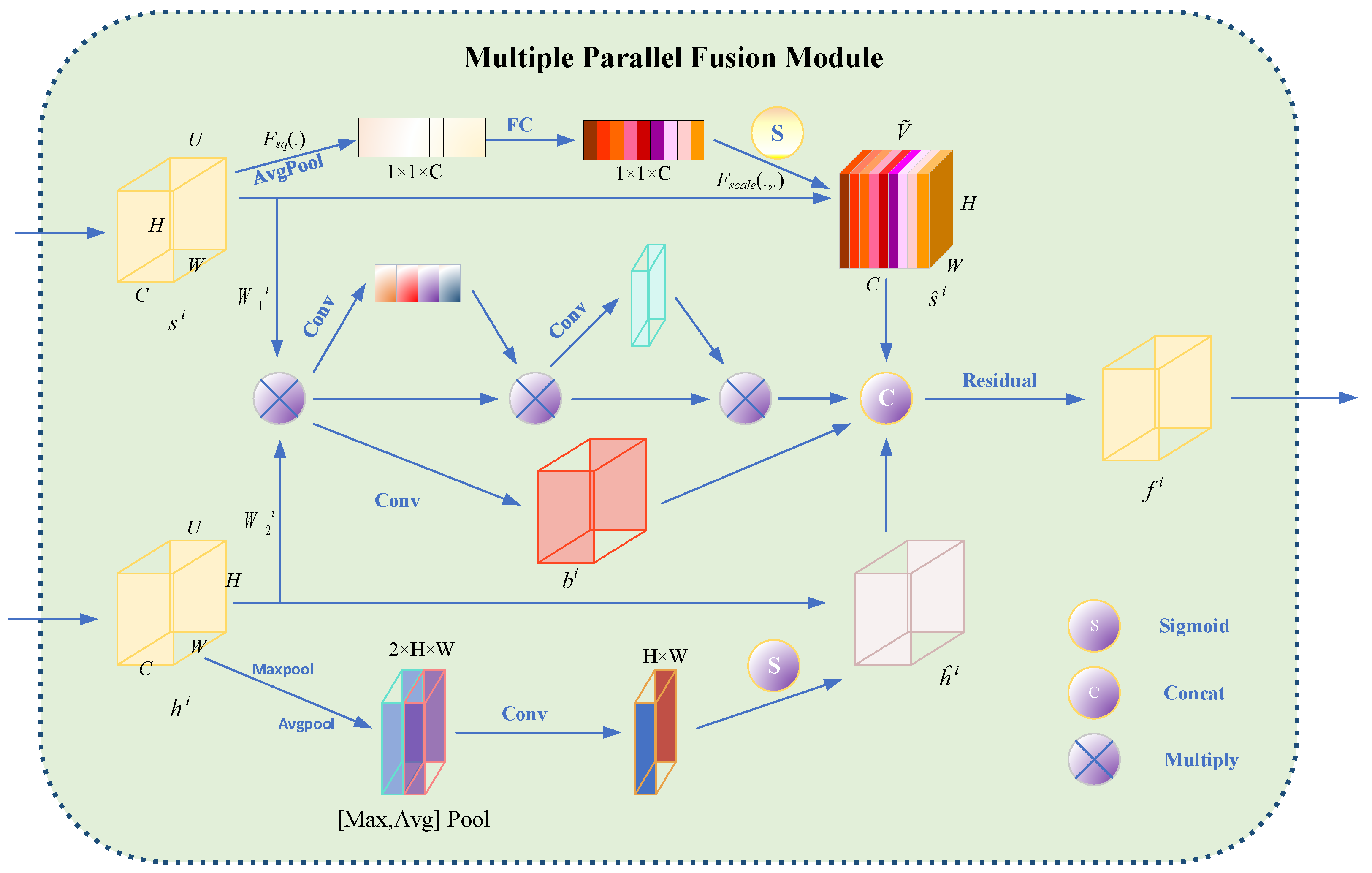
4.3.3. Multiple Parallel Fusion Module
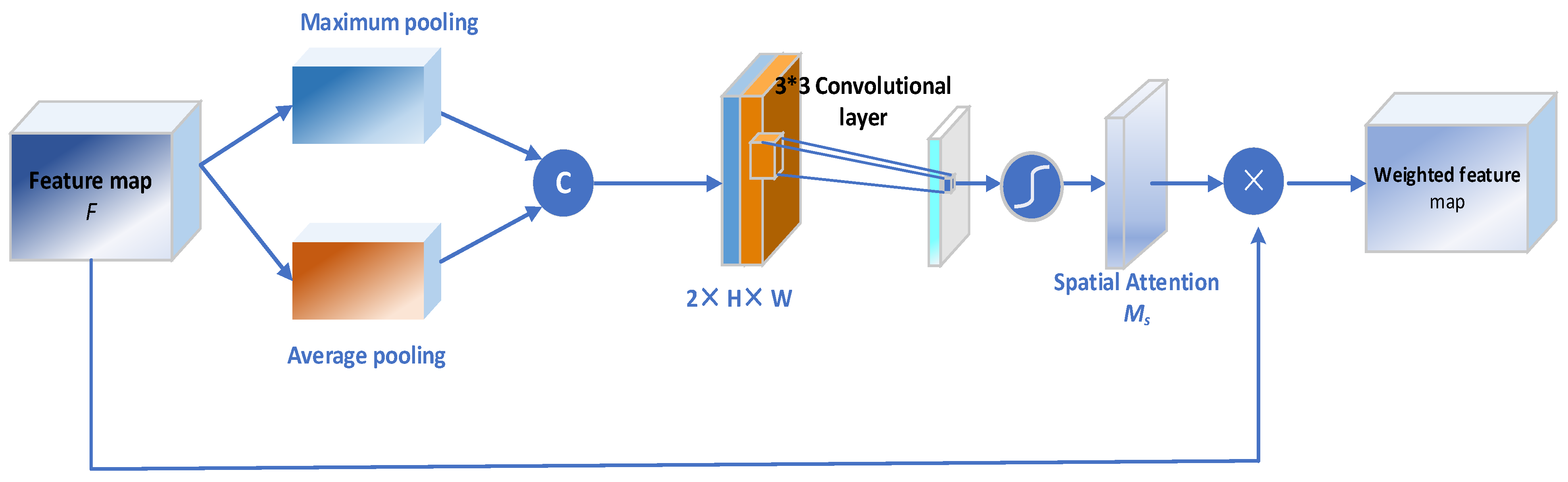
4.3.4. Spatial Attention Block
4.3.5. Channel-Wise Attention
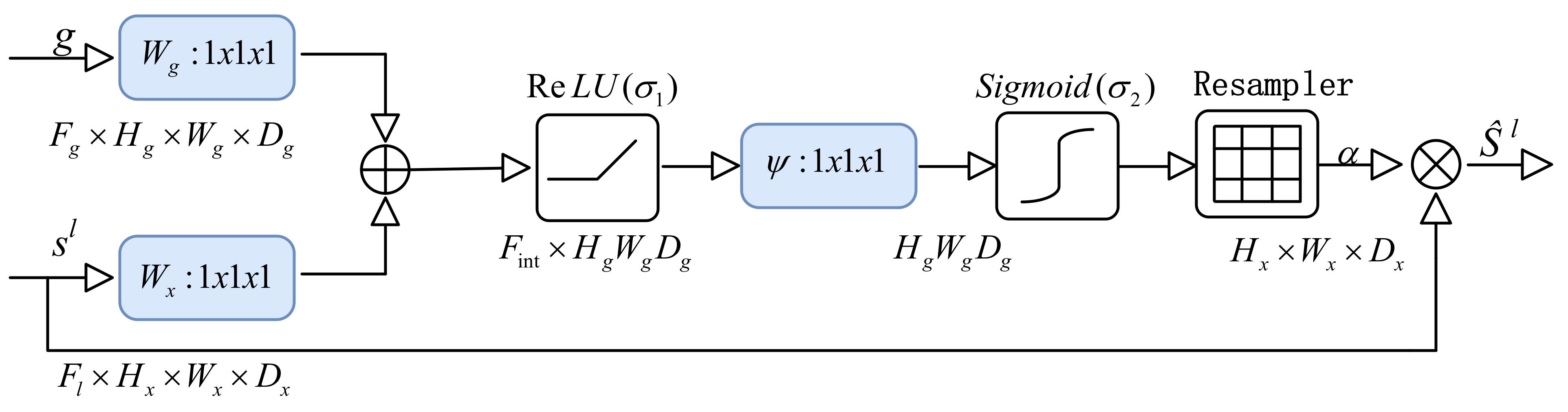
4.3.6. Attention Gate Block
4.4. Dataset
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parlakgül, G.; Arruda, A.P.; Pang, S.; Erika, C.; Nina, M.; Ekin, G.; Grace, Y.L.; Karen, I.; Hess, H.F.; Shan, X.C.; et al. Regulation of liver subcellular architecture controls metabolic homeostasis. Nature 2022, 603, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Mottis, A.; Herzig, S.; Auwerx, J. Mitocellular communication: Shaping health and disease. Science 2019, 366, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Cai, L.; Wang, Z.Y.; Hu, X.; Zhang, S.T.; Ji, S.W. Computational Modeling of Cellular Structures Using Conditional Deep Generative Networks. Bioinformatics 2019, 35, 2141–2149. [Google Scholar] [CrossRef] [PubMed]
- Koenig, F.; Knittel, J.; Stepp, H. Diagnosing cancer in vivo. Science 2001, 292, 401–1403. [Google Scholar] [CrossRef]
- Szabo, V.; Ventalon, C.; De Sars, V.; Bradley, J.; Emiliani, V. Spatially selective holographic photoactivation and functional fluorescence imaging in freely behaving mice with a fiberscope. Neuron 2014, 84, 1157–1169. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Recent progress in protein subcellular location prediction. Anal. Biochem. 2007, 370, 1–16. [Google Scholar] [CrossRef]
- Xu, Y.Y.; Yang, F.; Zhang, Y.; Shen, H.B. An image-based multi-label human protein subcellular localization predictor (iLocator) reveals protein mislocalizations in cancer tissues. Bioinformatics 2013, 29, 2032–2040. [Google Scholar] [CrossRef]
- Hung, M.C.; Link, W. Protein localization in disease and therapy. J. Cell Sci. 2011, 124, 3381–3392. [Google Scholar] [CrossRef]
- Guo, X.; Liu, F.; Ju, Y.; Wang, Z.; Wang, C. Human Protein Subcellular Localization with Integrated Source and Multi-label Ensemble Classifier. Sci. Rep. 2016, 6, 28087. [Google Scholar] [CrossRef]
