Engineering Nanofiber Scaffolds with Biomimetic Cues for Differentiation of Skin-Derived Neural Crest-like Stem Cells to Schwann Cells
Abstract
:1. Introduction
2. Results
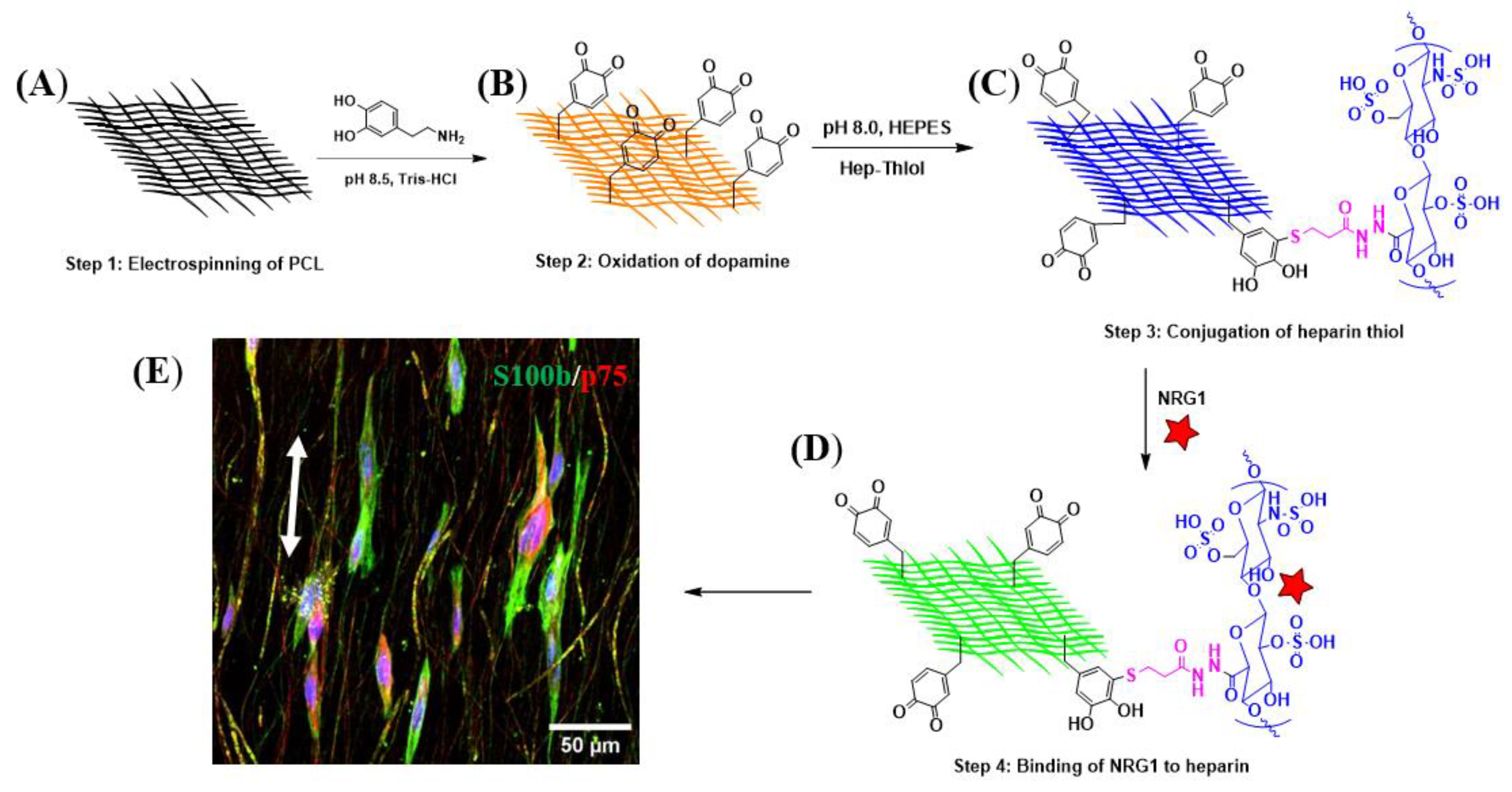
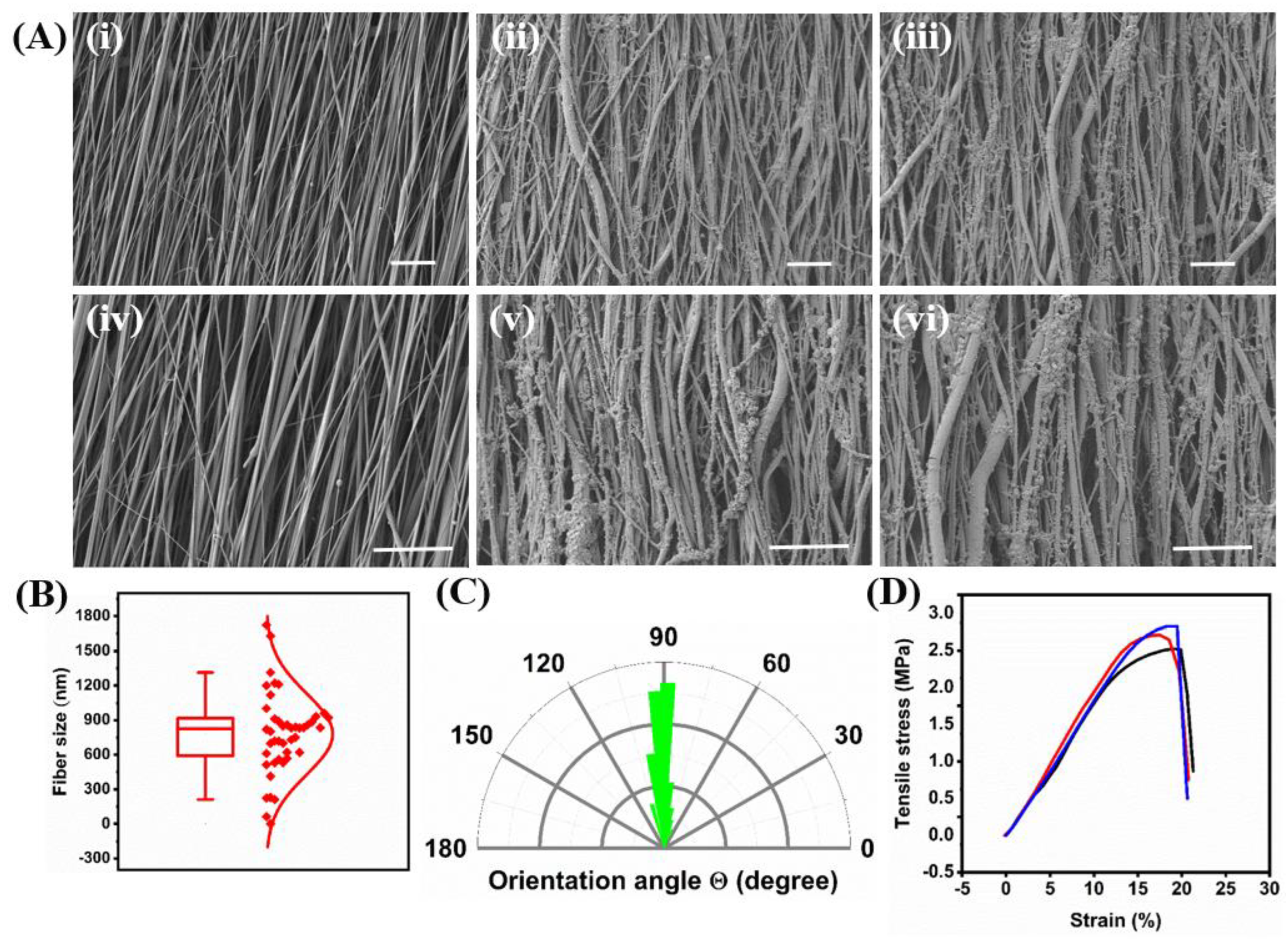
2.1. Fiber Formation and Decoration with Biological Cues
2.2. Effect of Biomolecules on Adhesion and Spreading of KC-NC
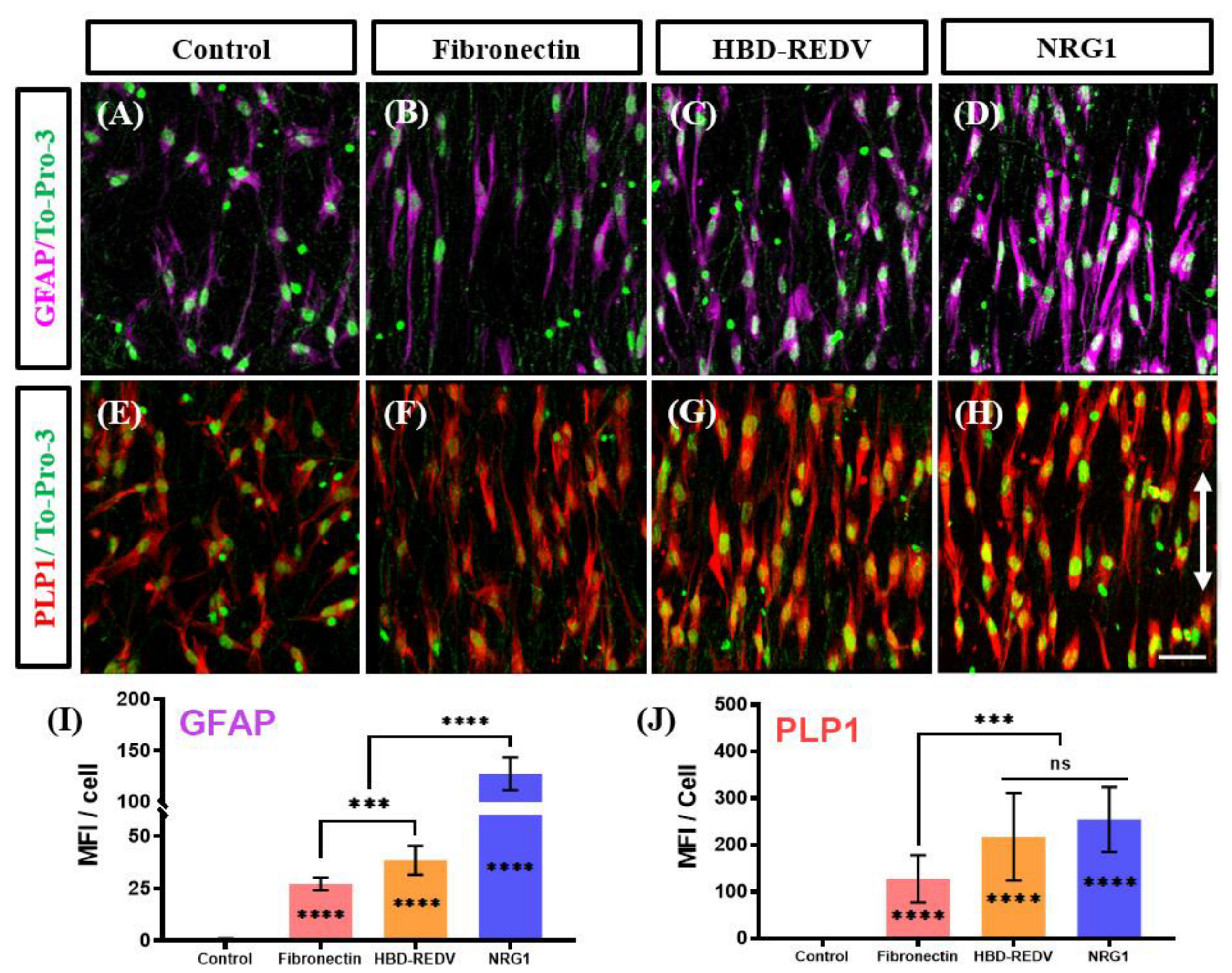
2.3. Effect of the Biological Cues on Differentiation of KC-NC Cells to SC Phenotype
3. Discussion
4. Materials and Methods
4.1. Materials
4.1.1. Development of Anisotropically Aligned PCL-PDA
4.1.2. Synthesis of Hep-Thiol
4.1.3. Surface Functionalization with Hep-Thiol and Conjugation of the Biological Cues
4.1.4. Fiber Characterization Measurements
4.1.5. Cloning and Production of the Recombinant Fusion Proteins
4.1.6. Release Kinetics of the Immobilized NRG1 from PCL Fiber Surface
4.2. Cell Culture
4.2.1. Epidermal Cell Isolation
4.2.2. Induction of KC into KC-NC
4.2.3. Proliferation and Differentiation of KC-NC
4.2.4. Immunocytochemistry
4.2.5. Fluorescence Microscopy and Image Analysis
4.2.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BSA | Bovine serum albumin |
| BDNF | Brain-derived neurotrophic factor |
| DI water | Deionized Water |
| DIV | Day in-vitro |
| DMEM | Dulbecco’s Modified Eagle Medium |
| DOPA | 3,4-dihydroxy-L-phenylalanine |
| DTPDH | Dithiobis (propanoic dihydrazide) |
| ECM | Extracellular matrix |
| EDC | Ethylcarbodiimide hydrochloride |
| EGF | Epidermal growth factor |
| FGF2 | Fibroblast growth factor 2 |
| FIB-SEM | Focused ion beam scanning electron microscopy |
| FTIR | Fourier transform infrared |
| GFAP | Glial fibrillary acidic protein |
| HBD | Heparin-binding domain |
| Hep | Heparin |
| HFIP | Hexafluoro-2-propanol |
| iPSC | Induced pluripotent stem cells |
| IPTG | isopropyl thiogalactopyranoside |
| KC | Keratinocyte |
| KCM | Keratinocyte growth medium |
| KC-NC | Keratinocyte-derived Neural Crest |
| KSFM | KC serum-free growth medium |
| MAG | Myelin-associated glycoprotein |
| MPG | Myelin protein zero |
| NCAM | Neural cell adhesion molecule |
| NC | Neural crest |
| NCSC | Neural crest stem cell |
| NRG1 | Neuregulin beta1 |
| OD | Optical density |
| p75 NGFR | p75 Nerve growth factor receptor |
| PCL | poly-ε-caprolactone |
| PDA | Polydopamine |
| PDGF-BB | Platelet-derived growth factor-BB |
| PGA | Polyglycolic acid |
| PLA | Polylactic acid |
| PLP1 | Proteolipid protein 1 |
| PMSF | Phenylmethanesulfonyl fluoride |
