Photosynthetic Efficiency and Glyco-Metabolism Changes in Artificial Triploid Loquats Contribute to Heterosis Manifestation
Abstract
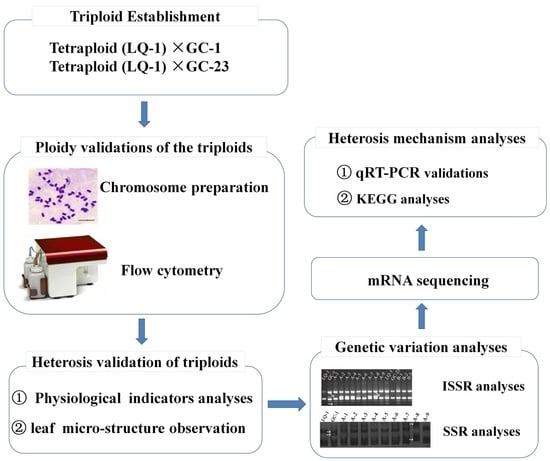
:1. Introduction
2. Results
2.1. Ploidy Validation of the Hybrids Obtained with the Two Cross Combinations
2.2. Leaves of Triploid Loquat Exhibited Significant Heterosis Compared with Diploid and Tetraploid Parents
2.3. Screening Suitable ISSR and SSR Markers for the Two Cross Lines
2.4. Polymorphism Analysis of ISSR and SSR Markers
2.5. Genetic Variation Pattern Analyses of Triploid Loquats
2.6. Genetic Diversity Analysis
2.7. Correlation Analysis of ISSR and SSR Markers Using Mantel Test
2.8. Transcriptome Analyses of the Parental and Triploid Leaf Tissues
2.9. qRT-PCR Validation of Carbohydrate- and Protein-Related DEGs
3. Discussion
3.1. ISSR and SSR Markers Were Two Effective Molecular Markers for Loquat Breeding
3.2. Hybridization Was an Effective Way for Triploid Loquat Breeding
3.3. The Formation of Triploid Loquats Was Accompanied by Extensive Genomic Variation
3.4. Alterations in Photosynthetic Efficiency and Glyco-Metabolism Resulted in Heterosis Manifestation in Triploid Loquats
4. Materials and Methods
4.1. Plant Lines
4.2. Ploidy Validation
4.3. DNA Extraction and Examination
4.4. ISSR and SSR Primers
4.5. PCR and Electrophoreses
4.6. MRNA-Seq Analyses of the Leaf Tissues in Different Ploidy Loquats
4.7. Quantitative Real-Time PCR Validation
4.8. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kolar, F. Tracing evolutionary history and admixture in mixed-ploidy systems. Mol. Ecol. Resour. 2021, 21, 1413–1415. [Google Scholar] [CrossRef]
- Adams, K.L.; Wendel, J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005, 8, 135–141. [Google Scholar] [CrossRef]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef]
- Leitch, I.J.; Bennett, M. Polyploidy in angiosperms. Trends Plant Sci. 1997, 2, 470–476. [Google Scholar] [CrossRef]
- Stebbins, G.L. Chromosomal Evolution in Higher Plants; Edward Arnold: London, UK, 1971. [Google Scholar]
- Masterson, J. Stomatal size in fossil plants: Evidence for polyploidy in majority of angiosperms. Science 1994, 264, 421–424. [Google Scholar] [CrossRef]
- Wang, Q.; Yan, T.; Long, Z.; Huang, L.Y.; Zhu, Y.; Xu, Y.; Chen, X.; Pak, H.; Li, J.; Wu, D.; et al. Prediction of heterosis in the recent rapeseed (Brassica napus) polyploid by pairing parental nucleotide sequences. PLoS Genet. 2021, 17, e1009879. [Google Scholar] [CrossRef]
- Carlson, C.H.; Choi, Y.; Chan, A.P.; Town, C.D.; Smart, L.B. Nonadditive gene expression is correlated with nonadditive phenotypic expression in interspecific triploid hybrids of willow (Salix spp.). G3 2022, 12, jkab436. [Google Scholar] [CrossRef]
- Madlung, A. Polyploidy and its effect on evolutionary success: Old questions revisited with new tools. Heredity 2013, 110, 99–104. [Google Scholar] [CrossRef]
- Leal-Bertioli, S.C.M.; Nascimento, E.; Chavarro, M.C.F.; Custodio, A.R.; Hopkins, M.S.; Moretzsohn, M.C.; Bertioli, D.J.; Araujo, A.C.G. Spontaneous generation of diversity in Arachis neopolyploids (Arachis ipaensis x Arachis duranensis)4x replays the early stages of peanut evolution. G3 2021, 11, jkab289. [Google Scholar] [CrossRef]