- Vicar, T.; Balvan, J.; Jaros, J.; Jug, F.; Kolar, R.; Masarik, M.; Gumulec, J. Cell segmentation methods for label-free contrast microscopy: Review and comprehensive comparison. BMC Bioinform. 2019, 20, 360. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, Y.; Zheng, B.; Su, L.; Chen, Y.; Ma, S.; Hu, Q.; Zou, X.; Yao, L.; Yang, Y.; et al. Rapid histology of laryngeal squamous cell carcinoma with deep-learning based stimulated Raman scattering microscopy. Theranostics 2019, 9, 2541–2554. [Google Scholar] [CrossRef] [PubMed]
- Ounkomol, C.; Seshamani, S.; Maleckar, M.M.; Collman, F.; Johnson, G.R. Label-free prediction of three- dimensional fluorescence images from transmitted-light microscopy. Nat. Methods 2018, 15, 917–920. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Cell-PLoc 2.0: An improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Sci. 2010, 2, 1090–1103. [Google Scholar] [CrossRef]
- Chou, K.C.; Cai, Y.D. Prediction of protein subcellular locations by GO-FunD-PseAA predicor. Biochem. Biophys. Res. Commun. 2004, 320, 1236–1239. [Google Scholar] [CrossRef] [PubMed]
- Falk, T.; Mai, D.; Bensch, R.; Çiçek, Ö.; Abdulkadir, A.; Marrakchi, Y.; Böhm, A.; Deubner, J.; Jäckel, Z.; Seiwald, K.; et al. U-Net: Deep learning for cell counting, detection, and morphometry. Nat. Methods 2019, 16, 67–70. [Google Scholar] [CrossRef]
- Wang, A.; Zhang, Q.; Han, Y.; Megason, S.; Hormoz, S.; Mosaliganti, K.R.; Lam, J.C.K.; Li, V.O.K. A novel deep learning-based 3D cell segmentation framework for future image-based disease detection. Sci. Rep. 2022, 12, 342. [Google Scholar] [CrossRef]
- Wei, L.; Ding, Y.; Ran, S.; Tang, J.; Zou, Q. Prediction of human protein subcellular localization using deep learning. J. Parallel Distrib. Comput. 2018, 117, 212–217. [Google Scholar] [CrossRef]
- Jing, S.; Attila, T.; Su, X.T. Deep Learning-Based Single-Cell Optical Image Studies. Cytom. A 2020, 97, 226–240. [Google Scholar]
- Buggenthin, F.; Buettner, F.; Hoppe, F.; Endele, P.S.; Kroiss, M.; Strasser, M.; Schwarzfischer, M.; Loeffler, M.; Kokkaliaris, D.; Hilsenbeck, K.D.; et al. Prospective identification of hematopoietic lineage choice by deep learning. Nat. Methods 2017, 14, 403–406. [Google Scholar] [CrossRef]