| PNI | Peripheral nerve injury |
| PNS | Peripheral nervous system |
| SC | Schwann cell |
| TCEP | Tris(2-carboxyethyl) phosphine |
| TCP | Tissue Culture Plate |
| TMS | Tetramethylsilane |
References
- Mohamed, M.A.; Fallahi, A.; El-Sokkary, A.M.; Salehi, S.; Akl, M.A.; Jafari, A.; Tamayol, A.; Fenniri, H.; Khademhosseini, A.; Andreadis, S.T.; et al. Stimuli-responsive hydrogels for manipulation of cell microenvironment: From chemistry to biofabrication technology. Prog. Polym. Sci. 2019, 98, 101147. [Google Scholar] [CrossRef]
- Brown, T.E.; Anseth, K.S. Spatiotemporal hydrogel biomaterials for regenerative medicine. Chem. Soc. Rev. 2017, 46, 6532–6552. [Google Scholar] [CrossRef] [PubMed]
- Doyle, A.D.; Carvajal, N.; Jin, A.; Matsumoto, K.; Yamada, K.M. Local 3D matrix microenvironment regulates cell migration through spatiotemporal dynamics of contractility-dependent adhesions. Nat. Commun. 2015, 6, 8720. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ma, P.X. Phase separation, pore structure, and properties of nanofibrous gelatin scaffolds. Biomaterials 2009, 30, 4094–4103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, X.; Yang, M.; Zhu, L. Silk fibroin/sodium alginate composite nano-fibrous scaffold prepared through thermally induced phase-separation (TIPS) method for biomedical applications. Mater. Sci. Eng. C 2015, 55, 8–13. [Google Scholar] [CrossRef]
- Stapelfeldt, K.; Stamboroski, S.; Mednikova, P.; Brüggemann, D. Fabrication of 3D-nanofibrous fibrinogen scaffolds using salt-induced self assembly. Biofabrication 2019, 11, 025010. [Google Scholar] [CrossRef]
- Sant, S.; Coutinho, D.F.; Gaharwar, A.K.; Neves, N.M.; Reis, R.L.; Gomes, M.E.; Khademhosseini, A. Self-Assembled Hydrogel Fiber Bundles from Oppositely Charged Polyelectrolytes Mimic Micro-/Nanoscale Hierarchy of Collagen. Adv. Funct. Mater. 2017, 27, 1606273. [Google Scholar] [CrossRef]
- Hartgerink, J.D.; Beniash, E.; Stupp, S.I. Self-Assembly and Mineralization of Peptide-Amphiphile Nanofibers. Science 2001, 294, 1684–1688. [Google Scholar] [CrossRef]
- Sun, X.; Lang, Q.; Zhang, H.; Cheng, L.; Zhang, Y.; Pan, G.; Zhao, X.; Yang, H.; Zhang, Y.; Santos, H.A.; et al. Electrospun Photocrosslinkable Hydrogel Fibrous Scaffolds for Rapid In Vivo Vascularized Skin Flap Regeneration. Adv. Funct. Mater. 2017, 27, 1604617. [Google Scholar] [CrossRef]
- Wu, J.; Hong, Y. Enhancing cell infiltration of electrospun fibrous scaffolds in tissue regeneration. Bioact. Mater. 2016, 1, 56–64. [Google Scholar] [CrossRef] [Green Version]