- Fort, A.; Ryder, P.; McKeown, P.C.; Wijnen, C.; Aarts, M.G.; Sulpice, R.; Spillane, C. Disaggregating polyploidy, parental genome dosage and hybridity contributions to heterosis in Arabidopsis thaliana. New Phytol. 2016, 209, 590–599. [Google Scholar] [CrossRef] [Green Version]
- Levin, D.A. The role of chromosomal change in plant evolution. Bioessays 2003, 29, 460–461. [Google Scholar]
- Vitali, V.; Rothering, R.; Catania, F. Fifty Generations of Amitosis: Tracing Asymmetric Allele Segregation in Polyploid Cells with Single-Cell DNA Sequencing. Microorganisms 2021, 9, 1979. [Google Scholar] [CrossRef]
- Messing, J.; Bharti, A.K.; Karlowski, W.M.; Gundlach, H.; Kim, H.R.; Yu, Y.; Wei, F.; Fuks, G.; Soderlund, C.A.; Mayer, K.F.; et al. Sequence composition and genome organization of maize. Proc. Natl. Acad. Sci. USA 2004, 101, 14349–14354. [Google Scholar] [CrossRef]
- Song, K.; Lu, P.; Tang, K.; Osborn, T.C. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 1995, 92, 7719–7723. [Google Scholar] [CrossRef]
- Tate, J.A.; Ni, Z.; Scheen, A.C.; Koh, J.; Gilbert, C.A.; Lefkowitz, D.; Chen, Z.J.; Soltis, P.S.; Soltis, D.E. Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid. Genetics 2006, 173, 1599–1611. [Google Scholar] [CrossRef]
- Darwin, C.R. The Effects of Cross- and Self-Fertilization in the Vegetable Kingdom; John Murray: London, UK, 1876. [Google Scholar]
- Xiao, Y.; Jiang, S.; Cheng, Q.; Wang, X.; Yan, J.; Zhang, R.; Qiao, F.; Ma, C.; Luo, J.; Li, W.; et al. The genetic mechanism of heterosis utilization in maize improvement. Genome Biol. 2021, 22, 148. [Google Scholar] [CrossRef]
- Lin, T.; Zhou, C.; Chen, G.; Yu, J.; Wu, W.; Ge, Y.; Liu, X.; Li, J.; Jiang, X.; Tang, W.; et al. Heterosis-associated genes confer high yield in super hybrid rice. Theor. Appl. Genet. 2020, 133, 3287–3297. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, M.; Chen, A.; Hong, H.; Li, D.; Li, J.; Zhao, D.; Wang, W.; Wang, X.; Qiu, L. GmMs1 encodes a kinesin-like protein essential for male fertility in soybean (Glycine max L.). J. Integr. Plant Biol. 2021, 63, 1054–1064. [Google Scholar] [CrossRef]
- Zhang, Q. Utilization of crop heterosis: A review. Euphytica 2014, 197, 161–173. [Google Scholar]
- Jones, D.F. Dominance of Linked Factors as a Means of Accounting for Heterosis. Genetics 1917, 2, 466–479. [Google Scholar] [CrossRef]
- East, E.M. Heterosis. Genetics 1936, 21, 375–397. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.B.; Li, J.X.; Xu, C.G.; Tan, Y.F.; Gao, Y.J.; Li, X.H.; Zhang, Q.; Saghai Maroof, M.A. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA 1997, 94, 9226–9231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.J. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010, 15, 57–71. [Google Scholar] [CrossRef]
- Li, X.; Wei, Y.; Nettleton, D.; Brummer, E.C. Comparative gene expression profiles between heterotic and non-heterotic hybrids of tetraploid Medicago sativa. BMC Plant Biol. 2009, 9, 107. [Google Scholar] [CrossRef]
