MicroRNAs of Milk in Cells, Plasma, and Lipid Fractions of Human Milk, and Abzymes Catalyzing Their Hydrolysis
Abstract
:1. Introduction
2. Results
2.1. Isolation and Quantification of MicroRNAs
2.2. Purification and Characterizing of IgGs and sIgAs
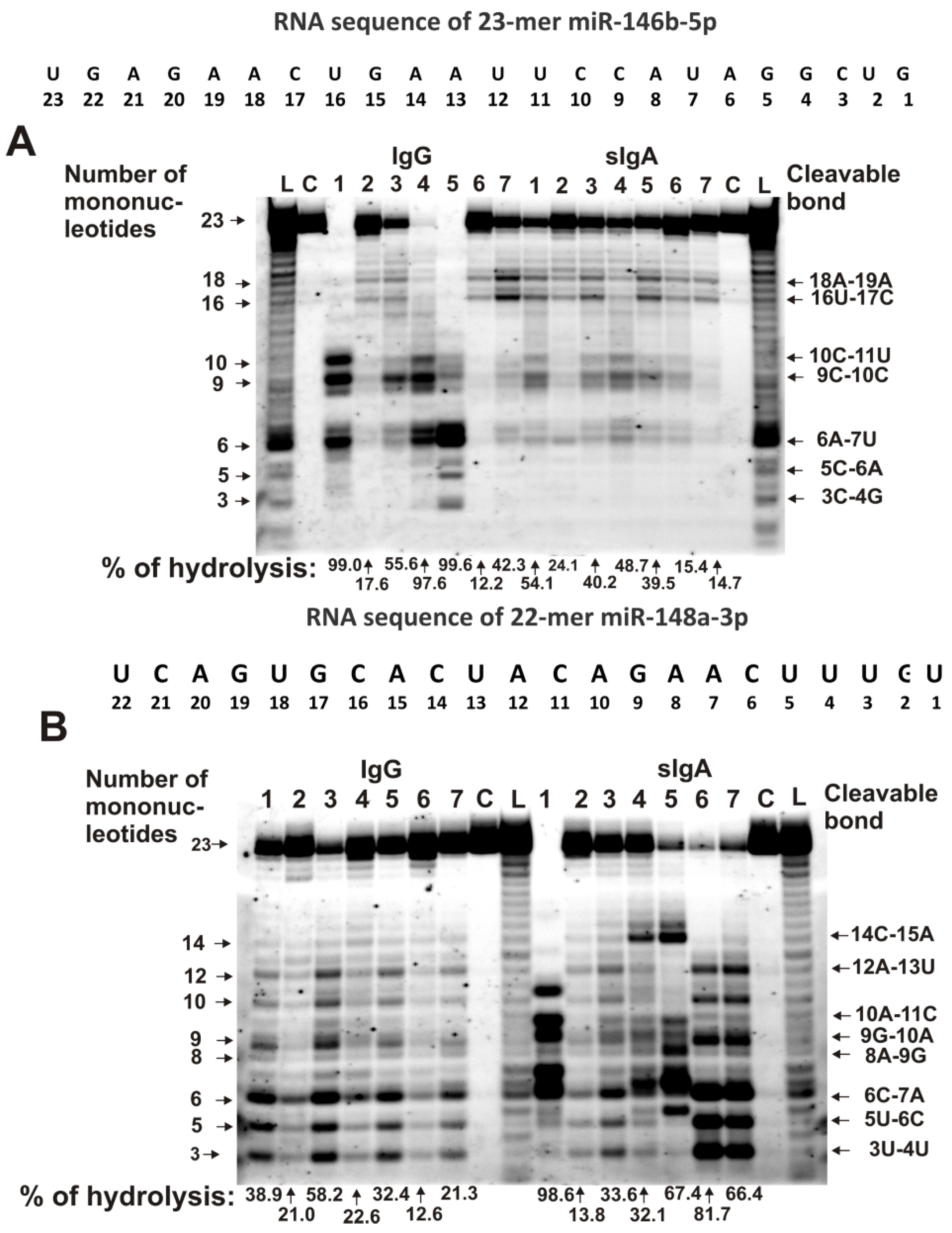
2.3. Hydrolysis of MicroRNAs
2.4. Spatial Structures of Five miRNAs
3. Discussion
4. Materials and Methods
4.1. Chemicals and Donors
4.2. Purification and Analysis of RNAs
4.3. RNA Amplification
4.4. Purification and Analysis of Antibodies
4.5. Analysis of MicroRNAs Hydrolysis by Abs
4.6. Spatial Model of MicroRNAs
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Matsuyama, H.; Suzuki, H.I. Systems and synthetic microRNA biology: From biogenesis to disease pathogenesis. Int. J. Mol. Sci. 2019, 2, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferro, E.C.; Enrico Bena, S.; Grigolon, S.; Bosia, C. From endogenous to synthetic microRNA-mediated regulatory circuits: An overview. Cells 2019, 8, 1540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, B.H.; Wahlestedt, C. MicroRNA dysregulation in psychiatric disease. Brain Res. 2010, 1338, 89–99. [Google Scholar] [CrossRef] [Green Version]
- Melnik, B.C.; Schmitz, G. MicroRNAs: Milk’s epigenetic regulators. Best. Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 427–442. [Google Scholar] [CrossRef]
- Alsaweed, M.; Hepworth, A.R.; Lefèvre, C.; Hartmann, P.E.; Geddes, D.T.; Hassiotou, F. Human milk microRNA and total RNA differ depending on milk fractionation. J. Cell. Biochem. 2015, 116, 2397–2407. [Google Scholar] [CrossRef] [Green Version]
- Alsaweed, M.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. MicroRNAs in breast milk and the lactating breast: Potential immunoprotectors and developmental regulators for the Infant and the mother. Int. J. Environ. Res. Public Health 2015, 12, 13981–14020. [Google Scholar] [CrossRef] [Green Version]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk cells contain numerous mirnas that may change with milk removal and regulate multiple physiological processes. Int. J. Mol. Sci. 2016, 17, 956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk cells and lipids conserve numerous known and novel mirnas, some of which are differentially expressed during lactation. PLoS ONE 2016, 11, e0152610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk miRNAs primarily originate from the mammary gland resulting in unique miRNA profiles of fractionated milk. Sci. Rep. 2016, 6, 20680. [Google Scholar] [CrossRef] [Green Version]
- Golan-Gerstl, R.; Elbaum Shiff, Y.; Moshayoff, V.; Schecter, D.; Leshkowitz, D.; Reif, S. Characterization and biological function of milk-derived miRNAs. Mol. Nutr. Food Res. 2017, 61, 1700009. [Google Scholar] [CrossRef]
- Keinan, E. (Ed.) Catalytic Antibodies; Wiley-VCH Verlag GmbH and Co. KgaA: Weinheim, Germany, 2005; pp. 1–586. [Google Scholar]
- Nevinsky, G.A.; Buneva, V.N. Catalytic antibodies in healthy humans and patients with autoimmune and viral pathologies. J. Cell. Mol. Med. 2003, 7, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Nevinsky, G.A.; Buneva, V.N. Natural catalytic antibodies–abzymes. In Catalytic Antibodies; Keinan, E., Ed.; VCH-Wiley Press: Weinheim, Germany, 2005; pp. 503–567. [Google Scholar]
- Nevinsky, G.A. Natural catalytic antibodies in norm and in autoimmune diseases. In Autoimmune Diseases: Symptoms, Diagnosis and Treatment; Brenner, K.J., Ed.; Nova Science Publishers: New York, NY, USA, 2010; pp. 1–107. [Google Scholar]
