MDM2-Based Proteolysis-Targeting Chimeras (PROTACs): An Innovative Drug Strategy for Cancer Treatment
Abstract
:1. Introduction
1.1. Targeted Protein Degradation
1.2. PROteolysis-TArgeting Chimeras—PROTACs
- The activation of Ub by the ubiquitin-activating E1 enzyme, which forms the ubiquitin-adenylate intermediate. Subsequently, a thioester bond is generated between the active site cysteine residue of the E1 enzyme and the glycine residue at the C-terminus of Ub in an ATP-dependent process [19,21,22,24];
- A ligand that allows connecting to the POI;
- A ligand that allows binding to the E3 ligase;
- A spacer between the two ligands—linker.
2. Evolutionary Perspective of PROTACs
2.1. First Generation—Peptide-Based PROTACs (2001–2008)
2.2. Second Generation—Small Molecule-Based PROTACs
2.3. Third Generation—Spatiotemporal Controllable PROTACs
3. Discovery of PROTACs
3.1. Targets
3.2. E3 Ligase
4. PROTACs That Recruit MDM2 E3 Ligase
4.1. MDM2/p53 Cycle and Cancer
4.2. MDM2-Based PROTACs—Classification
- The target they degrade, for example, if it is a nuclear receptor or a transcription factor [18];
- The type of cancer they want to treat [18];
- Their origin, that is, if they are peptides with aa chains in their composition or if they use chemically-synthesized molecules, or a mixture of both, for example [18];
- The nature of the linker used to join the two ligands, which can be, among other factors, a polyethylene glycol (PEG) based linker or with aliphatic chains to establish the link [18];
- Their mechanism of action, considering whether they degrade a target different than the chosen E3 ligase or if, for example, the target is the E3 ligase itself (HOMO PROTACs) [65];
- Other features of MDM2-based PROTACS.
| Name | Year | Cancer Type | POI | POI Ligand | Linker | MDM2 Ligand | Results | [Ref] |
|---|---|---|---|---|---|---|---|---|
| MDM2-BASED PROTACs | ||||||||
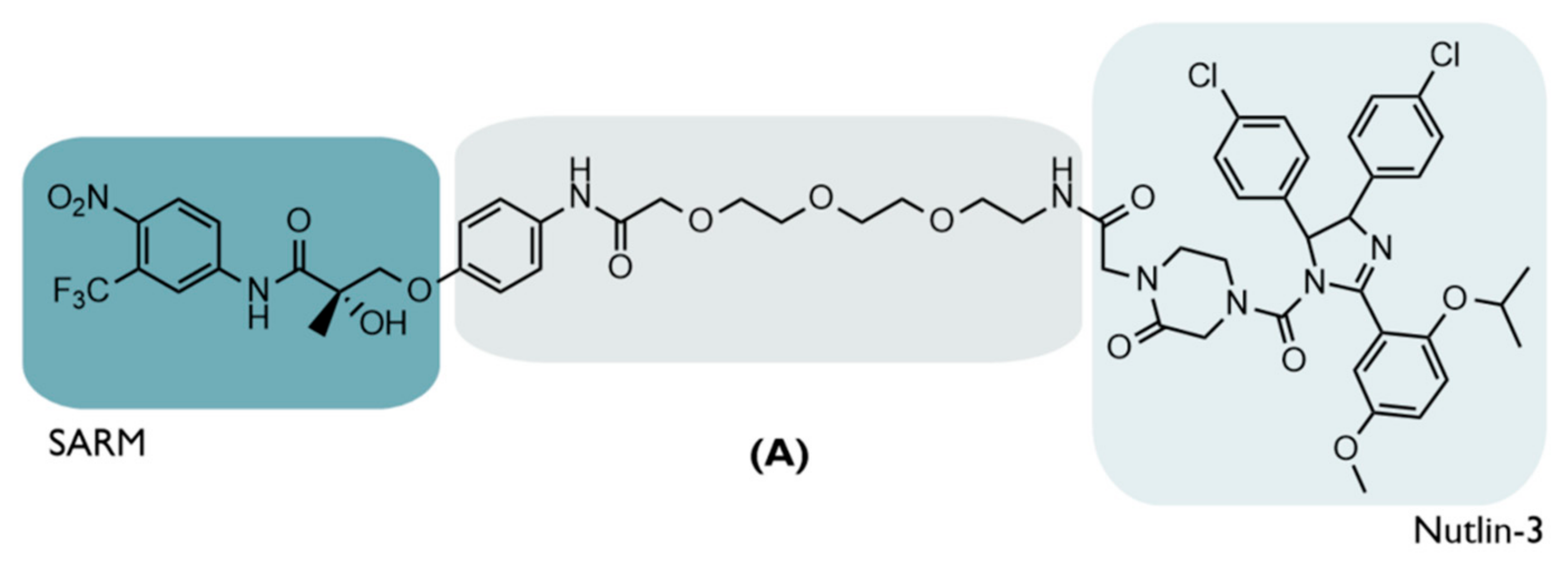
| A | 2008 | PC | AR | Selective Androgen Receptor Modulator (SARM)—hydroxyflutamide | PEG-based linker | Nutlin-3 | ↓AR Micromolar Potency | [20] |
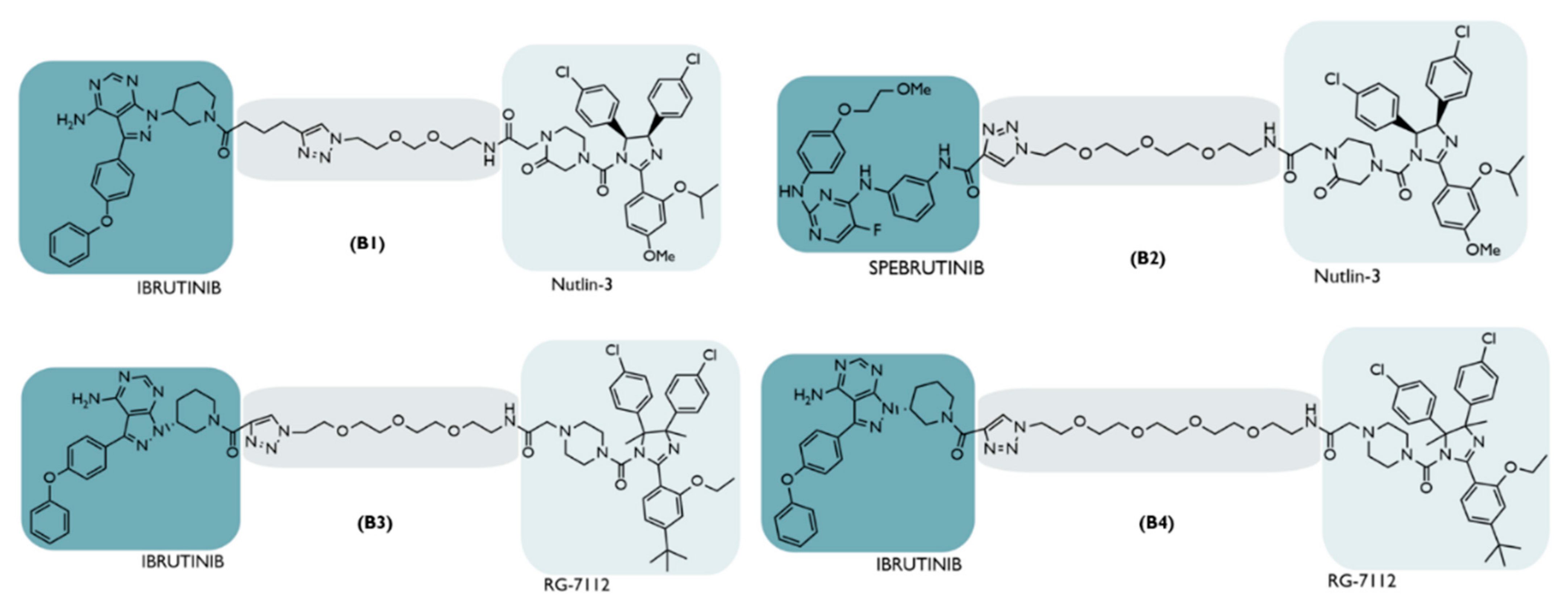
| B | 2018 | NHL | BTK | Inhibitor Ibrutinib/spebrutinib | Distinct types | Nutlin-3 or RG-7112 | No significant degradation | [66] |
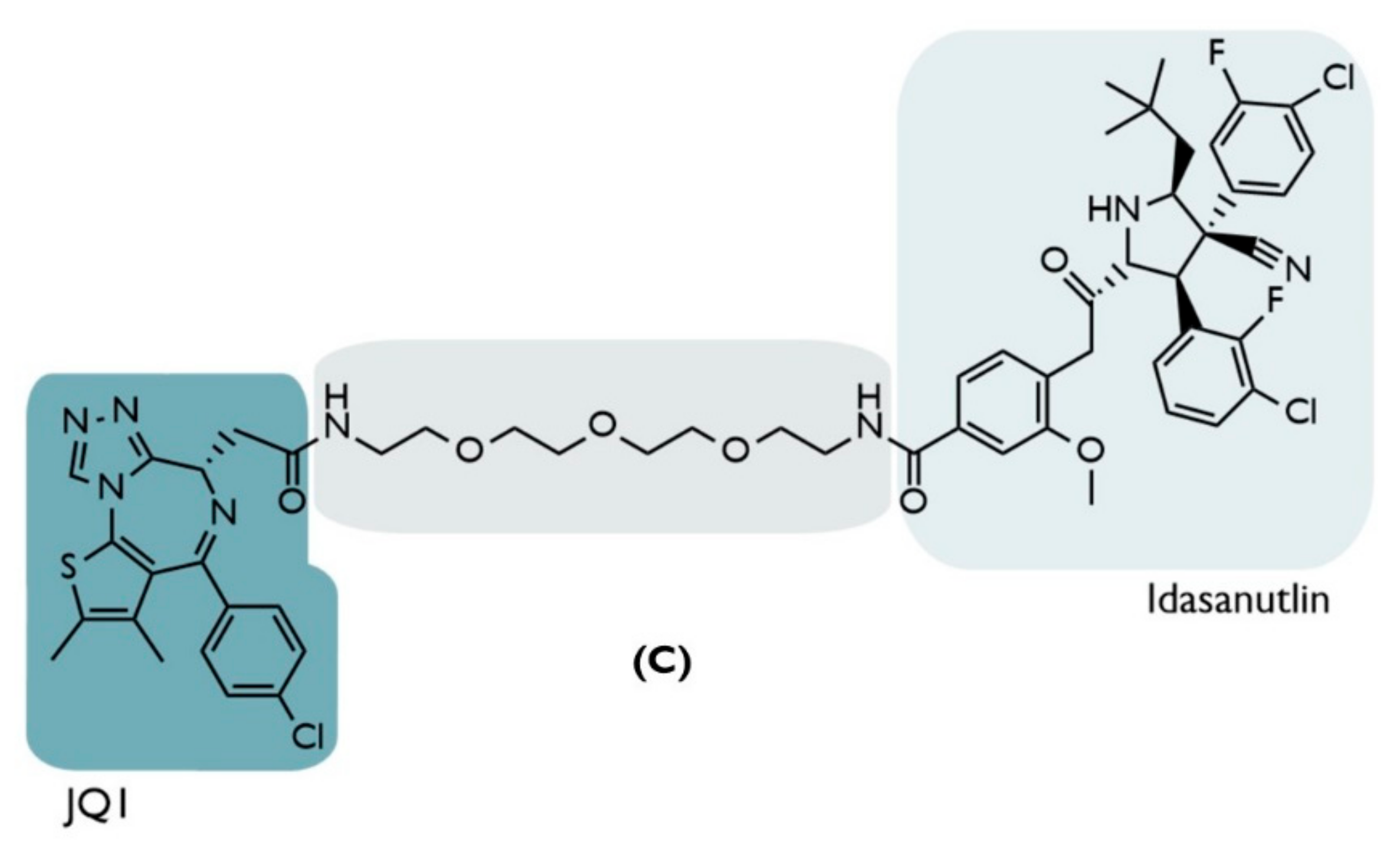
| C | 2019 | Hematologic and solid tumors | BRD4 | Inhibitor JQ1 | PEG-based linker | Idasanutlin | ↓BRD4 (Dmax = ~98%) Nanomolar Potency: (Cmax= 100 nmol/L) ↓c-Myc (~85%) ↑p53/p21 | [67] |