- Hasan, M.; Ahmad, S.; Molla, M. Protein Subcellular Localization Prediction Using Multiple Kernel Learning Based Support Vector Machine. Mol. BioSyst. 2017, 13, 785–795. [Google Scholar] [CrossRef]
- Juan, J.; Armenteros, A.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar]
- Wang, Y.; Xu, Y.; Zang, Z.; Wu, L.; Li, Z. Panoramic Manifold Projection (Panoramap) for Single-Cell Data Dimensionality Reduction and Visualization. Int. J. Mol. Sci. 2022, 23, 7775. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Roeffaers, M.; Basu, S.; Daniele, J.; Fu, D.; Freudiger, C.; Holtom, G.; Xie, X. Label-Free Live-Cell Imaging of Nucleic Acids Using Stimulated Raman Scattering Microscopy. ChemPhysChem 2012, 13, 1054–1059. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Lei, C.; Yasumoto, A.; Kobayashi, H.; Aisaka, Y.; Ito, T.; Guo, B.; Nitta, N.; Kutsuna, N.; Ozeki, Y.; et al. Label-free detection of aggregated platelets in blood by machine-learning-aided optofluidic time-stretch microscopy. Lab Chip 2017, 17, 2426–2434. [Google Scholar] [CrossRef]
- Kobayashi, H.; Lei, C.; Wu, Y.; Mao, A.; Jiang, Y.; Guo, B.; Ozeki, Y.; Goda, K. Label-free detection of cellular drug responses by high-throughput bright-field imaging and machine learning. Sci. Rep. 2017, 7, 12454. [Google Scholar] [CrossRef]
- Wei, H.; Xie, L.; Liu, Q.; Shao, C.; Wang, X.; Su, X. Automatic Classification of Label-Free Cells from Small Cell Lung Cancer and Poorly Differentiated Lung Adenocarcinoma with 2D Light Scattering Static Cytometry and Machine Learning. Cytom. Part A 2019, 95A, 302–308. [Google Scholar] [CrossRef]
- Li, J.T.; Chen, J.; Bai, H.; Wang, H.W.; Hao, S.P.; Ding, Y.; Peng, B.; Zhang, J.; Li, L.; Huang, W. An Overview of Organs-on-Chips Based on Deep Learning. Research 2022, 2022, 9869518. [Google Scholar] [CrossRef]