- Nevinsky, G.A. Natural catalytic antibodies in norm and in HIV-infected patients. In Understanding HIV/AIDS Management and Care—Pandemic Approaches the 21st Century; Kasenga, F.H., Ed.; InTech: Rijeka, Croatia, 2011; pp. 151–192. [Google Scholar]
- Nevinsky, G.A. Autoimmune processes in multiple sclerosis: Production of harmful catalytic antibodies associated with significant changes in the hematopoietic stem cell differentiation and proliferation. In Multiple Sclerosis; Conzalez-Quevedo, A., Ed.; InTech: Rijeka, Croatia, 2016; pp. 100–147. [Google Scholar]
- Nevinsky, G.A. Catalytic antibodies in norm and systemic lupus erythematosus. In Lupus; Khan, W.A., Ed.; InTech: Rijeka, Croatia, 2017; pp. 41–101. [Google Scholar]
- Jerne, N.K. Towards a network theory of the immune system. Ann. Immunol. 1974, 125, 373–398. [Google Scholar]
- Shuster, A.M.; Gololobov, G.V.; Kvashuk, O.A.; Bogomolova, A.E.; Smirnov, I.V.; Gabibov, A.G. DNA hydrolyzing autoantibodies. Science 1992, 256, 665–667. [Google Scholar] [CrossRef] [PubMed]
- Andrievskaya, O.A.; Buneva, V.N.; Naumov, V.A.; Nevinsky, G.A. Catalytic heterogenity of polyclonal RNA-hydrolyzing IgM from sera of patients with lupus erythematosus. Med. Sci. Monit. 2000, 6, 460–470. [Google Scholar]
- Andrievskaya, O.A.; Buneva, V.N.; Baranovskii, A.G.; Gal’vita, A.V.; Benzo, E.S.; Naumov, V.A.; Nevinsky, G.A. Catalytic diversity of polyclonal RNA-hydrolyzing IgG antibodies from the sera of patients with systemic lupus erythematosus. Immunol. Lett. 2002, 81, 191–198. [Google Scholar] [CrossRef]
- Baranovskii, A.G.; Kanyshkova, T.G.; Mogelnitskii, A.S.; Naumov, V.A.; Buneva, V.N.; Gusev, E.I.; Boiko, A.N.; Zargarova, T.A.; Favorova, O.O.; Nevinsky, G.A. Polyclonal antibodies from blood and cerebrospinal fluid of patients with multiple sclerosis effectively hydrolyze DNA and RNA. Biochemistry 1998, 63, 1239–1248. [Google Scholar]
- Baranovsky, A.G.; Matushin, V.G.; Vlassov, A.V.; Zabara, V.G.; Naumov, V.A.; Giege, R.; Buneva, V.N.; Nevinsky, G.A. DNA- and RNA-hydrolyzing antibodies from the blood of patients with various forms of viral hepatitis. Biochemistry 1997, 62, 1358–1366. [Google Scholar]
- Savel’ev, A.N.; Eneyskaya, E.V.; Shabalin, K.A.; Filatov, M.V.; Neustroev, K.N. Antibodies with amylolytic activity. Protein Pept. Lett. 1999, 6, 179–181. [Google Scholar]
- Savel’ev, A.N.; Kanyshkova, T.G.; Kulminskaya, A.A.; Buneva, V.N.; Eneyskaya, E.V.; Filatov, M.V.; Nevinsky, G.A.; Neustroev, K.N. Amylolytic activity of IgG and sIgA immunoglobulins from human milk. Clin. Chim. Acta 2001, 314, 141–152. [Google Scholar] [CrossRef]
- Neustoev, K.N.; Ivanen, D.R.; Kulminskaya, A.A.; Brumer, I.H.; Saveliev, A.N.; Nevinsky, G.A. Amylolytic activity and catalytic properties of IgM and IgG antibodies from patients with systemic lupus erythematosus. Hum. Antibodies 2003, 12, 31–34. [Google Scholar]
- Paul, S.; Volle, D.J.; Beach, C.M.; Johnson, D.R.; Powell, M.J.; Massey, R.J. Catalytic hydrolysis of vasoactive intestinal peptide by human autoantibody. Science 1989, 244, 1158–1162. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Paul, S.; Tyutyulkova, S.; Kazatchkine, M.D.; Kaveri, S. Catalytic activity of anti-thyroglobulin antibodies. J. Immunol. 1995, 154, 3328–3332. [Google Scholar]
- Kalaga, R.; Li, L.; O’Dell, J.R.; Paul, S. Unexpected presence of polyreactive catalytic antibodies in IgG from unimmunized donors and decreased levels in rheumatoid arthritis. J. Immunol. 1995, 155, 2695–2702. [Google Scholar]