- Yeo, M.; Kim, G.H. Anisotropically Aligned Cell-Laden Nanofibrous Bundle Fabricated via Cell Electrospinning to Regenerate Skeletal Muscle Tissue. Small 2018, 14, e1803491. [Google Scholar] [CrossRef] [PubMed]
- Davidson, M.D.; Ban, E.; Schoonen, A.C.; Lee, M.H.; D’Este, M.; Shenoy, V.B.; Burdick, J.A. Mechanochemical Adhesion and Plasticity in Multifiber Hydrogel Networks. Adv. Mater. 2020, 32, 1905719. [Google Scholar] [CrossRef] [PubMed]
- Park, D.Y.; Mun, C.H.; Kang, E.; No, D.Y.; Ju, J.; Lee, S.H. One-stop microfiber spinning and fabrication of a fibrous cell-encapsulated scaffold on a single microfluidic platform. Biofabrication 2014, 6, 024108. [Google Scholar] [CrossRef]
- Qing, H.; Ji, Y.; Li, W.; Zhao, G.; Yang, Q.; Zhang, X.; Luo, Z.; Lu, T.J.; Jin, G.; Xu, F. Microfluidic Printing of Three-Dimensional Graphene Electroactive Microfibrous Scaffolds. ACS Appl. Mater. Interfaces 2019, 12, 2049–2058. [Google Scholar] [CrossRef] [PubMed]
- Sharifi, F.; Patel, B.B.; Dzuilko, A.K.; Montazami, R.; Sakaguchi, D.S.; Hashemi, N. Polycaprolactone Microfibrous Scaffolds to Navigate Neural Stem Cells. Biomacromolecules 2016, 17, 3287–3297. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhang, J.; Li, J.; Li, D.; Xiao, C.; Xiao, H.; Yang, H.; Zhuang, X.; Chen, X. Electrospun polymer biomaterials. Prog. Polym. Sci. 2019, 90, 1–34. [Google Scholar] [CrossRef]
- Megelski, S.; Stephens, J.S.; Chase, A.D.B.; Rabolt, J.F. Micro- and Nanostructured Surface Morphology on Electrospun Polymer Fibers. Macromolecules 2002, 35, 8456–8466. [Google Scholar] [CrossRef]
- Ramakrishna, S.; Fujihara, K.; Teo, W.-E.; Yong, T.; Ma, Z.; Ramaseshan, R. Electrospun nanofibers: Solving global issues. Mater. Today 2006, 9, 40–50. [Google Scholar] [CrossRef]
- Agarwal, S.; Wendorff, J.H.; Greiner, A. Progress in the Field of Electrospinning for Tissue Engineering Applications. Adv. Mater. 2009, 21, 3343–3351. [Google Scholar] [CrossRef]
- Nemati, S.; Kim, S.-J.; Shin, Y.M.; Shin, H. Current progress in application of polymeric nanofibers to tissue engineering. Nano Converg. 2019, 6, 36. [Google Scholar] [CrossRef] [Green Version]
- Schaub, N.J.; Johnson, C.D.; Cooper, B.; Gilbert, R.J. Electrospun Fibers for Spinal Cord Injury Research and Regeneration. J. Neurotrauma 2016, 33, 1405–1415. [Google Scholar] [CrossRef] [PubMed]