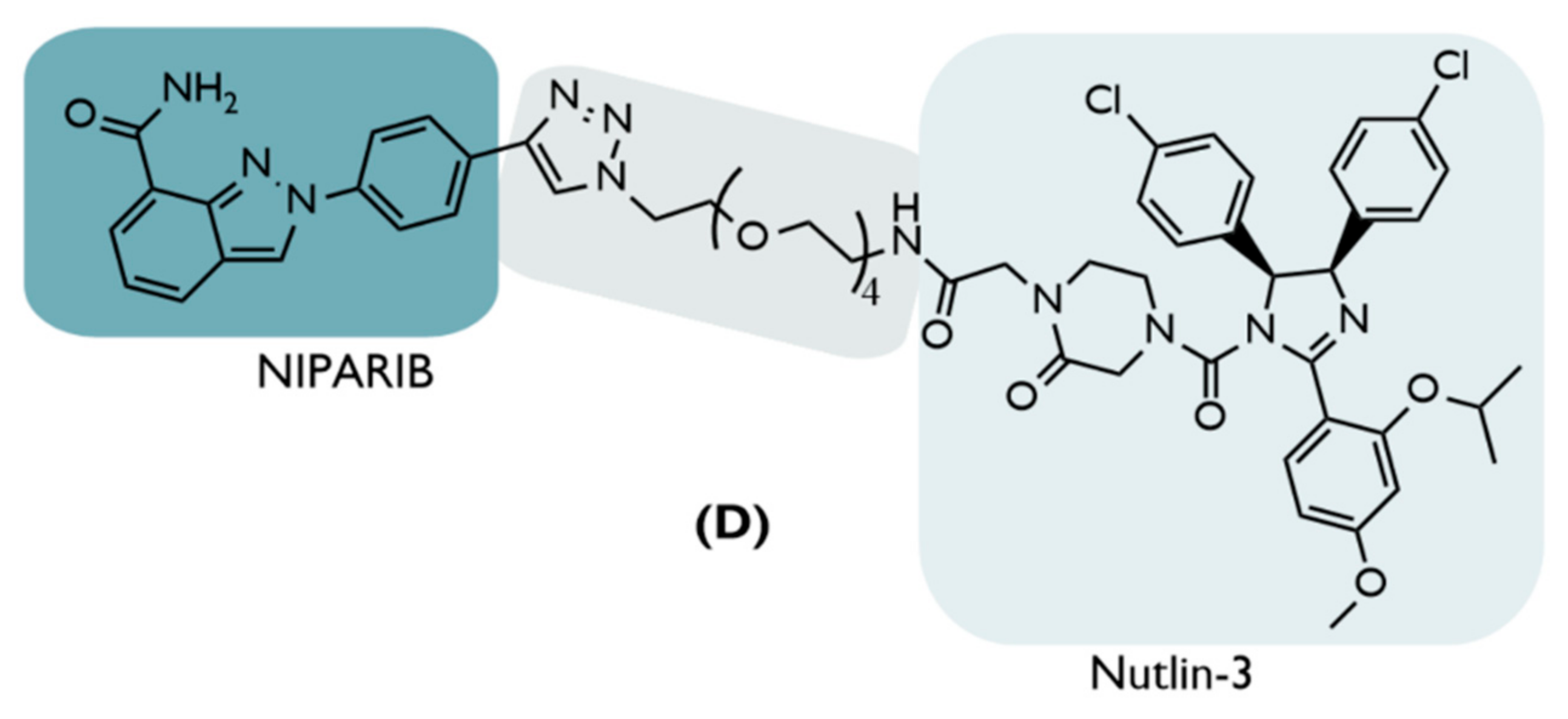
| D | 2019 | BC | PARP1 | Inhibitor Niraparib | PEG-based linkers | Nutlin-3 | ↓PARP1(52%,24 h) Inhib. cell growth (80–90%,48 h) Cellular selectivity | [68] |
| E | 2019 | BC | TrkC | Inhibitor Dasatinib | PEG-based linkers | Nutlin-3a | No Significant degradation | [69] |
| F | 2019 | BC | ERRα | Inhibitor XCT790 | Alkyl chains | Nutlin-3b | No significant degradation | [70] |
| G | 2019 | BC Lymphoma MM | CDK6 | Inhibitor Palbociclib | PEG-based linkers | Nutlin-3b | No significant degradation | [71] |
| H | 2022 | BC | HSP 90 | Inhibitor BIIB021 | Distinct types | Idasanutlin | No significant Cell growth inhibition | [72] |
| I1 | 2020 | NSCLC | EGFR mutant | Inhibitor XTF-262 | Alkyl chain (No carbonyl, 12C) | Idasanutlin | No Degradation (DC50 > 2000 nM) | [73] |
| I2 | Alkyl chain (12C) | ↓EGFRMUT Nanomolar potency: (H1975 cells: DC50 = 264 nM) Target Selectivity | [73] | |||||
| I3 | Alkyl chain (10C) | ↓EGFRMUT Nanomolar potency: (H1975 cells: DC50 = 77 nM) Target Selectivity | [73] | |||||
| I4 | Alkyl chain (8C) | No Degradation (DC50 > 2000 nM) | [73] | |||||
| I5 | Alkyl chain (6C) | No Degradation (DC50 > 2000 nM) | [73] | |||||
| HOMO-MDM2-BASED PROTACs | ||||||||
| J | 2021 | NSCLC | MDM2 | Derivative of Nutlin-3 | Distinct types | Derivative ofNutlin-3 | 11a: + potency ↓MDM2 (>95%, 24h) ↑p53 (DC50 = 1.0 µM) Enantiomer 11a-1: potent in vivo antitumor activity | [65] |
| PEPTIDIC-MDM2-BASED PROTACs | ||||||||
| PMIBCR/Abl-R6 | 2022 | Philadelphia chromosome-positive (Ph+) leukemia | Bcr/Abl | N-terminal helical region of the tetra-merizetion domain of Bcr/Abl | - | PMI—peptide inhibitor of the p53-MDM2 Interaction | ↓BCR/Abl ↑p53 OBS: C-terminal Arg-repeating hexapeptide | [74,75] |
4.3. The First MDM2-Based PROTAC
4.4. MDM2-Based PROTAC—Hematological and Colorectal Cancers
- The reduction of target protein levels without inducing protein up-regulation and impacting the signaling pathway. The first cell line used was the HCT116 colon cancer cells, given the importance of the BRD4/c-Myc signaling pathway in this type of tumor [67]. When subjected to increasing concentrations of PROTAC C over a 24 h period, it was possible to observe a reduction in the BRD4 levels in a dose-dependent way, with a maximum of around 100 nmol/L, which corresponds to a maximum degradation of 98% relative to the control levels, as well as a DC50 value of 32 nmol/L, lower