- Lacroix-Desmazes, S.; Moreau, A.; Sooryanarayana; Bonnemain, C.; Stieltjes, N.; Pashov, A.; Sultan, Y.; Hoebeke, J.M.; Kazatchkine, D.; Kaveri, S.V. Catalytic activity of antibodies against factor VIII in patients with hemophilia A. Nat. Med. 1999, 5, 1044–1047. [Google Scholar] [CrossRef]
- Polosukhina, D.I.; Kanyshkova, T.G.; Doronin, B.M.; Tyshkevich, O.B.; Buneva, V.N.; Boiko, A.N.; Gusev, E.I.; Favorova, O.O.; Nevinsky, G.A. Hydrolysis of myelin basic protein by polyclonal catalytic IgGs from the sera of patients with multiple sclerosis. J. Cell. Mol. Med. 2004, 8, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Polosukhina, D.I.; Kanyshkova, T.G.; Doronin, B.M.; Tyshkevich, O.B.; Buneva, V.N.; Boiko, A.N.; Gusev, E.I.; Nevinsky, G.A.; Favorova, O.O. Metal-dependent hydrolysis of myelin basic protein by IgGs from the sera of patients with multiple sclerosis. Immunol. Lett. 2006, 103, 75–81. [Google Scholar] [CrossRef]
- Baranova, S.V.; Buneva, V.N.; Kharitonova, M.A.; Sizyakina, L.P.; Calmels, C.; Andreola, M.L.; Parissi, V.; Nevinsky, G.A. HIV-1 integrase-hydrolyzing antibodies from sera of HIV-infected patients. Biochimie 2009, 91, 1081–1086. [Google Scholar] [CrossRef]
- Mestecky, J.; Russell, M.W.; Jackson, S.; Brown, T.A. The human IgA system: A reassessment. Clin. Immunol. Immunopathol. 1986, 40, 105–114. [Google Scholar] [CrossRef]
- Hanson, L.A.; Hahn-Zoric, M.; Berndes, M.; Ashraf, R.; Herias, V.; Jalil, F.; Bhutta, T.I.; Laeeq, A.; Mattsby-Baltzer, I. Breast feeding: Overview and breast milk immunology. Acta Paediatr. Jpn. 1994, 36, 557–561. [Google Scholar] [CrossRef]
- Bakos, M.A.; Kurosky, A.; Goldblum, R.M. Characterization of a critical binding site for human polymeric Ig on secretory component. J. Immunol. 1991, 147, 3419–3426. [Google Scholar]
- Roux, M.E.; McWilliams, M.; Phillips-Quagliata, J.M.; Weisz-Carrington, P.; Lamm, M.E. Origin of IgA secreting plasma cells in the mammary gland. J. Exp. Med. 1977, 146, 1311–1322. [Google Scholar] [CrossRef]
- Weisz-Carrington, P.; Roux, M.E.; McWilliams, M.; Phillips-Quagliata, J.M.; Lamm, M.E. Hormonal induction of the secretory immune system in the mammary gland. Proc. Natl. Acad. Sci. USA 1978, 75, 2928–2932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, L.A.; Carlsson, B.; Cruz, J.R. T cell regulation of IgA synthesis. In Immunology of Breast Milk; Ogra, P.L., Dayton, D.H., Eds.; Raven Press: New York, NY, USA, 1979; pp. 145–157. [Google Scholar]
- Rechtman, D.J.; Ferry, B.L.; Lee, M.L.; Chapel, H. Immunoglobulin A (IgA) content of human breast milk over time. Int. J. Infect. Dis. 2002, 6, S58. [Google Scholar]
- Amino, N.; Mori, H.; Iwatani, Y.; Tanizawa, O.; Kawashima, M.; Tsuge, I.; Ibaragi, K.; Kumahara, Y.; Miyai, K. High prevalence of transient post-partum thyrotoxicosis and hypothyroidism. N. Engl. J. Med. 1986, 306, 849–852. [Google Scholar] [CrossRef] [PubMed]
- Hazes, J.M. Pregnancy and its effect on the risk of developing rheumatoid arthritis. Ann. Rheum. Dis. 1991, 50, 71–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dayan, C.M.; Daniels, G.H. Chronic autoimmune thyroiditis. N. Engl. J. Med. 1996, 35, 99–107. [Google Scholar] [CrossRef]
- Tanaka, A.; Lindor, K.; Ansari, A.; Gershwin, M.E. Fetal microchimerisms in the mother: Immunologic implications. Liver Transpl. 2000, 6, 138–143. [Google Scholar] [CrossRef]
- Freeman, R.; Rosen, H.; Thysen, B. Incidence of thyroid dysfunction in an unselected postpartum population. Arch. Intern. Med. 1986, 146, 1361–1364. [Google Scholar] [CrossRef] [PubMed]