- Shirani, A.; Ganji, F.; Golmohammadi, M.; Hashemi, S.M.; Mozafari, M.; Amoabediny, G.; Osguei, N.K.; Samadikuchaksaraei, A. Cross-linked acellular lung for application in tissue engineering: Effects on biocompatibility, mechanical properties and immunological responses. Mater. Sci. Eng. C 2021, 122, 111938. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, R.; Palat, A.; Punnoose, A.M.; Joshi, S.; Ponraju, D.; Paul, S.F. Electrospun cellulose acetate phthalate nanofibrous scaffolds fabricated using novel solvent combinations biocompatible for primary chondrocytes and neurons. Tissue Cell 2016, 48, 634–643. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-H.; Tsai, W.-B. In situ UV-crosslinking gelatin electrospun fibers for tissue engineering applications. Biofabrication 2013, 5, 035008. [Google Scholar] [CrossRef]
- Madhavan, K.; Belchenko, D.; Motta, A.; Tan, W. Evaluation of composition and crosslinking effects on collagen-based composite constructs. Acta Biomater. 2010, 6, 1413–1422. [Google Scholar] [CrossRef]
- Hajzamani, D.; Shokrollahi, P.; Najmoddin, N.; Shokrolahi, F. Effect of engineered PLGA-gelatin-chitosan/PLGA-gelatin/PLGA-gelatin-graphene three-layer scaffold on adhesion/proliferation of HUVECs. Polym. Adv. Technol. 2020, 31, 1896–1910. [Google Scholar] [CrossRef]
- Dulnik, J.; Sajkiewicz, P. Crosslinking of Gelatin in Bicomponent Electrospun Fibers. Materials 2021, 14, 3391. [Google Scholar] [CrossRef]
- Shepherd, D.V.; Shepherd, J.H.; Ghose, S.; Kew, S.J.; Cameron, R.E.; Best, S.M. The process of EDC-NHS cross-linking of reconstituted collagen fibres increases collagen fibrillar order and alignment. APL Mater. 2015, 3, 014902. [Google Scholar] [CrossRef]
- Yang, X.; Wang, X.; Yu, F.; Ma, L.; Pan, X.; Luo, G.; Lin, S.; Mo, X.; He, C.; Wang, H. Hyaluronic acid/EDC/NHS-crosslinked green electrospun silk fibroin nanofibrous scaffolds for tissue engineering. RSC Adv. 2016, 6, 99720–99728. [Google Scholar] [CrossRef]
- Hajiabbas, M.; Alemzadeh, I.; Vossoughi, M. A porous hydrogel-electrospun composite scaffold made of oxidized alginate/gelatin/silk fibroin for tissue engineering application. Carbohydr. Polym. 2020, 245, 116465. [Google Scholar] [CrossRef]
- Martins, A.; Pinho, E.D.; Faria, S.; Pashkuleva, I.; Marques, A.; Reis, R.L.; Neves, N.M. Surface Modification of Electrospun Polycaprolactone Nanofiber Meshes by Plasma Treatment to Enhance Biological Performance. Small 2009, 5, 1195–1206. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, W.; Luo, B.; Chen, X.; Wen, W.; Zhou, C. Icariin immobilized electrospinning poly(l-lactide) fibrous membranes via polydopamine adhesive coating with enhanced cytocompatibility and osteogenic activity. Mater. Sci. Eng. C 2017, 79, 399–409. [Google Scholar] [CrossRef]