- Thiemann, A.; Fu, J.; Schrag, T.A.; Melchinger, A.E.; Frisch, M.; Scholten, S. Correlation between parental transcriptome and field data for the characterization of heterosis in Zea mays L. Theor. Appl. Genet. 2010, 120, 401–413. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Chen, Z.J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 2007, 58, 377–406. [Google Scholar] [CrossRef]
- Carrera, L.; Sanzol, J.; Soler, E.; Herrero, M.; Hormaza, J.I. Molecular S-genotyping and determination of S-RNase-based incompatibility groups in loquat [Eriobotrya japonica (Thunb.) Lindl.]. Euphytica 2011, 181, 267–275. [Google Scholar] [CrossRef]
- Kahramanoglu, I. Preserving postharvest storage quality of fresh loquat fruits by using different bio-materials. J. Food Sci. Technol. 2020, 57, 3004–3012. [Google Scholar] [CrossRef]
- Wu, D.; Fan, W.G.; He, Q.; Guo, Q.G.; Spano, A.J.; Wang, Y.; Timko, M.P.; Liang, G.L. Genetic Diversity of Loquat [Eriobotrya japonica (Thunb.) Lindl.] Native to Guizhou Province (China) and Its Potential in the Genetic Improvement of Domesticated Cultivars. Plant Mol. Biol. Rep. 2015, 33, 952–961. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, H.; Shi, L.; Li, Y.; Tuerhong, M.; Abudukeremu, M.; Cui, J.; Li, Y.; Jin, D.Q.; Xu, J.; et al. Structure features, selenylation modification, and improved anti-tumor activity of a polysaccharide from Eriobotrya japonica. Carbohydr. Polym. 2021, 273, 118496. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Sonam, T.; Shimizu, K. The Potential of Triterpenoids from Loquat Leaves (Eriobotrya japonica) for Prevention and Treatment of Skin Disorder. Int. J. Mol. Sci. 2017, 18, 1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikuchi, S.; Iwasuna, M.; Kobori, A.; Tsutaki, Y.; Yoshida, A.; Murota, Y.; Nishino, E.; Sassa, H.; Koba, T. Seed formation in triploid loquat (Eriobotrya japonica) through cross-hybridization with pollen of diploid cultivars. Breed Sci. 2014, 64, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Huang, R.; Wang, L.; Liang, G. Functional Identification of EjGIF1 in Arabidopsis and Preliminary Analysis of Its Regulatory Mechanisms in the Formation of Triploid Loquat Leaf Heterosis. Front. Plant Sci. 2020, 11, 612055. [Google Scholar] [CrossRef]
- Liu, C.; Liu, T.; Ohlson, E.W.; Wang, L.; Wu, D.; Guo, Q.; Timko, M.P.; Liang, G. Loquat (Eriobotrya japonica (Thunb.) circadian clock gene cloning and heterosis studies of artificial triploid loquat. Sci. Hortic. 2019, 246, 328–337. [Google Scholar] [CrossRef]
- Birchler, J.A. Genetic Consequences of Polyploidy in Plants; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Mertten, D.; Tsang, G.K.; Manako, K.I.; McNeilage, M.A.; Datson, P.M. Meiotic chromosome pairing in Actinidia chinensis var. deliciosa. Genetica 2012, 140, 455–462. [Google Scholar] [CrossRef]
- Wang, W. Genome Variation and DNA Methylation Analysis of Natural and Artificial Triploid Loquats. Ph.D. Thesis, Southwest University, Chongqing, China, 2008. [Google Scholar]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Kao, Y.T.; Gonzalez, K.L.; Bartel, B. Peroxisome Function, Biogenesis, and Dynamics in Plants. Plant Physiol. 2018, 176, 162–177. [Google Scholar] [CrossRef]