- Petri, M. Systemic lupus erythematosus and pregnancy. Rheum. Dis. Clin. N. Am. 1994, 20, 87–118. [Google Scholar] [CrossRef]
- Kattah, A.G.; Garovic, V.D. Pregnancy and Lupus Nephritis. Semin. Nephrol. 2015, 35, 487–499. [Google Scholar] [CrossRef]
- Eudy, A.M.; Siega-Riz, A.M.; Engel, S.M.; Franceschini, N.; Howard, A.G.; Clowse, E.B.M.; Petri, M. Effect of pregnancy on disease flares in patients with systemic lupus erythematosus. Ann. Rheum. Dis. 2018, 77, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Baalbaki, S.J.; Szychowski, M.; Tang, Y.; Wetta, L.; Subramaniam, A.; Ochsner, J. Systemic Lupus Erythematosus: Perinatal Outcomes in Patients Treated With and Without Hydroxychloroquine. Winter 2020, 20, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, L.V.; Alves, L.P.; Sim-Simc, M.M.F.; Surita, F.G. Perceptions of women with systemic lupus erythematosus undergoing high-risk prenatal care: A qualitative study. Midwifery 2020, 87, 102715. [Google Scholar] [CrossRef]
- Tincani, A.; Nalli, C.; Khizroeva, J.; Bitsadze, V.; Lojacono, A.; Andreoli, L.; Shoenfeld, Y.; Makatsariya, A. Autoimmune diseases and pregnancy. Best Pract. Res. Clin. Endocrinol. Metab. 2019, 33, 101322. [Google Scholar] [CrossRef]
- Alroughani, R.; Alowayesh, M.S.; Ahmed, S.F.; Behbehani, R.; Al-Hashel, J. Relapse occurrence in women with multiple sclerosis during pregnancy in the new treatment era. Neurology 2018, 90, e840–e846. [Google Scholar] [CrossRef] [PubMed]
- De Leo, S.; Pearce, E.N. Autoimmune thyroid disease during pregnancy. Lancet Diabetes Endocrinol. 2018, 6, 575–586. [Google Scholar] [CrossRef]
- Buneva, V.N.; Kanyshkova, T.G.; Vlassov, A.V.; Semenov, D.V.; Khlimankov, D.; Breusova, L.R.; Nevinsky, G.A. Catalytic DNA- and RNA-hydrolyzing antibodies from milk of healthy human mothers. Appl. Biochem. Biotechnol. 1998, 75, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Buneva, V.N.; Kudryavtseva, A.N.; Gal’vita, A.V.; Dubrovskaya, V.V.; Khokhlova, O.V.I.; Kalinina, A.; Galenok, V.A.; Nevinsky, G.A. Dynamics of antibody nuclease activity in blood of women during pregnancy and lactation. Biochemistry 2003, 68, 890–900. [Google Scholar] [PubMed]
- Semenov, D.V.; Kanyshkova, T.G.; Karotaeva, N.A.; Krasnorutskii, M.A.; Kuznetsova, I.A.; Buneva, V.N.; Nevinsky, G.A. Catalytic nucleotide-hydrolyzing antibodies in milk and serum of clinically healthy human mothers. Med. Sci. Monit. 2004, 10, BR23–BR33. [Google Scholar]
- Watanabe-Fukunada, R.; Brannan, C.I.; Copeland, N.G.; Jenkins, N.A.; Nagata, S. Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature 1992, 356, 314–317. [Google Scholar] [CrossRef]
- Andryushkova, A.A.; Kuznetsova, I.A.; Orlovskaya, I.A.; Buneva, V.N.; Nevinsky, G.A. Antibodies with amylase activity from the sera of autoimmune-prone MRL/MpJ-lpr mice. FEBS Lett. 2006, 580, 5089–5095. [Google Scholar] [CrossRef]
- Andryushkova, A.S.; Kuznetsova, I.A.; Buneva, V.N.; Toporkova, L.B.; Sakhno, L.V.; Tikhonova, M.A.; Chernykh, E.R.; Orlovskaya, I.A.; Nevinsky, G.A. Formation of different abzymes in autoimmune-prone MRL-lpr/lpr mice is associated with changes in colony formation of haematopoetic progenitors. J. Cell. Mol. Med. 2007, 11, 531–551. [Google Scholar] [CrossRef]
- Andryushkova, A.A.; Kuznetsova, I.A.; Orlovskaya, I.A.; Buneva, V.N.; Nevinsky, G.A. Nucleotide- hydrolyzing antibodies from the sera of autoimmune-prone MRL-lpr/lpr mice. Int. Immunol. 2009, 21, 935–945. [Google Scholar] [CrossRef] [Green Version]