- Siu, D.M.D.; Lee, K.C.M.; Lo, M.C.K.; Stassen, S.V.; Wang, M.L.; Zhang, I.Z.Q.; So, H.K.H.; Chan, G.C.F.; Cheah, K.S.E.; Wong, K.K.Y.; et al. Deep-learning-assisted biophysical imaging cytometry at massive throughput delineates cell population heterogeneity. Lab Chip 2020, 20, 3696–3708. [Google Scholar] [CrossRef]
- Yao, K.; Rochman, N.D.; Sun, S.X. Cell Type Classification and Unsupervised Morphological Phenotyping From Low-Resolution Images Using Deep Learning. Sci. Rep. 2019, 9, 13467. [Google Scholar] [CrossRef]
- Chen, C.; Mahjoubfar, A.; Tai, L.C.; Blaby, I.K.; Huang, A.; Niazi, K.R.; Jalali, B. Deep Learning in Label-free Cell Classification. Sci. Rep. 2016, 6, 21471. [Google Scholar] [CrossRef]
- Wang, F.S.; Wei, L.Y. Multi-scale deep learning for the imbalanced multi-label protein subcellular localization prediction based on immunohistochemistry images. Bioinformatics 2022, 38, 2602–2611. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, J.; Lin, H.; Tan, Y.; Cheng, J.X. High-Speed Chemical Imaging by Dense-Net Learning of Femtosecond Stimulated Raman Scattering. J. Phys. Chem. Lett. 2020, 11, 8573–8578. [Google Scholar] [CrossRef] [PubMed]
- Imboden, S.; Liu, X.; Lee, B.S.; Payne, M.C.; Hsieh, C.J.; Lin, N.Y. Investigating heterogeneities of live mesenchymal stromal cells using AI-based label-free imaging. Sci. Rep. 2021, 11, 6728. [Google Scholar] [CrossRef] [PubMed]
- Lynch, A.W.; Theodoris, C.V.; Long, H.W.; Brown, M.; Liu, X.S.; Meyer, C.A. MIRA: Joint regulatory modeling of multimodal expression and chromatin accessibility in single cells. Nat Methods 2022, 19, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Melanthota, S.K.; Gopal, D.; Chakrabarti, S.; Kashyap, A.A.; Radhakrishnan, R.; Mazumder, N. Deep learning-based image processing in optical microscopy. Biophys. Rev. 2022, 14, 463–481. [Google Scholar] [CrossRef] [PubMed]
- Manifold, B.; Men, S.; Hu, R.; Fu, D. A versatile deep learning architecture for classification and label-free prediction of hyper- spectral images. Nat. Mach. Intell. 2021, 3, 306–315. [Google Scholar] [CrossRef]
- Kobayashi, H.; Cheveralls, K.C.; Leonetti, M.D.; Royer, L.A. Self-supervised deep learning encodes high-resolution features of protein subcellular localization. Nat. Methods 2022, 19, 995–1003. [Google Scholar] [CrossRef]
