Monodelphis domestica Induced Pluripotent Stem Cells Reveal Metatherian Pluripotency Architecture
Abstract
:1. Introduction
2. Results
2.1. monoiPSC Reprogramming and Validation
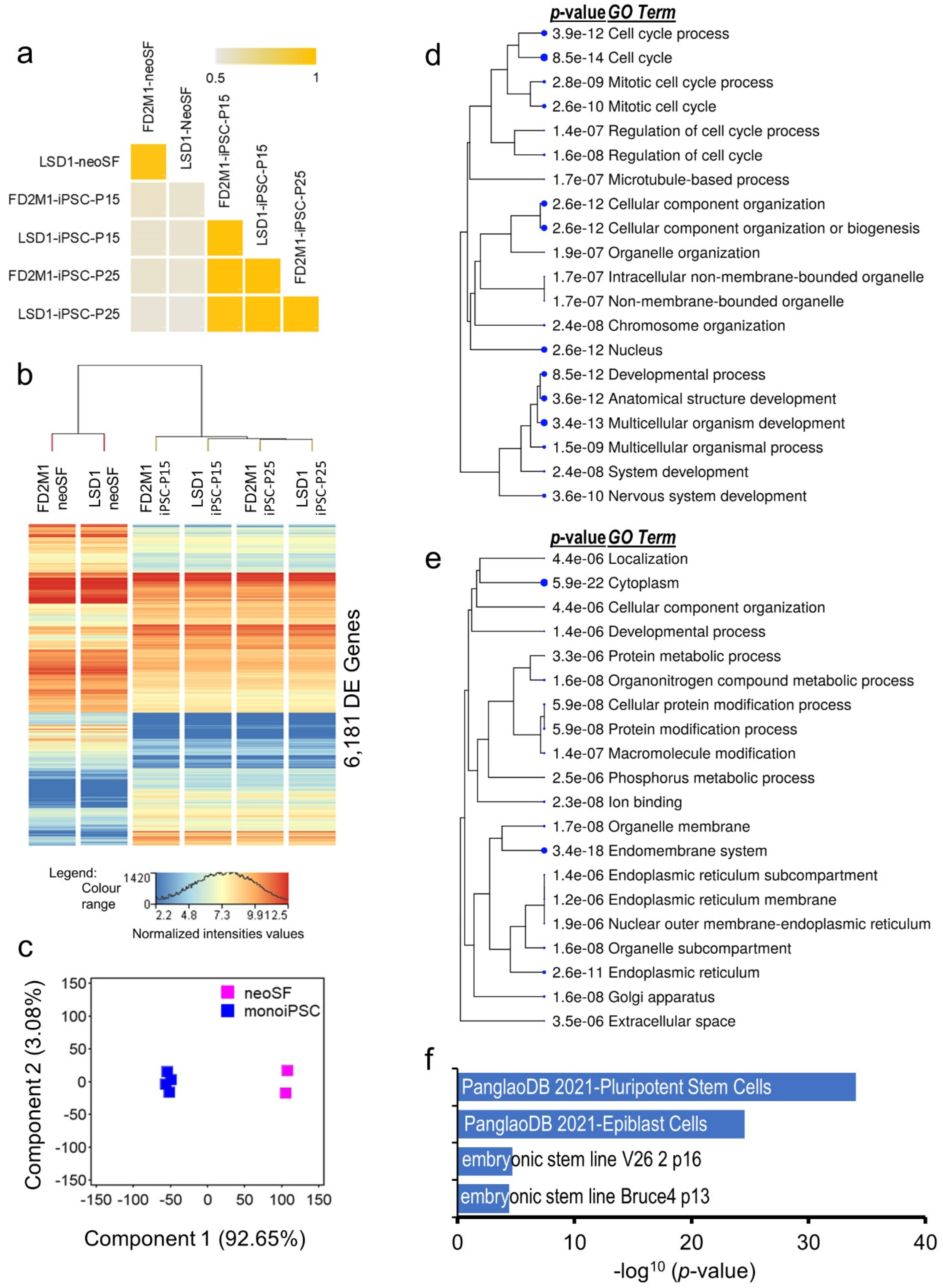
2.2. Transcriptomic and Functional Architecture of monoiPSC
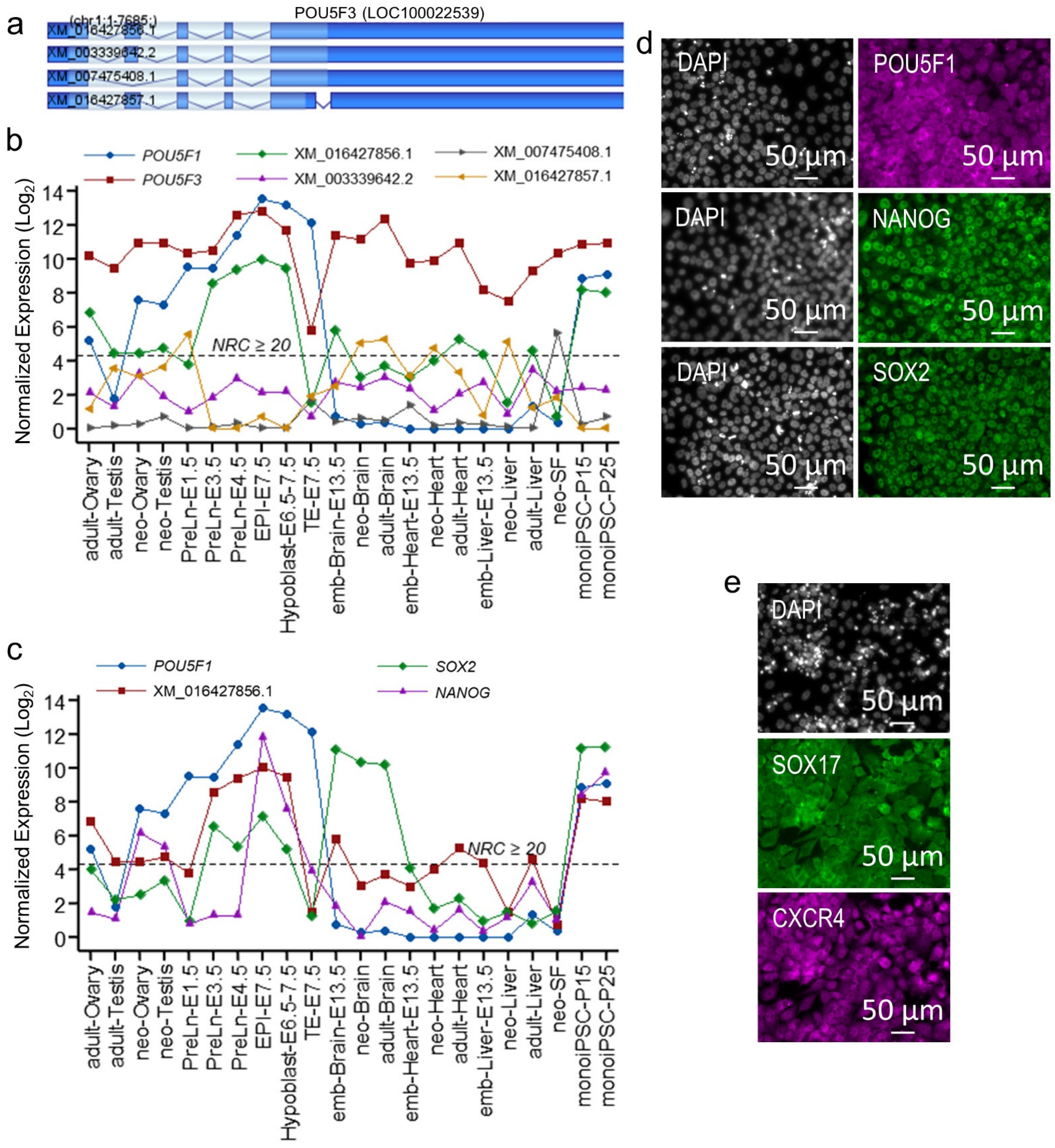
2.3. POU5F1 and POU5F3 Synergistically Regulate Metatherian Pluripotency
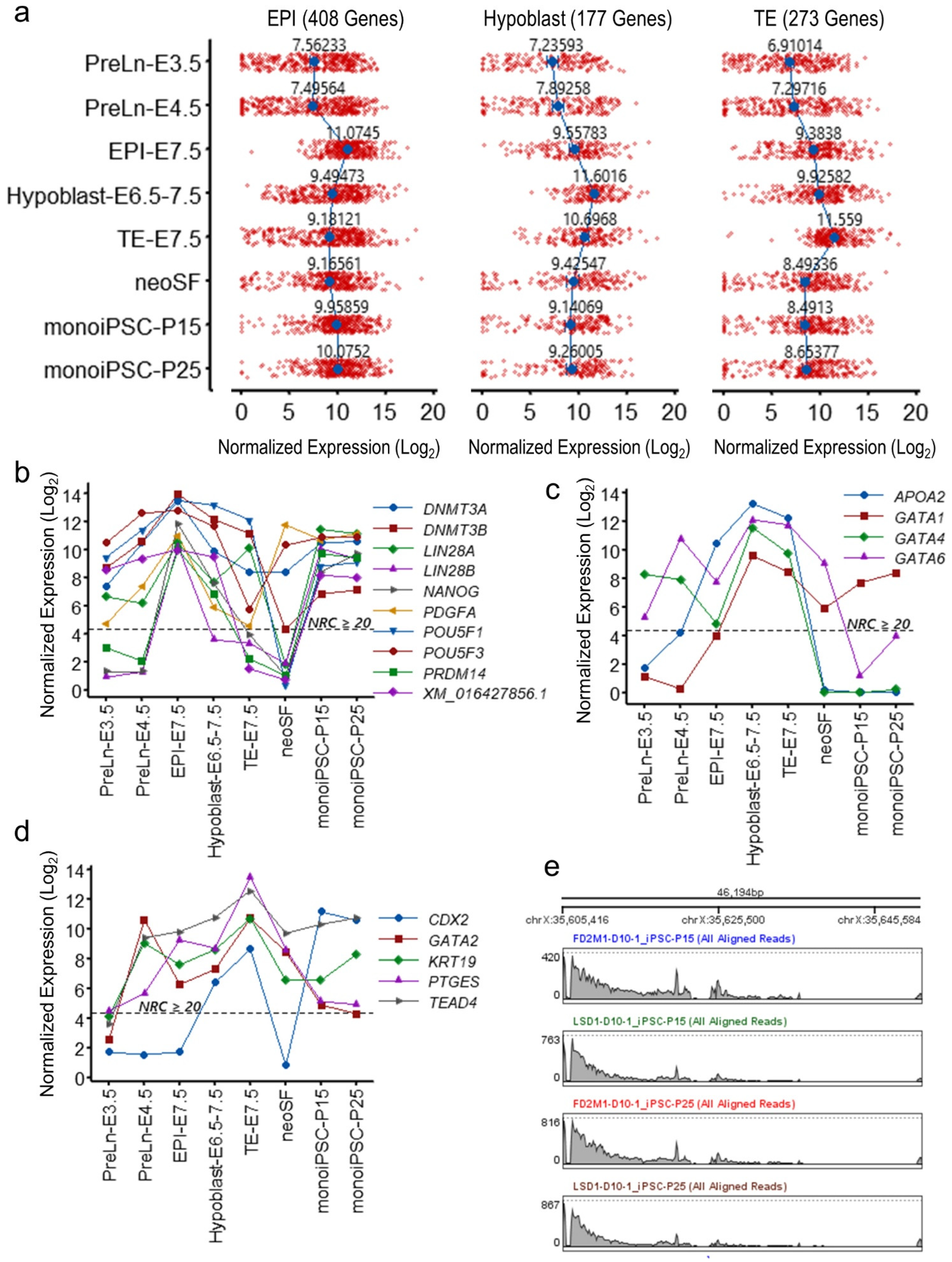
2.4. The monoiPSC Embryonic Lineage Marker Repertoire Is Akin to That of EPI
2.5. Transcriptional Architectures of monoiPSCs and Eutherian iPSCs Are Highly Similar
3. Discussion
4. Materials and Methods
4.1. Data Reporting
4.2. Animal Maintenance and Sample Collection
4.3. Neonatal Skin Fibroblast Culture
4.4. monoiPSC Reprogramming
4.5. monoiPSC Differentiation
4.6. Immunocytochemistry Analysis
4.7. RNA Sequencing
4.8. Additional Published RNA Sequencing Data