than that obtained by previous MDM2-based PROTACs [67]. Under equivalent conditions, treatment with the JQ1 inhibitor alone did not lead to a decrease in BRD4 levels [67]. This could be attributed to the fact that high concentrations of JQ1 induce up-regulation of the target protein [67]. This was not detected with higher concentrations of PROTAC C, which is also an advantage even in relation to other types of PROTAC, in which the hook effect* is verified [67]. Furthermore, the present PROTAC has been shown to reduce the expression of c-Myc (85%), which is directly related to BRD4 levels; in a way, this is superior to the results achieved with JQ1 (70%) without genetically modifying the cell [67];
- Synergistic Effect—increase in p53 suppressor levels, with the inhibition of cell development. The treatment of HCT116 with PROTAC C demonstrated (in addition to degrading BRD4 and decreasing c-Myc expression) that it is capable of increasing p53 values by 5.9 times compared to the control level, activating the subsequent signaling pathway and resulting in reduced cell viability by about 97% [67]. Compared with the use of idasanutlin, the increase in p53 levels with this inhibitor was slightly higher but with no effect on the target protein, reducing cell viability by 62% [67]. With JQ1, p53 levels remained unchanged; yet, since it reduces the c-Myc pathway, it ends up achieving a 25% loss of cell viability [67]. In summary, only PROTAC C is able to combine the actions of the two inhibitors in a way that is superior to the sum of their isolated effects, allowing it to be used with a single compound to obtain an anti-proliferative synergism with nanomolar concentrations (>100 nmol/L) [67];
- The synergistic effect is dependent on cellular context. The same characteristics above were confirmed when melanoma cell lines (A375) were treated with PROTAC C, which caused a 98% decrease in cell viability, a value that is also greater than the sum of the isolated effects of JQ1 (15%), and idasanutlin (64%), showing a synergistic effect [67]. However, both cell lines studied have wild-type p53. When PROTAC C was evaluated in cell lines with mutations in the p53 gene (Daudi cells: hematological cancer cells; colon cancer cells: HT-29; p53-/- HCT116), it showed a decrease in its ability to reduce cell viability, which was going to be in the order of 20–30% [67]. In this type of cell, VHL-based PROTAC proved to be the best option [67].