- Pan, R.; Liu, J.; Hu, J. Peroxisomes in plant reproduction and seed-related development. J. Integr. Plant Biol. 2019, 61, 784–802. [Google Scholar] [CrossRef]
- Löpez-Aljorna, A.; Bueno, M.A.; Aguinagalde, I.; Martín, J.P. Fingerprinting and genetic variability in cork oak (Quercus suber L.) elite trees using ISSR and SSR markers. Ann. For. Sci. 2007, 64, 773–779. [Google Scholar] [CrossRef]
- Xu, W.; Virmani, S.S.; Hernandez, J.E.; Sebastian, L.S.; Redoña, E.D.; Li, Z. Genetic diversity in the parental lines and heterosis of the tropical rice hybrids. Euphytica 2002, 127, 139–148. [Google Scholar] [CrossRef]
- Kono, A.; Ban, Y.; Mitani, N.; Fujii, H.; Sato, S.; Suzaki, K.; Azuma, A.; Onoue, N.; Sato, A. Development of SSR markers linked to QTL reducing leaf hair density and grapevine downy mildew resistance in Vitis vinifera. Mol. Breed. 2018, 38, 138. [Google Scholar] [CrossRef]
- Najafi Zarini, H.; Jafari, H.; Darzi Ramandi, H.; Bolandi, A.R.; Karimishahri, M.R. A comparative assessment of DNA fingerprinting assays of ISSR and RAPD markers for molecular diversity of Saffron and other Crocus spp. in Iran. Nucleus 2019, 62, 39–50. [Google Scholar] [CrossRef]
- Rhimi, A.; Mnasri, S.; Ayed, R.B.; Ali, I.; Boussaid, M. Genetic relationships among subspecies of Capparis spinosa L. from Tunisia by using ISSR markers. Mol. Biol. Rep. 2019, 46, 2209–2219. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Q.; Fu, Y.; Yang, Q.; Deng, Q.X.; Lu, X.L.; Luo, N.; Yan, J. Analysis of a germplasm collection of loquat using ISSR markers. J. Hortic. Sci. Biotechnol. 2010, 85, 113–118. [Google Scholar] [CrossRef]
- Wang, Y.; Yan, J.; Qin, Y.; Yan, F.; Nan, L.; Lv, X. Taxonomic Status of Daduhe Loquat (Eriobotrya prinoides var. dadunensis). J. Agric. Sci. Technol. B 2012, 2, 659–664. [Google Scholar]
- He, Q.; Li, X.W.; Liang, G.L.; Ji, K.; Guo, Q.G.; Yuan, W.M.; Zhou, G.Z.; Chen, K.S.; van de Weg, W.E.; Gao, Z.S. Genetic Diversity and Identity of Chinese Loquat Cultivars/Accessions (Eriobotrya japonica) Using Apple SSR Markers. Plant Mol. Biol. Rep. 2011, 29, 197–208. [Google Scholar] [CrossRef]
- Li, X.Y.; Xu, H.X.; Chen, J.W. Genetic diversity and relationships among 47 loquat varieties revealed by EST-SSR markers. Sci. Hortic. 2013, 160, 375–382. [Google Scholar] [CrossRef]
- Blaine Marchant, D.; Soltis, D.E.; Soltis, P.S. Patterns of abiotic niche shifts in allopolyploids relative to their progenitors. New Phytol. 2016, 212, 708–718. [Google Scholar] [CrossRef]
- Wang, H.; Dang, J.; Wu, D.; Xie, Z.; Yan, S.; Luo, J.; Guo, Q.; Liang, G. Genotyping of polyploid plants using quantitative PCR: Application in the breeding of white-fleshed triploid loquats (Eriobotrya japonica). Plant Methods 2021, 17, 93. [Google Scholar] [CrossRef]
- Grosser, J.W.; Gmitter, F.G. Protoplast fusion for production of tetraploids and triploids: Applications for scion and rootstock breeding in citrus. Plant Cell Tissue Organ Cult. PCTOC 2011, 104, 343–357. [Google Scholar] [CrossRef]