- Doronin, V.B.; Parkhomenko, T.A.; Korablev, A.; Toporkova, L.B.; Lopatnikova, J.A.; Alshevskaja, A.A.; Sennikov, S.V.; Buneva, V.N.; Budde, T.; Meuth, S.G.; et al. Changes in different parameters, lymphocyte proliferation and hematopoietic progenitor colony formation in EAE mice treated with myelin oligodendrocyte glycoprotein. J. Cell. Mol. Med. 2016, 20, 81–94. [Google Scholar] [CrossRef]
- Aulova, K.S.; Toporkova, L.B.; Lopatnikova, J.A.; Alshevskaya, A.A.; Sennikov, S.V.; Buneva, V.N.; Budde, T.; Meuth, S.G.; Popova, N.A.; Orlovskaya, I.A.; et al. Changes in haematopoietic progenitor colony differentiation and proliferation and the production of different abzymes in EAE mice treated with DNA. J. Cell. Mol. Med. 2017, 21, 3795–3809. [Google Scholar] [CrossRef]
- Gorbunov, D.V.; Semenov, D.V.; Shipitsin, M.V.; Kit, Y.Y.; Kanyshkova, T.G.; Buneva, V.N.; Nevinsky, G.A. Phosphorylation of minor lipids of human milk tightly bound to secretory immunoglobulin A. Rus. J. Immunol. 2000, 5, 267–278. [Google Scholar]
- Gorbunov, D.V.; Karataeva, N.A.; Buneva, V.N.; Nevinsky, G.A. Lipid kinase activity of antibodies from milk of clinically healthy human mothers. Biochim. Biophys. Acta 2005, 1735, 153–166. [Google Scholar] [CrossRef]
- Nevinsky, G.A.; Kit, Y.; Semenov, D.V.; Khlimankov, D.; Buneva, V.N. Secretory immunoglobulin A from human milk catalyses milk protein phosphorylation. Appl. Biochem. Biotechnol. 1998, 75, 77–91. [Google Scholar] [CrossRef]
- Kit, Y.Y.; Semenov, D.V.; Nevinsky, G.A. Phosphorylation of different human milk proteins by human catalytic secretory immunoglobulin A. Biochem. Mol. Biol. Int. 1996, 39, 521–527. [Google Scholar] [CrossRef]
- Karataeva, N.A.; Gorbunov, D.; Prokudin, I.V.; Buneva, V.N.; Kulminskaya, A.A.; Neustroev, K.N.; Nevinsky, G.A. Human milk antibodies with polysaccharide kinase activity. Immunol. Lett. 2006, 103, 58–67. [Google Scholar] [CrossRef]
- Karataeva, N.A.; Buneva, V.N.; Nevinsky, G.A. Polysaccharide kinase activity of human milk IgG antibodies. Biochemistry 2006, 71, 1207–1221. [Google Scholar] [CrossRef] [PubMed]
- Ermakov, E.A.; Kabirova, E.M.; Buneva, V.N.; Nevinsky, G.A. IgGs-Abzymes from the Sera of Patients with Multiple Sclerosis Recognize and Hydrolyze miRNAs. Int. J. Mol. Sci. 2021, 22, 2812. [Google Scholar] [CrossRef] [PubMed]
- Ermakov, E.A.; Kabirova, E.M.; Sizikov, A.E.; Buneva, V.N.; Nevinsky, G.A. IgGs-Abzymes from the Sera of Patients with Systemic Lupus Erythematosus Hydrolyzed miRNAs. J. Inflamm. Res. 2020, 13, 681–699. [Google Scholar] [CrossRef]
- Ermakov, E.A.; Ivanova, S.A.; Buneva, V.N.; Nevinsky, G.A. Hydrolysis by catalytic IgGs of microRNA specific for patients with schizophrenia. IUBMB Life 2018, 70, 153–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ermakov, E.A.; Ivanova, S.A.; Buneva, V.N.; Nevinsky, G.A. Blood-derived RNA- and microRNA-hydrolyzing IgG antibodies in schizophrenia patients. Biochemistry 2018, 83, 507–526. [Google Scholar] [CrossRef]
- Kompaneets, I.Y.; Ermakov, E.A.; Sedykh, S.E.; Buneva, V.N.; Nevinsky, G.A. IgGs from Human Milk Hydrolyze microRNAs. Molecules 2020, 25, 2366. [Google Scholar] [CrossRef] [PubMed]
- Kompaneets, I.Y.; Ermakov, E.A.; Sedykh, S.E.; Buneva, V.N.; Nevinsky, G.A. Secretory immunoglobulin A from human milk hydrolyzes microRNA. J. Dairy Sci. 2020, 103, 6782–6797. [Google Scholar] [CrossRef]