- More difficulties in developing drug resistance. Given that PROTAC presents a bifunctional interaction with two distinct mechanisms of action (on the one hand, the degradation of BRD4 with the reduction of cell proliferation, on the other hand, the increase of the tumor suppressor p53), even if a mutation occurs in one of the proteins at which PROTAC binds, it ends up not compromising its entire therapeutic activity, which is an advantage relative to the inhibitors, in which all its activity is annulled [67].
4.5. MDM2-Based PROTAC—Breast Cancers
4.5.1. Poly (ADP-Ribose) Polymerase-1 (PARP1)
4.5.2. Tropomyosin Receptor Kinase C (TrkC)
4.5.3. Estrogen-Related Receptor α (ERRα):
4.5.4. Cyclin-Dependent Kinase 6 (CDK6)
4.5.5. Heat Shock Protein 90 (HSP90)

4.6. MDM2-Based PROTAC—Lung Cancer
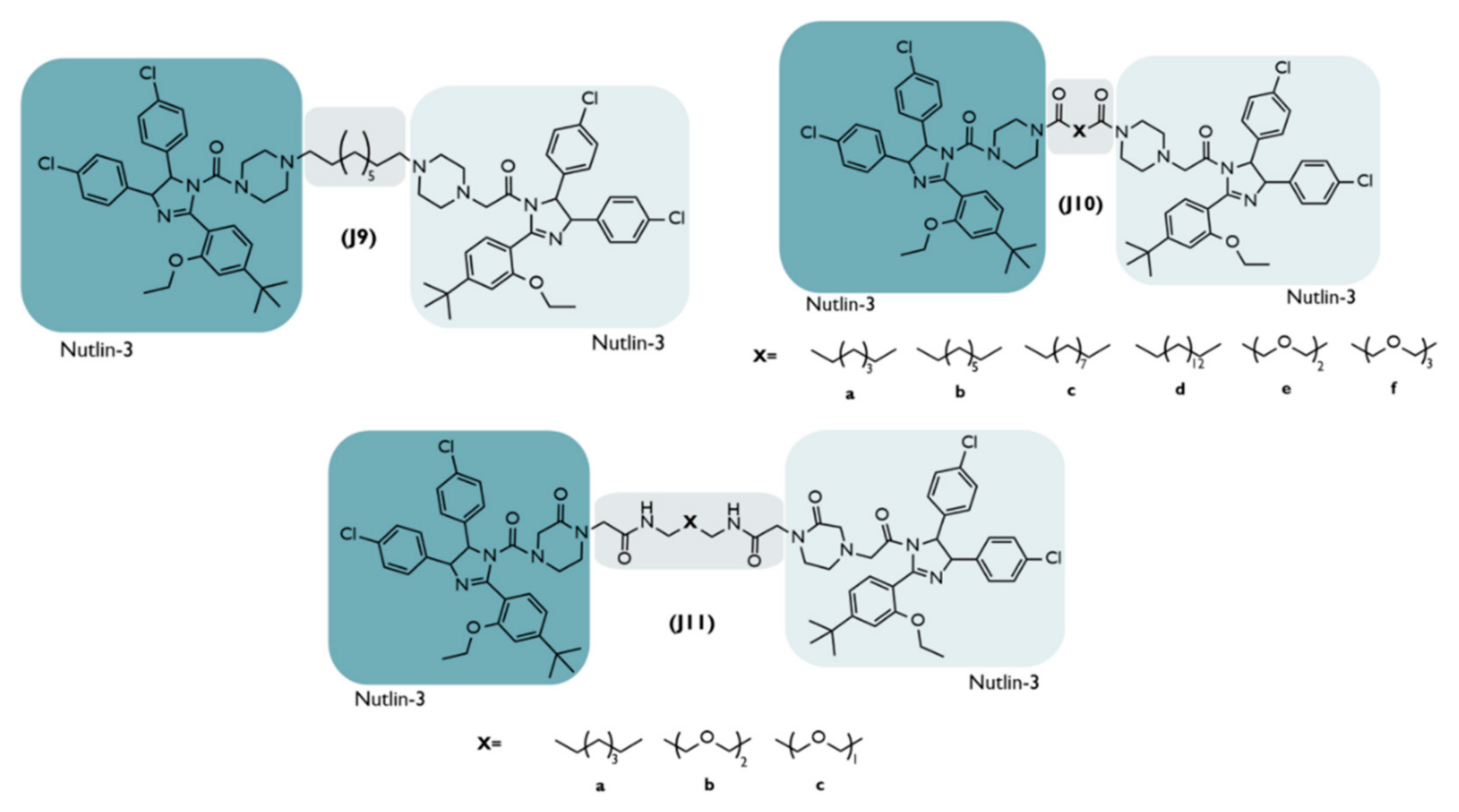
4.7. HOMO-MDM2-Based PROTACs
4.8. Peptidic-MDM2-Based PROTACs
5. Design of MDM2-Based PROTACs
6. Advantages of MDM2-Based PROTACs
- PROTACs can display cooperativity—binding to a second protein in PROTACs can lead to the formation of the ternary complex being favored (positive cooperativity), or it can lead to the formation of binary complexes (negative cooperativity) or simply remain not affected (neutral cooperativity). If positive cooperativity exists, a stable ternary complex can be formed even in the presence of low affinities with the POI [19,22];
- PROTACs are less susceptible to the development of resistance—the resistance me-chanism will have to encompass both of the pathways of the PROTAC, given that, in the absence of the degradation capacity, the inhibition of the target or MDM2 by itself may still have anticancer action [67].