- Uehlin, A.F.; Vines, J.B.; Feldman, D.S.; Nyairo, E.; Dean, D.R.; Thomas, V. Uni-Directionally Oriented Fibro-Porous PLLA/Fibrin Bio-Hybrid Scaffold: Mechano-Morphological and Cell Studies. Pharmaceutics 2022, 14, 277. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Zhang, X.; Pi, Y.; Wang, X.; Jia, Z.; Zhu, J.; Dai, L.; Chen, W.; Yin, L.; Chen, H.; et al. Polycaprolactone electrospun mesh conjugated with an MSC affinity peptide for MSC homing in vivo. Biomaterials 2012, 33, 3375–3387. [Google Scholar] [CrossRef] [PubMed]
- Truong, Y.B.; Glattauer, V.; Briggs, K.L.; Zappe, S.; Ramshaw, J.A. Collagen-based layer-by-layer coating on electrospun polymer scaffolds. Biomaterials 2012, 33, 9198–9204. [Google Scholar] [CrossRef] [PubMed]
- Sauka-Spengler, T.; Bronner-Fraser, M. A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 2008, 9, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Achilleos, A.; Trainor, P.A. Neural crest stem cells: Discovery, properties and potential for therapy. Cell Res. 2012, 22, 288–304. [Google Scholar] [CrossRef]
- Arthur-Farraj, P.J.; Latouche, M.; Wilton, D.K.; Quintes, S.; Chabrol, E.; Banerjee, A.; Woodhoo, A.; Jenkins, B.; Rahman, M.; Turmaine, M.; et al. c-Jun Reprograms Schwann Cells of Injured Nerves to Generate a Repair Cell Essential for Regeneration. Neuron 2012, 75, 633–647. [Google Scholar] [CrossRef]
- Zhou, Y.; Notterpek, L. Promoting peripheral myelin repair. Exp. Neurol. 2016, 283, 573–580. [Google Scholar] [CrossRef]
- Sullivan, R.; Dailey, T.; Duncan, K.; Abel, N.; Borlongan, C.V. Peripheral Nerve Injury: Stem Cell Therapy and Peripheral Nerve Transfer. Int. J. Mol. Sci. 2016, 17, 2101. [Google Scholar] [CrossRef]
- Grinsell, D.; Keating, C.P. Peripheral Nerve Reconstruction after Injury: A Review of Clinical and Experimental Therapies. BioMed Res. Int. 2014, 2014, 698256. [Google Scholar] [CrossRef] [PubMed]
- Sakaue, M.; Sieber-Blum, M. Human epidermal neural crest stem cells as a source of Schwann cells. Development 2015, 142, 3188–3197. [Google Scholar]
- Ren, Y.-J.; Zhang, S.; Mi, R.; Liu, Q.; Zeng, X.; Rao, M.; Hoke, A.; Mao, H.-Q. Enhanced differentiation of human neural crest stem cells towards the Schwann cell lineage by aligned electrospun fiber matrix. Acta Biomater. 2013, 9, 7727–7736. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, P.; Tseropoulos, G.; Bronner, M.E.; Andreadis, S.T. Adult tissue-derived neural crest-like stem cells: Sources, regulatory networks, and translational potential. Stem Cells Transl. Med. 2020, 9, 328–341. [Google Scholar] [CrossRef]