- Haas, G.; Cetin, S.; Messmer, M.; Chane-Woon-Ming, B.; Terenzi, O.; Chicher, J.; Kuhn, L.; Hammann, P.; Pfeffer, S. Identification of factors involved in target RNA-directed microRNA degradation. Nucleic Acids Res. 2016, 44, 2873–2887. [Google Scholar] [CrossRef] [Green Version]
- Ghini, F.; Rubolino, C.; Climent, M.; Simeone, I.; Marzi, M.J.; Nicassio, F. Endogenous transcripts control miRNA levels and activity in mammalian cells by target-directed miRNA degradation. Nat. Commun. 2018, 9, 3119. [Google Scholar] [CrossRef]
- Sheu-Gruttadauria, J.; Pawlica, P.; Klum, S.M.; Wang, S.; Yario, T.A.; Schirle Oakdale, N.T.; Steitz, J.A.; MacRae, I.J. Structural Basis for Target-Directed MicroRNA Degradation. Mol. Cell 2019, 75, 1243–1255.e7. [Google Scholar] [CrossRef]
- Pawlica, P.; Sheu-Gruttadauria, J.; MacRae, I.J.; Steitz, J.A. How Complementary Targets Expose the microRNA 3’ End for Tailing and Trimming during Target-Directed microRNA Degradation. Cold Spring Harb. Symp. Quant. Biol. 2019, 84, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Mei, J.; Jiang, N.; Ren, G.J. The F-box protein HAWAIIAN SKIRT is required for mimicry target-induced microRNA degradation in Arabidopsis. J. Integr. Plant. Biol. 2019, 61, 1121–1127. [Google Scholar] [CrossRef]
- Shi, C.Y.; Kingston, E.R.; Kleaveland, B.; Lin, D.H.; Stubna, M.W.; Bartel, D.P. The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science 2020, 370, eabc9359. [Google Scholar] [CrossRef] [PubMed]
- Simeone, I.C.; Rubolino, T.; Noviello, M.R.; Farinello, D.; Cerulo, L.; Marzi, M.; Nicassio, J.F. Prediction and pan-cancer analysis of mammalian transcripts involved in target directed miRNA degradation. Nucleic Acids Res. 2022, 50, 2019–2035. [Google Scholar] [CrossRef] [PubMed]
- Bryzgunova, O.; Konoshenko, M.; Zaporozhchenko, I.; Yakovlev, A.; Laktionov, P. Isolation of Cell-Free miRNA from Biological Fluids: Influencing Factors and Methods. Diagnostics 2021, 11, 865. [Google Scholar] [CrossRef]
- Blochm, S.; Węgrzyn, A.; Węgrzyn, G.; Nejman-Faleńczyk, B. Small and Smaller—sRNAs and MicroRNAs in the Regulation of Toxin Gene Expression in Prokaryotic Cells: A Mini-Review. Toxins 2017, 9, 181. [Google Scholar] [CrossRef]
- Chernikov, I.V.; Gladkikh, D.V.; Meschaninova, M.I.; Ven’yaminova, A.G.; Zenkova, M.A.; Vlassov, V.V.; Chernolovskaya, E.L. Cholesterol-containing nuclease-resistant siRNA. Accumulates in tumors in a carrier-free mode and silences MDR1 gene. Mol. Ther. Nucleic Acids 2017, 6, 209–220. [Google Scholar] [CrossRef]





| Number of Donor | MicroRNA Content (Number of Copies in 1 mg of Milk Plasma) | ||||
|---|---|---|---|---|---|
| miR-148a-3p | let-7f-5p | miR-146b-5p | miR-200c-3p | miR-378a-3p | |
| Number of Parameter | |||||
| p1 | p2 | p3 | p4 | p5 | |
| 1 | 1.2 × 108 * | 1.4 × 1010 | 1.2 × 109 | 3.7 × 109 | 6.5 × 107 |
| 2 | 8.0 × 106 | 6.2 × 109 | 1.1 × 109 | 1.4 × 108 | 2.8 × 107 |
| 3 | 1.8 × 107 | 5.2 × 109 | 8.6 × 108 | 1.5 × 109 | 7.0 × 106 |
| 4 | 6.0 × 106 | 1.0 × 1010 | 1.1 × 109 | 1.2 × 108 | 3.0 × 106 |
| 5 | 1.3 × 108 | 4.8 × 109 | 8.7 × 108 | 4.2 × 109 | 1.4 × 108 |
| 6 | 1.1 × 108 | 2.5 × 109 | 4.2 × 108 | 3.8 × 109 | 9.8 × 107 |
| 7 | 1.3 × 107 | 4.9 × 109 | 9.6 × 108 | 5.7 × 107 | 4.0 × 106 |
| CCs between parameters | 1–2 (0.20); 1–3 (−0.005); 1–4 (0.67); 1–5 (0.92); 2–3 (0.81); 2–4 (−0.19); 2–5 (−0.07); 3–4 (0.58); 3–5 (0.14); 4–5 (0.66) | ||||
| Number of donor | MicroRNA content in cells (number of copies in 1 mg of milk cells) | ||||
| miR-148a-3p | let-7f-5p | miR-146b-5p | miR-200c-3p | miR-378a-3p | |