7. Disadvantages of MDM2-Based PROTACs
8. Future Perspectives and Challenges
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| aa | Amino acid residue |
| ADP | Adenosine diphosphate |
| ALL | Acute lymphoblastic leukemia |
| APC | Antibody-PROTAC conjugate |
| AR | Androgen receptor |
| Arg | Arginine |
| BC | Breast cancer |
| BCR | B cell receptor |
| BET | Bromodomain and extra-terminal domain |
| BRD4 | Bromodomain-containing protein 4 |
| BTK | Bruton’s tyrosine kinase |
| CDK | Cyclin-dependent kinase complexes |
| CDK6 | Cyclin-dependent kinase 6 |
| CDK9 | Cyclin-dependent kinase 9 |
| CLIPTAC | In-cell click-formed Proteolysis Targeting Chimeras |
| CML | Chronic myeloid leukemia |
| CRBN | Cereblon |
| CRC | Colorectal cancer |
| DHT | Dihydroxytestosterone |
| DNA | Deoxyribonucleic acid |
| EGFR | Epidermal growth factor receptor |
| ERRα | Estrogen-related receptor α |
| FDA | Food and Drug Administration |
| HECT | Homologous to the E6AP carboxyl terminus |
| HER 2 | Human epidermal growth factor receptor 2 |
| HIF1α | Hypoxia-inducible factor-1α |
| HSP-90 | Heat shock protein 90 |
| IAP | Cellular inhibitor of apoptosis protein 1 |
| LK-Ph+ | Philadelphia chromosome positive chronic myeloid leukemia |
| mAb | Monoclonal antibodies |
| MCL1 | Induced myeloid leukemia cell differentiation protein |
| MDM2 | Mouse double minute 2 |
| MeBS | Bestatin |
| MetAP-2 | Methionine aminopeptidase-2 |
| MM | Multiple myeloma |
| mRNA | messenger RNA |
| Mut | Mutant |
| NAD+ | Nicotinamide-adenine dinucleotide |
| NHL | Non-Hodgkin’s lymphoma |
| NSCLC | Non-small cell lung cancer |
| PARP1 | Poly (ADP-ribose) polymerase-I |
| PARPs | Poly (ADP-ribose) polymerases enzyme |
| PC | Prostate cancer |
| PDP | Protein Degradation Probe |
| PEG | Polyethylene glycol |
| PIK3C3 | Phosphatidylinositol 3-Kinase Catalytic Subunit Type 3 |
| PKB | Protein kinase B |
| POI | Protein of interest |
| Poly-Ub | Polyubiquitin |
| PROTAC | PROteolysis-TArgeting Chimera |
| PROTAP | PROteolysis TArgeting Peptide |
| RARα | Retinoic acid receptor alpha |
| RBR | RING1-in-between-RING 2 |
| RING | Really Interesting New Gene |
| RNAi | RNA interference |
| Ro5 | Lipinski’s Rule of Five |
| SARM | Selective androgen receptor modulator |
| SMI | Small molecule inhibitors |
| SNIPPER | Specific and Non-genetic IAP-dependent Protein ERaseR |
| TNBC | Triple negative breast cancer |
| TPD | Targeted protein degradation |
| TrkC | Tropomyosin receptor kinase C |
| Ub | Ubiquitin |
| USP | Ubiquitin-proteasome system |
| VHL | Von Hippel-Lindau |
References
- National Cancer Institute. What is Cancer? 2021. Available online: https://www.cancer.gov/about-cancer/understanding/what-is-cancer (accessed on 1 July 2022).
- Basanta, D.; Anderson, A.R. Homeostasis Back and Forth: An Ecoevolutionary Perspective of Cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a028332. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- World Health Organisation (WHO) Media Centre. Cancer—Key Facts. 2011. Available online: http://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 1 July 2022).
- Cancer Research UK—What is Cancer? 2021. Available online: https://www.cancerresearchuk.org/about-cancer/what-is-cancer (accessed on 1 July 2022).
- World Health Organisation (WHO). Cancer. Available online: https://www.who.int/health-topics/cancer (accessed on 1 July 2022).
- Anifowose, A.; Agbowuro, A.A.; Yang, X.; Wang, B. Anticancer strategies by upregulating p53 through inhibition of its ubiquitination by MDM2. Med. Chem. Res. 2020, 29, 1105–1121. [Google Scholar] [CrossRef]
- Munisamy, M.; Mukherjee, N.; Thomas, L.; Pham, A.T.; Shakeri, A.; Zhao, Y.; Kolesar, J.; Rao, P.P.; Rangnekar, V.M.; Rao, M. Therapeutic opportunities in cancer therapy: Targeting the p53-MDM2/MDMX interactions. Am. J. Cancer Res. 2021, 11, 5762–5781. [Google Scholar]
- Dale, B.; Cheng, M.; Park, K.-S.; Kaniskan, H.; Xiong, Y.; Jin, J. Advancing targeted protein degradation for cancer therapy. Nat. Cancer 2021, 21, 638–654. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Crews, C.M. PROTACs: Past, present and future. Chem. Soc. Rev. 2022, 51, 5214–5236. [Google Scholar] [CrossRef]
- Cao, C.; He, M.; Wang, L.; He, Y.; Rao, Y. Chemistries of bifunctional PROTAC degraders. Chem. Soc. Rev. 2022, 51, 7066–7114. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ma, J.; Liu, Y.; Xia, J.; Li, Y.; Wang, Z.P.; Wei, W. PROTACs: A novel strategy for cancer therapy. Semin. Cancer Biol. 2020, 67, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.C.; Crews, C.M. Induced protein degradation: An emerging drug discovery paradigm. Nat. Rev. Drug Discov. 2017, 16, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Yan, C.; Gao, H.; Liu, T. Small-molecule PROTACs: Novel agents for cancer therapy. Futur. Med. Chem. 2020, 12, 915–938. [Google Scholar] [CrossRef]