- Soriano, L.; Francisco, A.; Camargo, L.; Cristofani-Yaly, M.; Latado, R.R.; Pacheco, C.A.; Azevedo, F.A.; Mendes, B. Regeneration and characterization of somatic hybrids combining sweet orange and mandarin/mandarin hybrid cultivars for citrus scion improvement. Plant Cell Tissue Organ Cult. 2012, 111, 385–392. [Google Scholar] [CrossRef]
- Hardigan, M.A.; Lorant, A.; Pincot, D.D.A.; Feldmann, M.J.; Famula, R.A.; Acharya, C.B.; Lee, S.; Verma, S.; Whitaker, V.M.; Bassil, N.; et al. Unraveling the Complex Hybrid Ancestry and Domestication History of Cultivated Strawberry. Mol. Biol. Evol. 2021, 38, 2285–2305. [Google Scholar] [CrossRef] [PubMed]
- Ćurčić, Ž.; Taški-Ajduković, K.; Nagl, N. Relationship between hybrid performance and genetic variation in self-fertile and self-sterile sugar beet pollinators as estimated by SSR markers. Euphytica 2017, 213, 108. [Google Scholar] [CrossRef]
- Hofmann, N.R. A global view of hybrid vigor: DNA methylation, small RNAs, and gene expression. Plant Cell 2012, 24, 841. [Google Scholar] [CrossRef] [PubMed]
- Gisbert, A.D.; Romero, C.; Martínez-Calvo, J.; Leida, C.; Llácer, G.; Badenes, M.L. Genetic diversity evaluation of a loquat (Eriobotrya japonica (Thunb) Lindl) germplasm collection by SSRs and S-allele fragments. Euphytica 2009, 168, 121–134. [Google Scholar] [CrossRef]
- Hu, X.; Wang, H.; Diao, X.; Liu, Z.; Li, K.; Wu, Y.; Liang, Q.; Wang, H.; Huang, C. Transcriptome profiling and comparison of maize ear heterosis during the spikelet and floret differentiation stages. BMC Genom. 2016, 17, 959. [Google Scholar] [CrossRef]
- Oyekunle, M.; Badu-Apraku, B.; Hearne, S.; Franco, J. Genetic diversity of tropical early-maturing maize inbreds and their performance in hybrid combinations under drought and optimum growing conditions. Field Crops Res. 2015, 170, 55–65. [Google Scholar] [CrossRef]
- Jesske, T.; Olberg, B.; Schierholt, A.; Becker, H.C. Resynthesized lines from domesticated and wild Brassica taxa and their hybrids with B. napus L.: Genetic diversity and hybrid yield. Theor. Appl. Genet. 2013, 126, 1053–1065. [Google Scholar] [CrossRef]
- Xiao, J.; Li, J.; Yuan, L.; McCouch, S.R.; Tanksley, S.D. Genetic diversity and its relationship to hybrid performance and heterosis in rice as revealed by PCR-based markers. Theor. Appl. Genet. 1996, 92, 637–643. [Google Scholar] [CrossRef]
- Wei, X.; Zhang, J. The optimal mating distance resulting from heterosis and genetic incompatibility. Sci. Adv. 2018, 4, eaau5518. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Zhang, X.; Liu, Q.; Luo, T.; Tang, Z.; Zhou, Y. The genetic diversity and relationships of cauliflower (Brassica oleracea var. botrytis) inbred lines assessed by using SSR markers. PLoS ONE 2018, 13, e0208551. [Google Scholar] [CrossRef]
- Zhang, H.; Peng, S.; Cai, L.; Fang, D. The germplasm resources of the genus Eriobotrya with special reference on the origin of E. Japonica lindl. Acta Hortic. Sin. 1990, 17, 5–12. [Google Scholar]