| Number of parameter | |||||
| p1 | p2 | p3 | p4 | p5 | |
| 1 | 8.1 × 107 | 5.0 × 1010 | 1.2 × 1010 | 9.2 × 109 | 1.1 × 108 |
| 2 | 1.2 × 108 | 4.4 × 1010 | 9.4 × 109 | 5.4 × 109 | 2.0 × 108 |
| 3 | 5.8 × 107 | 4.4 × 1010 | 1.2 × 1010 | 2.6 × 109 | 1.5 × 108 |
| 4 | 9.7 × 108 | 8.1 × 1010 | 2.3 × 1010 | 4.0 × 109 | 1.2 × 108 |
| 5 | 3.2 × 108 | 5.5 × 1010 | 2.0 × 1010 | 5.3 × 109 | 4.1 × 108 |
| 6 | 8.8 × 108 | 4.5 × 1010 | 1.1 × 1010 | 2.4 × 109 | 1.6 × 108 |
| 7 | 4.2 × 107 | 3.7 × 1010 | 1.0 × 1010 | 2.2 × 109 | 2.1 × 108 |
| CCs between parameters | 1–2 (0.24); 1–3 (0.23); 1–4 (0.28); 1–5 (−0.17); 2–3 (0.99); 2–4 (0.86); 2–5 (−0.37); 3–4 (0.86); 3–5 (0.33); 4–5 (0.04) | ||||
| MicroRNA content (number of copies in 1 mg of milk lipid fraction) | |||||
| Number of donor | miR-148a-3p | let-7f-5p | miR-146b-5p | miR-200c-3p | miR-378a-3p |
| Number of parameter | |||||
| p1 | p2 | p3 | p4 | p5 | |
| 1 | 7.2 × 107 | 4.1 × 1010 | 1.6 × 1010 | 3.1 × 109 | 4.1 × 107 |
| 2 | 1.3 × 107 | 7.2 × 1010 | 1.6 × 1010 | 3.4 × 109 | 3.7 × 107 |
| 3 | 4.0 × 106 | 2.4 × 1010 | 3.6 × 1010 | 1.6 × 109 | 8.3 × 107 |
| 4 | 6.2 × 108 | 3.1 × 1010 | 6.4 × 109 | 2.6 × 109 | 7.1 × 107 |
| 5 | 2.0 × 106 | 1.0 × 1010 | 4.6 × 109 | 5.9 × 108 | 4.9 × 107 |
| 6 | 4.1 × 108 | 6.6 × 109 | 2.0 × 109 | 2.1 × 108 | 2.3 × 107 |
| 7 | 2.6 × 107 | 7.4 × 109 | 2.3 × 109 | 1.9 × 109 | 1.6 × 107 |
| CCs between parameters | 1–2 (0.15); 1–3 (0.40); 1–4 (0.10); 1–5 (0.28); 2–3 (0.38); 2–4 (0.84); 2–5 (−0.32); 3–4 (0.29); 3–5 (0.26); 4–5 (0.48) | ||||
| CCs reflecting the content of each of the five RNAs (parameters p1–p5) in plasma, cells, and lipid fraction | |||||
| p1 cells | p1 lipid fraction | p2 cells | p2 lipid fraction | ||
| 1 plasma | 0.57 | 0.49 | 2 plasma | 0.18 | 0.30 |
| 1 cells | - | 0.94 | 2 cells | - | 0.26 |
| p3 cells | p3 lipid fraction | - | p4 cells | p4 lipid fraction | |
| 3 plasma | 0.4 | 0.62 | 4 plasma | −0.22 | 0.003 |
| 3 cells | - | 0.12 | 4 cells | - | 0.54 |
| p5 cells | p5 lipid fraction | - | - | - | |
| 5 plasma | 0.25 | 0.46 | - | - | - |
| 5 cells | 0.92 | - | - | - | |
| MicroRNAs | Number of Copies in 1 mg of the Analyzed Sample | |||||
|---|---|---|---|---|---|---|
| Cells | Lipid Fraction | Plasma | ||||
| Diapason * | Average Value (M and IQR) ** | Diapason | Average Value (M and IQR) | Diapason | Average Value (M and IQR) | |
| miR-148a-3p | 4.2 × 107–9.7 × 108 | 3.8 × 108 ± 1.3 × 108 (2.2 × 108; 5 × 108) | 2.0 × 106–6.0 × 108 | 2.0 × 108 ± 9.0 × 107 (5 × 107; 4 × 108) | 2 × 106–1 × 108 | 5.1 × 107 ± 2.0 × 107 (1.6 × 107; 1.0 × 108) |
| miR-200c-3p | 1.0 × 109–9.0 × 109 | 4.0 × 109 ± 9.0 × 108 (3.0 × 109; 4.2 × 109) | 2.1 × 108–3.4 × 109 | 1.8 × 109 ± 4.0 × 108 (1.7 × 109; 2.0 × 109) | 6.0 × 107–4.0 × 109 | 2.0 × 109 ± 7.0 × 108 (3.0 × 109; 3.6 × 109) |
| miR-378a-3p | 6.2 × 107–4.1 × 109 | 1.8 × 108 ± 3.7 × 107 (1.5 × 108; 5.0 × 108) | 2.0 × 107–8.0 × 107 | 4.0 × 107 ± 8.0 × 106 (4.0 × 107; 4.0 × 108) | 2.0 × 106–1.0 × 108 | 4.0 × 107 ± 2.0 × 107 (2.0 × 107; 1.0 × 108) |
| let-7f-5p | 1.8 × 1010–8.1 × 1010 | 4.6 × 1010 ± 6.0 × 109 (4.5 × 1010; 3.2 × 1010) | 4.1 × 109–7.2 × 1010 | 2.5 × 1010 ± 8.0 × 109 (1.7 × 1010; 2.4 × 1010) | 2.1 × 109–1.4 × 1010 | 6.2 × 109 ± 1.0 × 109 (5.0 × 109; 3.6 × 109) |
| miR-146b-5p | 4.6 × 109–2.3 × 1010 | 1.2 × 1010 ± 2.3 × 109 (1.2 × 1010; 7.8 × 109) | 1.1 × 109–3.6 × 1010 | 4.6 × 1010 ± 6.0 × 109 (5.4 × 109; 1.4 × 1010) | 3.3 × 108–1.2 × 109 | 8.5 × 108 ± 1.0 × 108 (9.2 × 108; 6.7 × 108) |