- Békés, M.; Langley, D.R.; Crews, C.M. PROTAC targeted protein degraders: The past is prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef]
- Pettersson, M.; Crews, C.M. PROteolysis TArgeting Chimeras (PROTACs)—Past, present and future. Drug Discov. Today Technol. 2019, 31, 15–27. [Google Scholar] [CrossRef]
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559. [Google Scholar] [CrossRef]
- Zou, Y.; Ma, D.; Wang, Y. The PROTAC technology in drug development. Cell Biochem. Funct. 2019, 37, 21–30. [Google Scholar] [CrossRef]
- Konstantinidou, M.; Li, J.; Zhang, B.; Wang, Z.; Shaabani, S.; Ter Brake, F.; Essa, K.; Dömling, A. PROTACs– a game-changing technology. Expert Opin. Drug Discov. 2019, 14, 1255–1268. [Google Scholar] [CrossRef]
- Schneekloth, A.R.; Pucheault, M.; Tae, H.S.; Crews, C.M. Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorganic Med. Chem. Lett. 2008, 18, 5904–5908. [Google Scholar] [CrossRef]
- Kleiger, G.; Mayor, T. Perilous journey: A tour of the ubiquitin–proteasome system. Trends Cell Biol. 2014, 24, 352–359. [Google Scholar] [CrossRef]
- Nath, D.; Shadan, S. The ubiquitin system. Nature 2009, 458, 421. [Google Scholar] [CrossRef]
- Mansour, M.A. Ubiquitination: Friend and foe in cancer. Int. J. Biochem. Cell Biol. 2018, 101, 80–93. [Google Scholar] [CrossRef]
- Hu, Z.; Crews, C.M. Recent Developments in PROTAC-Mediated Protein Degradation: From Bench to Clinic. ChemBioChem 2022, 23, e202100270. [Google Scholar] [CrossRef]
- Tomoshige, S.; Ishikawa, M. In vivo synthetic chemistry of proteolysis targeting chimeras (PROTACs). Bioorganic Med. Chem. 2021, 41, 116221. [Google Scholar] [CrossRef]
- Nalepa, G.; Rolfe, M.; Harper, J.W. Drug discovery in the ubiquitin–proteasome system. Nat. Rev. Drug Discov. 2006, 5, 596–613. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Dong, R.; Zhang, J.-Y.; Zheng, X.; Sun, L.-P. PROTAC: A promising technology for cancer treatment. Eur. J. Med. Chem. 2020, 203, 112539. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.M.; Kim, K.B.; Verma, R.; Ransick, A.; Stein, B.; Crews, C.M.; Deshaies, R. Development of Protacs to Target Cancer-promoting Proteins for Ubiquitination and Degradation. Mol. Cell. Proteom. 2003, 2, 1350–1358. [Google Scholar] [CrossRef] [PubMed]
- Schneekloth, J.S., Jr.; Fonseca, F.N.; Koldobskiy, M.; Mandal, A.; Deshaies, R.; Sakamoto, K.; Crews, C.M. Chemical Genetic Control of Protein Levels: Selective in Vivo Targeted Degradation. J. Am. Chem. Soc. 2004, 126, 3748–3754. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Gonzalez, A.; Cyrus, K.; Salcius, M.; Kim, K.; Crews, C.M.; Deshaies, R.J.; Sakamoto, K.M. Targeting steroid hormone receptors for ubiquitination and degradation in breast and prostate cancer. Oncogene 2008, 27, 7201–7211. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Ishikawa, M.; Naito, M.; Hashimoto, Y. Protein Knockdown Using Methyl Bestatin−Ligand Hybrid Molecules: Design and Synthesis of Inducers of Ubiquitination-Mediated Degradation of Cellular Retinoic Acid-Binding Proteins. J. Am. Chem. Soc. 2010, 132, 5820–5826. [Google Scholar] [CrossRef]
- Okuhira, K.; Ohoka, N.; Sai, K.; Nishimaki-Mogami, T.; Itoh, Y.; Ishikawa, M.; Hashimoto, Y.; Naito, M. Specific degradation of CRABP-II via cIAP1-mediated ubiquitylation induced by hybrid molecules that crosslink cIAP1 and the target protein. FEBS Lett. 2011, 585, 1147–1152. [Google Scholar] [CrossRef]
- Okuhira, K.; Shoda, T.; Omura, R.; Ohoka, N.; Hattori, T.; Shibata, N.; Demizu, Y.; Sugihara, R.; Ichino, A.; Kawahara, H.; et al. Targeted Degradation of Proteins Localized in Subcellular Compartments by Hybrid Small Molecules. Mol. Pharmacol. 2016, 91, 159–166. [Google Scholar] [CrossRef]
- Ohoka, N.; Okuhira, K.; Ito, M.; Nagai, K.; Shibata, N.; Hattori, T.; Ujikawa, O.; Shimokawa, K.; Sano, O.; Koyama, R.; et al. In Vivo Knockdown of Pathogenic Proteins via Specific and Nongenetic Inhibitor of Apoptosis Protein (IAP)-dependent Protein Erasers (SNIPERs). J. Biol. Chem. 2017, 292, 4556–4570. [Google Scholar] [CrossRef] [PubMed]
- Buckley, D.L.; Gustafson, J.L.; Van Molle, I.; Roth, A.G.; Tae, H.S.; Gareiss, P.C.; Jorgensen, W.L.; Ciulli, A.; Crews, C.M. Small-Molecule Inhibitors of the Interaction between the E3 Ligase VHL and HIF1α. Angew. Chem. Int. Ed. 2012, 51, 11463–11467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, G.E.; Buckley, D.L.; Paulk, J.; Roberts, J.M.; Souza, A.; Dhe-Paganon, S.; Bradner, J.E. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 2015, 348, 1376–1381. [Google Scholar] [CrossRef] [PubMed]
- ARVINAS®. Androgen Receptor. Available online: https://www.arvinas.com/pipeline-programs/androgen-receptor (accessed on 3 June 2022).
- ARVINAS®. Estrogen Receptor. Available online: https://www.arvinas.com/pipeline-programs/estrogen-receptor (accessed on 3 June 2022).
- ARVINAS®. Pipeline Programs. Available online: https://www.arvinas.com/pipeline-programs/pipeline (accessed on 3 June 2022).