- Donovan-Maiye, R.M.; Brown, J.M.; Chan, C.K.; Ding, L.; Yan, C.; Gaudreault, N.; Theriot, J.A.; Maleckar, M.M.; Knijnenburg, T.A.; Johnson, G.R. A deep generative model of 3D single-cell organization. PLoS Comput. Biol. 2018, 18, e1009155. [Google Scholar] [CrossRef]
- Gomariz, A.; Portenier, T.; Helbling, P.M.; Isringhausen, S.; Suessbier, U.; Nombela-Arrieta, C.; Goksel, O. Modality attention and sampling enables deep learning with heterogeneous marker combinations in fluorescence microscopy. Nat. Mach. Intell. 2021, 3, 799–811. [Google Scholar] [CrossRef]
- Chen, X.; Li, Y.; Wyman, N.; Zhang, Z.; Fan, H.; Le, M.; Gannon, S.; Rose, C.; Zhang, Z.; Mercuri, J.; et al. Deep Learning Provides High Accuracy in Automated Chondrocyte Viability Assessment in Articular Cartilage Using Nonlinear Optical Microscopy. Biomed. Opt. Express 2021, 12, 2759–2772. [Google Scholar] [CrossRef]
- Lu, A.X.; Kraus, O.Z.; Cooper, S.; Moses, A.M. Learning unsupervised feature representations for single cell microscopy images with paired cell inpainting. PLoS Comput. Biol. 2019, 15, e1007348. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, J.; Zhang, C.; Wang, S. SSGraphCPI: A Novel Model for Predicting Compound-Protein Interactions Based on Deep Learning. Int. J. Mol. Sci. 2022, 23, 3780. [Google Scholar] [CrossRef]
- Voronin, D.V.; Kozlova, A.A.; Verkhovskii, R.A.; Ermakov, A.V.; Makarkin, M.A.; Inozemtseva, O.A.; Bratashov, D.N. Detection of Rare Objects by Flow Cytometry: Imaging, Cell Sorting, and Deep Learning Approaches. Int. J. Mol. Sci. 2020, 21, 2323. [Google Scholar] [CrossRef] [PubMed]
- Calizo, R.C.; Bell, M.K.; Ron, A.; Hu, M.; Bhattacharya, S.; Wong, N.J.; Janssen, W.G.M.; Perumal, G.; Pederson, P.; Scarlata, S.; et al. Cell shape regulates subcellular organelle location to control early Ca2+ signal dynamics in vascular smooth muscle cells. Sci. Rep. 2020, 10, 17866. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. MPFnet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Fang, J.; Yang, C.; Shi, Y.T.; Wang, N.; Zhao, Y. External Attention Based MPFnet and Label Expansion Strategy for Crack Detection. IEEE Trans. Intell. Transp. Syst. 2022, 17, 1–10. [Google Scholar]