| Average content of all microRNAs | 1.25 × 1010 ± 1.93 × 1010 | 1.46 × 1010 ± 2.4 × 1010 | 0.18 × 1010 ± 0.04 × 1010 | |||
| Abs Number | miR-148a-3p | miR-200c-3p | miR-378a-3p | let-7f-5p | miR-146b-5p |
|---|---|---|---|---|---|
| Number of Parameter | |||||
| p1 | p2 | p3 | p4 | p5 | |
| IgG1 | 39.0 * | 62.1 | 69.8 | 72.1 | 99.5 |
| IgG2 | 21.0 | 3.9 | 8.2 | 34.6 | 17.6 |
| IgG3 | 58.2 | 25.2 | 7.2 | 61.5 | 55.6 |
| IgG4 | 22.6 | 15.1 | 8.5 | 63.1 | 97.6 |
| IgG5 | 32.4 | 28.3 | 76.1 | 66.3 | 99.7 |
| IgG6 | 12.7 | 4.7 | 12.1 | 40.21 | 12.3 |
| IgG7 | 21.3 | 82.9 | 17.9 | 76.0 | 42.3 |
| Average values | 29.6 ± 15.2 | 31.7 ± 29.9 | 28.5 ± 30.6 | 59.1 ± 15.7 | 60.7 ± 38.6 |
| M; (IQR) ** | 22.6 (18.0) | 25.2 (57.4) | 12.1 (61.6) | 63.1 (31.9) | 55.6 (81.9) |
| Correl. Coef. | 1–2 (0.17); 1–3 (0.21); 1–4 (0.39); 1–5 (0.40); 2–3 (0.37); 2–4 (0.83); 2–5 (0.29); 3–4 (0.48); 3–5 (0.66); 4–5 (0.71) | ||||
| Difference (p) | 1–2 (0.002); 1–3 (0.002); 1–4 (0.02); 1–5 (0.002); 2–3 (0.002); 2–4 (0.03); 2–5 (0.002); 3–4 (0.03); 3–5 (0.002); 4–5 (0.002) | ||||
| Number of parameter | |||||
| Group number | 6 | 7 | 8 | 9 | 10 |
| sIgA1 | 98.7 | 99.0 | 32.6 | 99.0 | 54.1 |
| sIgA2 | 13.8 | 4.9 | 17.2 | 26.5 | 24.0 |
| sIgA3 | 33.7 | 52.0 | 43.7 | 62.2 | 40.2 |
| sIgA4 | 32.1 | 77.1 | 12.9 | 83.0 | 48.7 |
| sIgA5 | 67.4 | 94.0 | 9.2 | 98.3 | 39.5 |
| sIgA6 | 81.8 | 4.1 | 21.7 | 24.0 | 15.3 |
| sIgA7 | 66.4 | 40.4 | 9.4 | 70.2 | 14.7 |
| Average values | 56.3 ± 30.5 | 53.1 ± 39.2 | 21.0 ± 12.9 | 66.2 ± 31.0 | 33.8 ± 15.9 |
| M and (IQR) | 66.4 (98.7) | 52.0 (99.0) | 17.2 (43.7) | 70.2 (99.0) | 39.5 (54.1) |
| Correl. Coef. | 6–7 (0.3); 6–8 (0.04); 6–9 (0.37); 6–10 (0.06); 7–8 (0.04); 7–9 (0.98); 7–10 (0.84); 8–9 (−0.08); 8–10 (0.36); 9–10 (0.73) | ||||
| Difference (p) *** | 6–7 (0.7); 6–8 (0.002); 6–9 (0.7); 6–10 (0.03); 7–8 (0.002); 7–9 (0.4); 7–10 (0.03); 8–9 (0.7); 8–10 (0.03); 9–10 (0.03) | ||||
| CC between IgGs and sIgAs | 1–6 (−0.07); 2–7 (−0.41); 3–8 (−0.09); 4–9 (0.87); 5–10 (0.87) | ||||
| Difference between IgGs and sIgAs (p) | 1–6 (0.002); 2–7 (0.70); 3–8 (0.002); 4–9 (0.50); 5–10 (0.03) | ||||
| MicroRNAs | miR-200c-3p | miR-378a-3p | let-7f-5p | miR-146b-5p |
|---|---|---|---|---|
| miR-148a-3p | 0.97 | 0.99 | 0.07 | 0.33 |
| miR-378a-3p | 0.98 | - | 0.07 | 0.33 |
| let-7f-5p | 0.007 | 0.07 | - | 0.79 |
| miR-146b-5p | 0.42 | 0.33 | 0.79 | - |
| Antibodies | MicroRNAs | ||||
|---|---|---|---|---|---|
| miR-148a-3p | miR-200c-3p | miR-378a-3p | let-7f-5p | miR-146b-5p | |
| IgGs | −0.01 | −0.05 | 0.79 | 0.41 | 0.53 |
| sIgAs | 0.8 | 0.33 | 0.03 | 0.59 | 0.59 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kompaneets, I.Y.; Ermakov, E.A.; Buneva, V.N.; Nevinsky, G.A. MicroRNAs of Milk in Cells, Plasma, and Lipid Fractions of Human Milk, and Abzymes Catalyzing Their Hydrolysis. Int. J. Mol. Sci. 2022, 23, 12070. https://doi.org/10.3390/ijms232012070
Kompaneets IY, Ermakov EA, Buneva VN, Nevinsky GA. MicroRNAs of Milk in Cells, Plasma, and Lipid Fractions of Human Milk, and Abzymes Catalyzing Their Hydrolysis. International Journal of Molecular Sciences. 2022; 23(20):12070. https://doi.org/10.3390/ijms232012070
Chicago/Turabian StyleKompaneets, Ivan Yu., Evgeny A. Ermakov, Valentina N. Buneva, and Georgy A. Nevinsky. 2022. "MicroRNAs of Milk in Cells, Plasma, and Lipid Fractions of Human Milk, and Abzymes Catalyzing Their Hydrolysis" International Journal of Molecular Sciences 23, no. 20: 12070. https://doi.org/10.3390/ijms232012070