- Yang, X.; Liu, C.; Lin, S. Genetic relationships in Eriobotrya species as revealed by amplified fragment length polymorphism (AFLP) markers. Sci. Hortic. 2009, 122, 264–268. [Google Scholar] [CrossRef]
- Vilanova, S.; Badenes, M.L.; Martinez-Calvo, J.; Llacer, G. Analysis of loquat germplasm (Eriobotrya japonica Lindl) by RAPD molecular markers. Euphytica 2001, 121, 25–29. [Google Scholar] [CrossRef]
- Kong, X.; Chen, L.; Wei, T.; Zhou, H.; Bai, C.; Yan, X.; Miao, Z.; Xie, J.; Zhang, L. Transcriptome analysis of biological pathways associated with heterosis in Chinese cabbage. Genomics 2020, 112, 4732–4741. [Google Scholar] [CrossRef]
- Yang, M.; Wang, X.; Ren, D.; Huang, H.; Xu, M.; He, G.; Deng, X.W. Genomic architecture of biomass heterosis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 8101–8106. [Google Scholar] [CrossRef]
- Ding, Y.; Zhang, R.; Zhu, L.; Wang, M.; Ma, Y.; Yuan, D.; Liu, N.; Hu, H.; Min, L.; Zhang, X. An enhanced photosynthesis and carbohydrate metabolic capability contributes to heterosis of the cotton (Gossypium hirsutum) hybrid ‘Huaza Mian H318’, as revealed by genome-wide gene expression analysis. BMC Genom. 2021, 22, 277. [Google Scholar] [CrossRef]
- Wang, D.; Mu, Y.; Hu, X.; Ma, B.; Wang, Z.; Zhu, L.; Xu, J.; Huang, C.; Pan, Y. Comparative proteomic analysis reveals that the Heterosis of two maize hybrids is related to enhancement of stress response and photosynthesis respectively. BMC Plant Biol. 2021, 21, 34. [Google Scholar] [CrossRef]
- Zhu, A.; Wang, A.; Zhang, Y.; Dennis, E.S.; Peacock, W.J.; Greaves, A.I.K. Early Establishment of Photosynthesis and Auxin Biosynthesis Plays a Key Role in Early Biomass Heterosis in Brassica napus (Canola) Hybrids. Plant Cell Physiol. 2020, 61, 1134–1143. [Google Scholar] [CrossRef]
- Wen, G.; Dang, J.; Xie, Z.; Wang, J.; Jiang, P.; Guo, Q.; Liang, G. Molecular karyotypes of loquat (Eriobotrya japonica) aneuploids can be detected by using SSR markers combined with quantitative PCR irrespective of heterozygosity. Plant Methods 2020, 16, 22. [Google Scholar] [CrossRef]
- Galbraith, D.W.; Harkins, K.R.; Maddox, J.M.; Ayres, N.M.; Sharma, D.P.; Firoozabady, E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 1983, 220, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Zhang, C.; Chen, Z.J. Ploidy and Hybridity Effects on Growth Vigor and Gene Expression in Arabidopsis thaliana Hybrids and Their Parents. G3 2012, 2, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.X.; Yang, X.H.; Lin, S.Q.; Gui-Bing, H.U.; Liu, C.M. An improved procedure for nuclear DNA isolation from Eriobotrya plants and its application. J. Fruit Sci. 2005, 22, 182–185. [Google Scholar]
- Bassam, B.J.; Caetano-Anolles, G.; Gresshoff, P.M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 1991, 196, 80–83. [Google Scholar] [CrossRef]