- Rezatofighi, H.; Tsoi, N.; Gwak, J.Y.; Sadeghian, A.; Reid, I.; Savarese, S. Generalized Intersection over Union: A Metric and A Loss for Bounding Box Regression. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2019, Long Beach, CA, USA, 15–20 June 2019; pp. 658–666. [Google Scholar]
- Boslaugh, S.; Watters, P.A. Statistics in a Nutshell: A Desktop Quick Reference; O’Reilly Media: Sebastopol, CA, USA, 2008; ISBN 9780596510497. [Google Scholar]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Esakkirajan, S.; Veerakumar, T.; Subramanyam, A.N.; PremChand, C.H. Removal of High Density Salt and Pepper Noise Through Modified Decision Based Unsymmetric Trimmed Median Filter. IEEE Signal Process. Lett. 2011, 18, 287–290. [Google Scholar] [CrossRef]
- Hsieh, M.h.; Cheng, F.C.; Shie, M.C.; Ruan, S.J. Fast and efficient median filter for removing 1–99% levels of salt-and-pepper noise in images. Eng. Appl. Artif. Intell. 2013, 26, 1333–1338. [Google Scholar] [CrossRef]
- Manifold, B.; Thomas, E.; Francis, A.T.; Hill, A.H.; Fu, D. Denoising of stimulated Raman scattering microscopy images via deep learning. Biomed. Opt. Express 2019, 10, 3860–3874. [Google Scholar] [CrossRef]
- Al-Kofahi, Y.; Zaltsman, A.; Graves, R.; Marshall, W.; Rusu, M. A deep learning-based algorithm for 2-D cell segmentation in microscopy images. BMC Bioinform. 2018, 19, 365. [Google Scholar] [CrossRef] [PubMed]





| Organelle/ Model | MFPnet Model | ||||
|---|---|---|---|---|---|
| NRMSE↓ | SSIM↑ | PCC↑ | Dice↑ | mIOU↑ | |
| Nucleus | 0.192 ± 0.013 | 0.913 ± 0.019 | 0.908 ± 0.018 | 0.935 ± 0.009 | 0.879 ± 0.014 |
| Endoplasmic Reticulum | 0.187 ± 0.015 | 0.886 ± 0.032 | 0.911 ± 0.019 | 0.936 ± 0.011 | 0.881 ± 0.016 |
| Mitochondrial | 0.206 ± 0.028 | 0.915 ± 0.028 | 0.903 ± 0.016 | 0.933 ± 0.005 | 0.876 ± 0.007 |
| Organelle\Model | UwUnet Model | ||||
|---|---|---|---|---|---|