- Cai, S.; Tsui, Y.-P.; Tam, K.-W.; Shea, G.K.-H.; Chang, R.S.-K.; Ao, Q.; Shum, D.K.-Y.; Chan, Y.-S. Directed Differentiation of Human Bone Marrow Stromal Cells to Fate-Committed Schwann Cells. Stem Cell Rep. 2017, 9, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Lu, F.; Han, J.; Tao, K.; Wang, H.; Simental, A.; Hu, D.; Yang, H. Efficient generation of functional Schwann cells from adipose-derived stem cells in defined conditions. Cell Cycle 2017, 16, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Spusta, S.C.; Mi, R.; Lassiter, R.N.; Stark, M.R.; Höke, A.; Rao, M.S.; Zeng, X. Human Neural Crest Stem Cells Derived from Human ESCs and Induced Pluripotent Stem Cells: Induction, Maintenance, and Differentiation into Functional Schwann Cells. STEM CELLS Transl. Med. 2012, 1, 266–278. [Google Scholar] [CrossRef]
- Lee, G.; Kim, H.; Elkabetz, Y.; Al Shamy, G.; Panagiotakos, G.; Barberi, T.; Tabar, V.; Studer, L. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat. Biotechnol. 2007, 25, 1468–1475. [Google Scholar] [CrossRef]
- Thoma, E.C.; Merkl, C.; Heckel, T.; Haab, R.; Knoflach, F.; Nowaczyk, C.; Flint, N.; Jagasia, R.; Zoffmann, S.J.; Truong, H.H.; et al. Chemical Conversion of Human Fibroblasts into Functional Schwann Cells. Stem Cell Rep. 2014, 3, 539–547. [Google Scholar] [CrossRef]
- Bajpai, V.K.; Kerosuo, L.; Tseropoulos, G.; Cummings, K.A.; Wang, X.; Lei, P.; Liu, B.; Liu, S.; Popescu, G.K.; Bronner, M.E.; et al. Reprogramming Postnatal Human Epidermal Keratinocytes Toward Functional Neural Crest Fates. Stem Cells 2017, 35, 1402–1415. [Google Scholar] [CrossRef]
- Boroujeni, S.M.; Koontz, A.; Tseropoulos, G.; Kerosuo, L.; Mehrotra, P.; Bajpai, V.K.; Selvam, S.R.; Lei, P.; Bronner, M.E.; Andreadis, S.T. Neural crest stem cells from human epidermis of aged donors maintain their multipotency in vitro and in vivo. Sci. Rep. 2019, 9, 9750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tseropoulos, G.; Moghadasi Boroujeni, S.; Bajpai, V.K.; Lei, P.; Andreadis, S.T. Derivation of neural crest stem cells from human epidermal keratinocytes requires FGF-2, IGF-1, and inhibition of TGF-β1. Bioeng. Transl. Med. 2018, 3, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-S.; Lee, J.; Lee, D.Y.; Kim, Y.-D.; Kim, J.Y.; Lim, H.J.; Lim, S.; Cho, Y.S. Schwann Cell Precursors from Human Pluripotent Stem Cells as a Potential Therapeutic Target for Myelin Repair. Stem Cell Rep. 2017, 8, 1714–1726. [Google Scholar] [CrossRef] [PubMed]