- Turner, J.H. A Study of Heterosis in Upland Cotton I. Yield of Hybrids Compared with Varieties1. Agron. J. 1953, 45, 484–486. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Dang, J.; Wang, L.; Zhi, C.; Liang, G. Segregation Correlation of SSR Markers and Flower Traits in Aneuploid Tobacco. World J. Eng. Technol. 2018, 6, 225–240. [Google Scholar] [CrossRef]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Wright, S.W. The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Nei, M. Genetic Distance between Populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Rohlf, F.; Rohlf, F.; Rohlf, F.; Rohlf, F.; Rohlf, F.J. NTSYS-pc, Numerical Taxonomy and Multivariate Analysis System; Version 2.1; Exeter Publishing: Setauket, NY, USA, 2000. [Google Scholar]
- Mantel, N. The Detection of Disease Clustering and a Generalized Regression Approach. Cancer Res. 1967, 59, 209–220. [Google Scholar]
- Shaked, H.; Kashkush, K.; Ozkan, H.; Feldman, M.; Levy, A.A. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 2001, 13, 1749–1759. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, H.; Levy, A.A.; Feldman, M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 2001, 13, 1735–1747. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Locus Name | TB | PB | PPB | I | He | Ho | F | PIC | ||
|---|---|---|---|---|---|---|---|---|---|---|
| ISSR | ||||||||||
| ISSR-807 | 6 | 2 | 33.33% | -a | -a | 0.810 | ||||
| ISSR-811 | 6 | 0 | 0.00% | -a | -a | 0.810 | ||||
| ISSR-823 | 4 | 2 | 50.00% | -a | -a | 0.703 | ||||
| ISSR-825 | 7 | 4 | 57.14% | -a | -a | 0.818 | ||||
| ISSR-834 | 7 | 4 | 57.14% | -a | -a | 0.811 | ||||
| ISSR-835 | 9 | 4 | 44.44% | -a | -a | 0.858 | ||||
| ISSR-836 | 7 | 2 | 28.57% | -a | -a | 0.835 | ||||
| ISSR-840 | 6 | 3 | 50.00% | -a | -a | 0.809 | ||||
| ISSR-845 | 6 | 4 | 66.67% | -a | -a | 0.780 | ||||
| ISSR-853 | 9 | 6 | 66.67% | -a | -a | 0.828 | ||||
| ISSR-858 | 2 | 0 | 0.00% | -a | -a | 0.375 | ||||
| ISSR-874 | 16 | 4 | 25.00% | -a | -a | 0.930 | ||||
| ISSR-876 | 7 | 3 | 42.86% | -a | -a | 0.839 | ||||
| ISSR-879 | 6 | 4 | 66.67% | -a | -a | 0.810 | ||||
| ISSR-880 | 5 | 2 | 40.00% | -a | -a | 0.737 | ||||
| ISSR-886 | 5 | 0 | 0.00% | -a | -a | 0.768 | ||||
| ISSR-887 | 5 | 2 | 40.00% | -a | -a | 0.755 | ||||
| ISSR-892 | 1 | 0 | 0.00% | -a | -a | monomorphic | ||||
| ISSR-899 | 6 | 5 | 83.33% | -a | -a | 0.562 | ||||
| ISSR-900 | 8 | 5 | 62.50% | -a | -a | 0.813 | ||||
| Mean | 6.4 | 2.8 | 43.75% | 0.733 | ||||||
| Total | 128 | 56 | 43.75% | |||||||
| Locus name | NAobs | NA Wu | I | He | Ho | F | PIC | |||
| SSR | ||||||||||
| ssrEJ005 | 5 | 2 | 40.00% | 5 | 2 | 0.693 | 0.524 | 0.273 | 0.479 | 0.822 |
| ssrEJ012 | 4 | 0 | 0.00% | 4 | 5 | -a | -a | -a | -a | 0.634 |
| ssrEJ014 | 4 | 0 | 0.00% | 4 | 3 | 0.185 | 0.091 | 0.091 | 0 | 0.525 |
| ssrEJ037 | 4 | 1 | 25.00% | 4 | 3 | 0.185 | 0.091 | 0.091 | 0 | 0.740 |
| ssrEJ039 | 5 | 3 | 60.00% | 5 | 2 | 0.185 | 0.091 | 0.091 | 0 | 0.678 |
| ssrEJ042 | 4 | 0 | 0.00% | 4 | 3 | 0.656 | 0.485 | -a | -a | 0.674 |
| ssrEJ046 | 6 | 0 | 0.00% | 6 | 6 | 0.690 | 0.520 | 0.909 | -0.748 | 0.810 |
| ssrEJ049 | 6 | 0 | 0.00% | 6 | 5 | 0.689 | 0.520 | 0.909 | -0.748 | 0.819 |
| ssrEJ061 | 4 | 0 | 0.00% | 4 | 6 | -a | -a | -a | -a | 0.457 |
| ssrEJ066 | 4 | 0 | 0.00% | 4 | 4 | -a | -a | -a | -a | 0.634 |
| ssrEJ075a | 6 | 0 | 0.00% | 6 | 3 | -a | -a | -a | -a | 0.697 |