| NRMSE↓ | SSIM↑ | PCC↑ | Dice↑ | mIOU↑ | |
| Nucleus | 0.201 ± 0.012 | 0.892 ± 0.037 | 0.892 ± 0.016 | 0.919 ± 0.009 | 0.852 ± 0.015 |
| Endoplasmic Reticulum | 0.225 ± 0.014 | 0.857 ± 0.030 | 0.903 ± 0.019 | 0.925 ± 0.005 | 0.861 ± 0.009 |
| Mitochondrial | 0.217 ± 0.017 | 0.886 ± 0.046 | 0.880 ± 0.024 | 0.921 ± 0.004 | 0.854 ± 0.008 |
| Model/Organelle | Unet Model | ||||
|---|---|---|---|---|---|
| NRMSE↓ | SSIM↑ | PCC↑ | Dice↑ | mIOU↑ | |
| Nucleus | 0.442 ± 0.024 | 0.757 ± 0.044 | 0.843 ± 0.024 | 0.855 ± 0.018 | 0.716 ± 0.008 |
| Endoplasmic Reticulum | 0.454 ± 0.022 | 0.761 ± 0.021 | 0.856 ± 0.022 | 0.873 ± 0.008 | 0.771 ± 0.013 |
| Mitochondrial | 0.511 ± 0.013 | 0.760 ± 0.053 | 0.835 ± 0.021 | 0.843 ± 0.015 | 0.731 ± 0.021 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Z.; Liu, W.; Yu, W.; Liu, X.; Yan, R.; Liu, Q.; Guo, Q. Multiple Parallel Fusion Network for Predicting Protein Subcellular Localization from Stimulated Raman Scattering (SRS) Microscopy Images in Living Cells. Int. J. Mol. Sci. 2022, 23, 10827. https://doi.org/10.3390/ijms231810827
Wei Z, Liu W, Yu W, Liu X, Yan R, Liu Q, Guo Q. Multiple Parallel Fusion Network for Predicting Protein Subcellular Localization from Stimulated Raman Scattering (SRS) Microscopy Images in Living Cells. International Journal of Molecular Sciences. 2022; 23(18):10827. https://doi.org/10.3390/ijms231810827
Chicago/Turabian StyleWei, Zhihao, Wu Liu, Weiyong Yu, Xi Liu, Ruiqing Yan, Qiang Liu, and Qianjin Guo. 2022. "Multiple Parallel Fusion Network for Predicting Protein Subcellular Localization from Stimulated Raman Scattering (SRS) Microscopy Images in Living Cells" International Journal of Molecular Sciences 23, no. 18: 10827. https://doi.org/10.3390/ijms231810827
APA StyleWei, Z., Liu, W., Yu, W., Liu, X., Yan, R., Liu, Q., & Guo, Q. (2022). Multiple Parallel Fusion Network for Predicting Protein Subcellular Localization from Stimulated Raman Scattering (SRS) Microscopy Images in Living Cells. International Journal of Molecular Sciences, 23(18), 10827. https://doi.org/10.3390/ijms231810827