- Olwin, B.B. Heparin-binding growth factors and their receptors. Cytotechnology 1989, 2, 351–365. [Google Scholar] [CrossRef]
- Ribeiro-Resende, V.T.; Koenig, B.; Nichterwitz, S.; Oberhoffner, S.; Schlosshauer, B. Strategies for inducing the formation of bands of Büngner in peripheral nerve regeneration. Biomaterials 2009, 30, 5251–5259. [Google Scholar] [CrossRef]
- Chew, S.Y.; Mi, R.; Hoke, A.; Leong, K.W. The effect of the alignment of electrospun fibrous scaffolds on Schwann cell maturation. Biomaterials 2008, 29, 653–661. [Google Scholar] [CrossRef]
- Lee, G.; Chambers, S.M.; Tomishima, M.J.; Studer, L. Derivation of neural crest cells from human pluripotent stem cells. Nat. Protoc. 2010, 5, 688–701. [Google Scholar] [CrossRef]
- Puhl, D.L.; Funnell, J.L.; Nelson, D.W.; Gottipati, M.K.; Gilbert, R.J. Electrospun Fiber Scaffolds for Engineering Glial Cell Behavior to Promote Neural Regeneration. Bioengineering 2020, 8, 4. [Google Scholar] [CrossRef]
- Daud, M.F.; Pawar, K.C.; Claeyssens, F.; Ryan, A.J.; Haycock, J.W. An aligned 3D neuronal-glial co-culture model for peripheral nerve studies. Biomaterials 2012, 33, 5901–5913. [Google Scholar] [CrossRef]
- Lee, H.; Dellatore, S.M.; Miller, W.M.; Messersmith, P.B. Mussel-Inspired Surface Chemistry for Multifunctional Coatings. Science 2007, 318, 426–430. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Z.; Lu, W.W.; Zhen, W.; Yang, D.; Peng, S. Novel biomaterial strategies for controlled growth factor delivery for biomedical applications. NPG Asia Mater. 2017, 9, e435. [Google Scholar] [CrossRef]
- Smith, C.L.; Tallquist, M.D. PDGF function in diverse neural crest cell populations. Cell Adhes. Migr. 2010, 4, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Previtali, S.C.; Feltri, M.; Archelos, J.J.; Quattrini, A.; Wrabetz, L.; Hartung, H.-P. Role of integrins in the peripheral nervous system. Prog. Neurobiol. 2001, 64, 35–49. [Google Scholar] [CrossRef]
- Ruoslahti, E. Fibronectin and Its Receptors. Annu. Rev. Biochem. 1988, 57, 375–413. [Google Scholar] [CrossRef] [PubMed]
- Strachan, L.R.; Condic, M.L. Cranial neural crest recycle surface integrins in a substratum-dependent manner to promote rapid motility. J. Cell Biol. 2004, 167, 545–554. [Google Scholar] [CrossRef]
- Testaz, S.; Duband, J.-L. Central role of the α4β1 integrin in the coordination of avian truncal neural crest cell adhesion, migration, and survival. Dev. Dyn. 2001, 222, 127–140. [Google Scholar] [CrossRef]