- Hines, J.; Gough, J.D.; Corson, T.W.; Crews, C.M. Posttranslational protein knockdown coupled to receptor tyrosine kinase activation with phosphoPROTACs. Proc. Natl. Acad. Sci. USA 2013, 110, 8942–8947. [Google Scholar] [CrossRef]
- Liu, J.; Chen, H.; Ma, L.; He, Z.; Wang, D.; Liu, Y.; Lin, Q.; Zhang, T.; Gray, N.; Kaniskan, H.Ü.; et al. Light-induced control of protein destruction by opto-PROTAC. Sci. Adv. 2020, 6, eaay5154. [Google Scholar] [CrossRef]
- Reynders, M.; Matsuura, B.S.; Bérouti, M.; Simoneschi, D.; Marzio, A.; Pagano, M.; Trauner, D. PHOTACs enable optical control of protein degradation. Sci. Adv. 2020, 6, eaay5064. [Google Scholar] [CrossRef]
- Pfaff, P.; Samarasinghe, K.T.G.; Crews, C.M.; Carreira, E.M. Reversible Spatiotemporal Control of Induced Protein Degradation by Bistable PhotoPROTACs. ACS Cent. Sci. 2019, 5, 1682–1690. [Google Scholar] [CrossRef]
- Lebraud, H.; Wright, D.J.; Johnson, C.N.; Heightman, T.D. Protein Degradation by In-Cell Self-Assembly of Proteolysis Targeting Chimeras. ACS Central Sci. 2016, 2, 927–934. [Google Scholar] [CrossRef]
- Maneiro, M.; Forte, N.; Shchepinova, M.M.; Kounde, C.S.; Chudasama, V.; Baker, J.R.; Tate, E.W. Antibody–PROTAC Conjugates Enable HER2-Dependent Targeted Protein Degradation of BRD. ACS Chem. Biol. 2020, 15, 1306–1312. [Google Scholar] [CrossRef]
- Itoh, Y.; Kitaguchi, R.; Ishikawa, M.; Naito, M.; Hashimoto, Y. Design, synthesis and biological evaluation of nuclear receptor-degradation inducers. Bioorganic Med. Chem. 2011, 19, 6768–6778. [Google Scholar] [CrossRef]
- You, I.; Erickson, E.C.; Donovan, K.A.; Eleuteri, N.A.; Fischer, E.S.; Gray, N.S.; Toker, A. Discovery of an AKT Degrader with Prolonged Inhibition of Downstream Signaling. Cell Chem. Biol. 2019, 27, 66–73.e7. [Google Scholar] [CrossRef]
- Shimokawa, K.; Shibata, N.; Sameshima, T.; Miyamoto, N.; Ujikawa, O.; Nara, H.; Ohoka, N.; Hattori, T.; Cho, N.; Naito, M. Targeting the Allosteric Site of Oncoprotein BCR-ABL as an Alternative Strategy for Effective Target Protein Degradation. ACS Med. Chem. Lett. 2017, 8, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Tinworth, C.P.; Lithgow, H.; Dittus, L.; Bassi, Z.I.; Hughes, S.E.; Muelbaier, M.; Dai, H.; Smith, I.E.D.; Kerr, W.J.; Burley, G.A.; et al. PROTAC-Mediated Degradation of Bruton’s Tyrosine Kinase Is Inhibited by Covalent Binding. ACS Chem. Biol. 2019, 14, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Steinebach, C.; Ng, Y.L.D.; Sosič, I.; Lee, C.-S.; Chen, S.; Lindner, S.; Vu, L.P.; Bricelj, A.; Haschemi, R.; Monschke, M.; et al. Systematic exploration of different E3 ubiquitin ligases: An approach towards potent and selective CDK6 degraders. Chem. Sci. 2020, 11, 3474–3486. [Google Scholar] [CrossRef]
- Cromm, P.M.; Crews, C.M. Targeted Protein Degradation: From Chemical Biology to Drug Discovery. Cell Chem. Biol. 2017, 24, 1181–1190. [Google Scholar] [CrossRef]
- Ryu, M.Y.; Cho, S.K.; Hong, Y.; Kim, J.; Kim, J.H.; Kim, G.M.; Chen, Y.-J.; Knoch, E.; Møller, B.L.; Kim, W.T.; et al. Classification of barley U-box E3 ligases and their expression patterns in response to drought and pathogen stresses. BMC Genom. 2019, 20, 326. [Google Scholar] [CrossRef]
- Yang, Q.; Zhao, J.; Chen, D.; Wang, Y. E3 ubiquitin ligases: Styles, structures and functions. Mol. Biomed. 2021, 2, 1–17. [Google Scholar] [CrossRef]
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef]
- Edmondson, S.D.; Yang, B.; Fallan, C. Proteolysis targeting chimeras (PROTACs) in ‘beyond rule-of-five’ chemical space: Recent progress and future challenges. Bioorganic Med. Chem. Lett. 2019, 29, 1555–1564. [Google Scholar] [CrossRef]
- Chen, J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb. Perspect. Med. 2016, 6, a026104. [Google Scholar] [CrossRef]
- Bieging, K.T.; Mello, S.S.; Attardi, L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 2014, 14, 359–370. [Google Scholar] [CrossRef]
- Koo, N.; Sharma, A.K.; Narayan, S. Therapeutics Targeting p53-MDM2 Interaction to Induce Cancer Cell Death. Int. J. Mol. Sci. 2022, 23, 5005. [Google Scholar] [CrossRef]
- DeLeo, A.B.; Jay, G.; Appella, E.; Dubois, G.C.; Law, L.W.; Old, L.J. Detection of a transformation-related antigen in chemically induced sarcomas and other transformed cells of the mouse. Proc. Natl. Acad. Sci. USA 1979, 76, 2420–2424. [Google Scholar] [CrossRef]
- Freedman, D.A.; Wu, L.; Levine, A.J. Functions of the MDM2 oncoprotein. Cell. Mol. Life Sci. CMLS 1999, 55, 96–107. [Google Scholar] [CrossRef]
- Karni-Schmidt, O.; Lokshin, M.; Prives, C. The Roles of MDM2 and MDMX in Cancer. Annu. Rev. Pathol. Mech. Dis. 2016, 11, 617–644. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.N.; Hancock, A.R.; Vogel, H.; Donehower, L.A.; Bradley, A. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc. Natl. Acad. Sci. USA 1998, 95, 15608–15612. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Jiménez, C.; Morafraile, E.C.; Alonso-Moreno, C.; Ocaña, A. Clinical considerations for the design of PROTACs in cancer. Mol. Cancer 2022, 21, 67. [Google Scholar] [CrossRef]
- He, S.; Ma, J.; Fang, Y.; Liu, Y.; Wu, S.; Dong, G.; Wang, W.; Sheng, C. Homo-PROTAC mediated suicide of MDM2 to treat non-small cell lung cancer. Acta Pharm. Sin. B 2020, 11, 1617–1628. [Google Scholar] [CrossRef]
- Sun, Y.; Zhao, X.; Ding, N.; Gao, H.; Wu, Y.; Yang, Y.; Zhao, M.; Hwang, J.; Song, Y.; Liu, W.; et al. PROTAC-induced BTK degradation as a novel therapy for mutated BTK C481S induced ibrutinib-resistant B-cell malignancies. Cell Res. 2018, 28, 779–781. [Google Scholar] [CrossRef]
- Hines, J.; Lartigue, S.; Dong, H.; Qian, Y.; Crews, C.M. MDM2-Recruiting PROTAC Offers Superior, Synergistic Antiproliferative Activity via Simultaneous Degradation of BRD4 and Stabilization of p53. Cancer Res. 2019, 79, 251–262. [Google Scholar] [CrossRef]
- Zhao, Q.; Lan, T.; Su, S.; Rao, Y. Induction of apoptosis in MDA-MB-231 breast cancer cells by a PARP1-targeting PROTAC small molecule. Chem. Commun. 2019, 55, 369–372. [Google Scholar] [CrossRef]
- Zhao, B.; Burgess, K. TrkC-Targeted Kinase Inhibitors and PROTACs. Mol. Pharm. 2019, 16, 4313–4318. [Google Scholar] [CrossRef]