| ssrEJ086 | 3 | 1 | 33.33% | 3 | 3 | 0.305 | 0.173 | -a | -a | 0.553 |
| ssrEJ088 | 4 | 0 | 0.00% | 4 | 4 | 0.305 | 0.173 | -a | -a | 0.752 |
| ssrEJ095b | 4 | 0 | 0.00% | 4 | 4 | 0.693 | 0.524 | 0.818 | -0.561 | 0.701 |
| ssrEJ104 | 4 | 0 | 0.00% | 4 | 4 | -a | -a | -a | -a | 0.702 |
| ssrEJ106 | 1 | 0 | 0.00% | 1 | 2 | -a | -a | -a | -a | monomorphic |
| ssrEJ271 | 5 | 3 | 60.00% | 5 | 6 | 0.185 | 0.091 | 0.091 | 0 | 0.743 |
| ssrEJ282 | 6 | 3 | 50.00% | 6 | 5 | 0.398 | 0.247 | 0.273 | -0.105 | 0.872 |
| ssrEJ324 | 4 | 1 | 25.00% | 4 | 6 | 0.689 | 0.520 | 0.909 | -0.748 | 0.670 |
| ssrEJ329b | 5 | 3 | 60.00% | 5 | 3 | 0.185 | 0.091 | 0.091 | 0 | 0.766 |
| Mean | 4.4 | 0.85 | 19.32% | 4 | 4 | 0.302 | 0.207 | 0.227 | -0.122 | 0.662 |
| Total | 88 | 17 | 19.32% | 88 | 79 |
| Comparison between Different Groups | Total DEGs | Number of the Annotated DEGs | Percentage a (%) | Number of KEGG-Mapped DEGs | Percentage b (%) |
|---|---|---|---|---|---|
| LQ-1 vs. GC-1 | 6970 | 6068 | 87.06 | 2029 | 29.11 |
| LQ-1 vs. Triploid-A | 3824 | 3619 | 94.64 | 1257 | 32.87 |
| GC-1 vs. Triploid-A | 2207 | 1799 | 81.51 | 407 | 18.44 |
| LQ-1 vs. GC-23 | 4384 | 3910 | 89.19 | 1279 | 29.17 |
| LQ-1 vs. Triploid-B | 5010 | 4608 | 91.98 | 1553 | 31.00 |
| GC-23 vs. Triploid-B | 3588 | 2855 | 79.57 | 978 | 27.26 |
| MPL a | HPL b | LPL c | AHP d | BLP e | |
|---|---|---|---|---|---|
| C41504 | N/A | A-2, A-3 | N/A | A-1, A-4, A-5, A-6, A-7, A-8, A-9, B-1, B-2, B-3 | N/A |
| C126732 | N/A | N/A | N/A | A-3, A-4, A-5, A-6, A-7, A-8, B-2 | A-1, A-2, A-9, B-1, B-3 |
| C126973 | A-4, A-7 | N/A | N/A | A-3, A-5, A-6, A-8, B-2 | A-1, A-2, A-9, B-1, B-3 |
| C110057 | N/A | A-7, A-8, A-9, B-3 | N/A | A-1, A-2, A-3, A-4, A-5, A-6, B-1, B-2 | N/A |
| C124069 | A-3, A-8 | A-1, A-4, A-5, A-7, A-9, B-1, B-2 | N/A | A-2, A-6, B-3 | N/A |
| C103155 | A-6, B-3 | A-1, A-2, A-3, A-9, B-1, B-2 | A-5 | A-4, A-7, A-8 | N/A |
| Cross Combinations | Serial Number of Triploid Loquats (Triploid-A) | ||||||||
| LQ-1× GC-1 | A-1 | A-2 | A-3 | A-4 | A-5 | A-6 | A-7 | A-8 | A-9 |
| Serial Number of Triploid Loquats (Triploid-B) | |||||||||
| LQ-1 × GC-23 | B-1 | B-2 | B-3 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Tu, M.; Li, J.; Sun, S.; Song, H.; Xu, Z.; Chen, D.; Liang, G. Photosynthetic Efficiency and Glyco-Metabolism Changes in Artificial Triploid Loquats Contribute to Heterosis Manifestation. Int. J. Mol. Sci. 2022, 23, 11337. https://doi.org/10.3390/ijms231911337
Wang L, Tu M, Li J, Sun S, Song H, Xu Z, Chen D, Liang G. Photosynthetic Efficiency and Glyco-Metabolism Changes in Artificial Triploid Loquats Contribute to Heterosis Manifestation. International Journal of Molecular Sciences. 2022; 23(19):11337. https://doi.org/10.3390/ijms231911337
Chicago/Turabian StyleWang, Lingli, Meiyan Tu, Jing Li, Shuxia Sun, Haiyan Song, Zihong Xu, Dong Chen, and Guolu Liang. 2022. "Photosynthetic Efficiency and Glyco-Metabolism Changes in Artificial Triploid Loquats Contribute to Heterosis Manifestation" International Journal of Molecular Sciences 23, no. 19: 11337. https://doi.org/10.3390/ijms231911337