- Coles, E.G.; Gammill, L.S.; Miner, J.H.; Bronner-Fraser, M. Abnormalities in neural crest cell migration in laminin α5 mutant mice. Dev. Biol. 2006, 289, 218–228. [Google Scholar] [CrossRef]
- Murphy, M.; Reid, K.; Ford, M.; Furness, J.B.; Bartlett, P. FGF2 regulates proliferation of neural crest cells, with subsequent neuronal differentiation regulated by LIF or related factors. Development 1994, 120, 3519–3528. [Google Scholar] [CrossRef]
- Birchmeier, C.; Nave, K.-A. Neuregulin-1, a key axonal signal that drives Schwann cell growth and differentiation. Glia 2008, 56, 1491–1497. [Google Scholar] [CrossRef]
- Nasiri, B.; Yi, T.; Wu, Y.; Smith, R.J.; Podder, A.K.; Breuer, C.K.; Andreadis, S.T. Monocyte Recruitment for Vascular Tissue Regeneration. Adv Healthc Mater. 2022; Accepted. [Google Scholar]



| Forward H2_BamHI | ATATGGATCCGCCATTCCTGCACCAACTGACCTGAAGTTCA |
| Reverse H2_HindIII | ATATAAGCTTATGTCCAATCAGGGGCTCGCTCTTCT |
| Forward Fragment-1 of HBD-RGD | TTGGACATAAGCTTGGCGGCGGAGGCAGTGGCCGTGGCGATTCCGGCGGCGGAGGCAGTGGCCGTGGCGATTCCGGCGGCGGAGGCAGTGGCCGTGG |
| Reverse Fragment-1 of HBD-RGD | TGGTGGTGCTCGAGATTATTAGGAATCGCCACGGCCACTGCCTCCGCCGCCGGAATCGCCACGGCCACTGCCTCCGCCGCCGGAATCG |
| Forward Fragment-2 of HBD-RGD | CGATTCCGGCGGCGGAGGCAGTGGCCGTGGCGATTCCGGCGGCGGAGGCAGTGGCCGTGGCGATTCCTAATAATCTCGAGCACCACCA |
| Reverse Fragment-2 of HBD-RGD | CCACGGCCACTGCCTCCGCCGCCGGAATCGCCACGGCCACTGCCTCCGCCGCCGGAATCGCCACGGCCACTGCCTCCGCCGCCAAGCTTATGTCCAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Podder, A.K.; Mohamed, M.A.; Tseropoulos, G.; Nasiri, B.; Andreadis, S.T. Engineering Nanofiber Scaffolds with Biomimetic Cues for Differentiation of Skin-Derived Neural Crest-like Stem Cells to Schwann Cells. Int. J. Mol. Sci. 2022, 23, 10834. https://doi.org/10.3390/ijms231810834
Podder AK, Mohamed MA, Tseropoulos G, Nasiri B, Andreadis ST. Engineering Nanofiber Scaffolds with Biomimetic Cues for Differentiation of Skin-Derived Neural Crest-like Stem Cells to Schwann Cells. International Journal of Molecular Sciences. 2022; 23(18):10834. https://doi.org/10.3390/ijms231810834
Chicago/Turabian StylePodder, Ashis Kumar, Mohamed Alaa Mohamed, Georgios Tseropoulos, Bita Nasiri, and Stelios T. Andreadis. 2022. "Engineering Nanofiber Scaffolds with Biomimetic Cues for Differentiation of Skin-Derived Neural Crest-like Stem Cells to Schwann Cells" International Journal of Molecular Sciences 23, no. 18: 10834. https://doi.org/10.3390/ijms231810834