- Peng, L.; Zhang, Z.; Lei, C.; Li, S.; Zhang, Z.; Ren, X.; Chang, Y.; Zhang, Y.; Xu, Y.; Ding, K. Identification of New Small-Molecule Inducers of Estrogen-related Receptor α (ERRα) Degradation. ACS Med. Chem. Lett. 2019, 10, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Yang, Z.; Gao, H.; Yang, H.; Zhu, S.; An, Z.; Wang, J.; Li, Q.; Chandarlapaty, S.; Deng, H.; et al. Potent and Preferential Degradation of CDK6 via Proteolysis Targeting Chimera Degraders. J. Med. Chem. 2019, 62, 7575–7582. [Google Scholar] [CrossRef]
- Liu, Q.; Tu, G.; Hu, Y.; Jiang, Q.; Liu, J.; Lin, S.; Yu, Z.; Li, G.; Wu, X.; Tang, Y.; et al. Discovery of BP3 as an efficacious proteolysis targeting chimera (PROTAC) degrader of HSP90 for treating breast cancer. Eur. J. Med. Chem. 2022, 228, 114013. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, F.; Tong, L.; Zhang, T.; Xie, H.; Lu, X.; Ren, X.; Ding, K. Design and synthesis of selective degraders of EGFRL858R/T790M mutant. Eur. J. Med. Chem. 2020, 192, 112199. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Niu, F.; Qu, X.; He, W.; Feng, C.; Wang, S.; Ouyang, Z.; Yan, J.; Wen, Y.; Xu, D.; et al. A tetrameric protein scaffold as a nano-carrier of antitumor peptides for cancer therapy. Biomaterials 2019, 204, 1–12. [Google Scholar] [CrossRef]
- Ma, B.; Feng, H.; Feng, C.; Liu, Y.; Zhang, H.; Wang, J.; Wang, W.; He, P.; Niu, F. Kill Two Birds with One Stone: A Multifunctional Dual-Targeting Protein Drug to Overcome Imatinib Resistance in Philadelphia Chromosome-Positive Leukemia. Adv. Sci. 2022, 9, 2104850. [Google Scholar] [CrossRef]
- Jamroze, A.; Chatta, G.; Tang, D.G. Androgen receptor (AR) heterogeneity in prostate cancer and therapy resistance. Cancer Lett. 2021, 518, 1–9. [Google Scholar] [CrossRef]
- Singh, S.P.; Dammeijer, F.; Hendriks, R.W. Role of Bruton’s tyrosine kinase in B cells and malignancies. Mol. Cancer 2018, 17, 57. [Google Scholar] [CrossRef]
- Qin, A.-C.; Jin, H.; Song, Y.; Gao, Y.; Chen, Y.-F.; Zhou, L.-N.; Wang, S.-S.; Lu, X.-S. The therapeutic effect of the BRD4-degrading PROTAC A1874 in human colon cancer cells. Cell Death Dis. 2020, 11, 805. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Tian, J.; Wu, T. BRD4 in physiology and pathology: ‘‘BET’’ on its partners. BioEssays News Rev. Mol. Cell. Dev. Biol. 2021, 43, 2100180. [Google Scholar] [CrossRef] [PubMed]
- Chaidos, A.; Caputo, V.; Karadimitris, A. Inhibition of bromodomain and extra-terminal proteins (BET) as a potential therapeutic approach in haematological malignancies: Emerging preclinical and clinical evidence. Ther. Adv. Hematol. 2015, 6, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Zhang, Z.; Liu, J.-J.; Jiang, N.; Zhang, J.; Ross, T.M.; Chu, X.-J.; Bartkovitz, D.; Podlaski, F.; Janson, C.; et al. Discovery of RG7388, a Potent and Selective p53–MDM2 Inhibitor in Clinical Development. J. Med. Chem. 2013, 56, 5979–5983. [Google Scholar] [CrossRef]
- U.S. National Library of Medicine. Search of: RG7388 OR RO-5503781—List Results—ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/results?intr=RG7388+OR+RO5503781&Search=Apply&age_v=&gndr=&type=&rslt= (accessed on 17 June 2022).
- Montesinos, P.; Beckermann, B.M.; Catalani, O.; Esteve, J.; Gamel, K.; Konopleva, M.Y.; Martinelli, G.; Monnet, A.; Papayannidis, C.; Park, A.; et al. MIRROS: A randomized, placebo-controlled, Phase III trial of cytarabine ± idasanutlin in relapsed or refractory acute myeloid leukemia. Futur. Oncol. 2020, 16, 807–815. [Google Scholar] [CrossRef]
- World Health Organization. Colorectal Cancer Awareness Month 2022—IARC. Available online: https://www.iarc.who.int/featured-news/colorectal-cancer-awareness-month-2022/ (accessed on 18 June 2022).
- Spiegel, J.O.; Van Houten, B.; Durrant, J.D. PARP1: Structural insights and pharmacological targets for inhibition. DNA Repair 2021, 103, 103125. [Google Scholar] [CrossRef]
- Li, X.; Pu, W.; Zheng, Q.; Ai, M.; Chen, S.; Peng, Y. Proteolysis-targeting chimeras (PROTACs) in cancer therapy. Mol. Cancer 2022, 21, 99. [Google Scholar] [CrossRef]
- An, S.; Fu, L. Small-molecule PROTACs: An emerging and promising approach for the development of targeted therapy drugs. eBioMedicine 2018, 36, 553–562. [Google Scholar] [CrossRef] [Green Version]






| Process | Mechanism | [Ref] |
|---|---|---|
| Damaged DNA | Genes, such as PUMA and BTG2, responsible for DNA repair, apoptosis, cell-cycle arrest and senescence, are transcribed with p53 activation. | [56] |
| Metabolism | Metabolic reprogramming is prevented by p53. | [57] |
| Autophagy | The p53, through the activation of genes such as ATG10, can promote the degradation of damaged cell organelles, inhibiting the process of tumor formation. | [57] |
| Domain | Mechanism | [Ref] |
|---|---|---|
| N-terminal p53 Binding domain | Interacts with p53. | [58] |
| Nuclear localization signal | Transport of MDM2 from the cytoplasm to the nucleus. | [58] |
| Nuclear export signal | Transport of MDM2 from the nucleus to the cytoplasm. | [58] |
| Acidic domain | It induces p53 degradation by being phosphorylated. | [58] |
| Zinc-finger domain | Regulation of p53 levels: interacts with ribosome proteins that bind to the acidic domain and inhibit p53 degradation; it induces suppressor ubiquitination after MDM2-p53 binding. | [58] |
| C-terminal RING-Finger domain | It induces suppressor ubiquitination after MDM2-p53 binding. | [58] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vicente, A.T.S.; Salvador, J.A.R. MDM2-Based Proteolysis-Targeting Chimeras (PROTACs): An Innovative Drug Strategy for Cancer Treatment. Int. J. Mol. Sci. 2022, 23, 11068. https://doi.org/10.3390/ijms231911068
Vicente ATS, Salvador JAR. MDM2-Based Proteolysis-Targeting Chimeras (PROTACs): An Innovative Drug Strategy for Cancer Treatment. International Journal of Molecular Sciences. 2022; 23(19):11068. https://doi.org/10.3390/ijms231911068
Chicago/Turabian StyleVicente, André T. S., and Jorge A. R. Salvador. 2022. "MDM2-Based Proteolysis-Targeting Chimeras (PROTACs): An Innovative Drug Strategy for Cancer Treatment" International Journal of Molecular Sciences 23, no. 19: 11068. https://doi.org/10.3390/ijms231911068