4.9. RNA Sequencing Analyses
4.10. eKaryotyping
4.11. Differential Gene Expression Analyses
4.12. Functional Annotations and Enrichment Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Halliday, T.J.; Upchurch, P.; Goswami, A. Eutherians experienced elevated evolutionary rates in the immediate aftermath of the Cretaceous-Palaeogene mass extinction. Proc. Biol. Sci. 2016, 283, 20153026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedges, S.B.; Kumar, S. Precision of molecular time estimates. Trends Genet. 2004, 20, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.X.; Yuan, C.X.; Meng, Q.J.; Ji, Q. A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature 2011, 476, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Samollow, P.B. The opossum genome: Insights and opportunities from an alternative mammal. Genome Res. 2008, 18, 1199–1215. [Google Scholar] [CrossRef] [Green Version]
- Das, R.; Anderson, N.; Koran, M.I.; Weidman, J.R.; Mikkelsen, T.S.; Kamal, M.; Murphy, S.K.; Linblad-Toh, K.; Greally, J.M.; Jirtle, R.L. Convergent and divergent evolution of genomic imprinting in the marsupial Monodelphis domestica. BMC Genom. 2012, 13, 394. [Google Scholar] [CrossRef] [Green Version]
- Graves, J.A.; Renfree, M.B. Marsupials in the age of genomics. Annu. Rev. Genom. Hum. Genet. 2013, 14, 393–420. [Google Scholar] [CrossRef]
- Mahadevaiah, S.K.; Royo, H.; VandeBerg, J.L.; McCarrey, J.R.; Mackay, S.; Turner, J.M. Key features of the X inactivation process are conserved between marsupials and eutherians. Curr. Biol. CB 2009, 19, 1478–1484. [Google Scholar] [CrossRef] [Green Version]
- Murchison, E.P.; Schulz-Trieglaff, O.B.; Ning, Z.; Alexandrov, L.B.; Bauer, M.J.; Fu, B.; Hims, M.; Ding, Z.; Ivakhno, S.; Stewart, C.; et al. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell 2012, 148, 780–791. [Google Scholar] [CrossRef] [Green Version]
- Saunders, N.R.; Noor, N.M.; Dziegielewska, K.M.; Wheaton, B.J.; Liddelow, S.A.; Steer, D.L.; Ek, C.J.; Habgood, M.D.; Wakefield, M.J.; Lindsay, H.; et al. Age-dependent transcriptome and proteome following transection of neonatal spinal cord of Monodelphis domestica (South American grey short-tailed opossum). PLoS ONE 2014, 9, e99080. [Google Scholar] [CrossRef]
- VandeBerg, J.; Williams-Blangero, S. The Laboratory Opossum. In The UFAW Handbook on the Care and Management of Laboratory and Other Research Animals, 8th ed.; Hubrecht, R., Kirkwood, J., Eds.; Wiley-Blackwell: Oxford, UK, 2010; pp. 246–261. [Google Scholar] [CrossRef]
- VandeBerg, J.L.; Robinson, E.S. The laboratory opossum (Monodelphis domestica) in laboratory research. ILAR J. 1997, 38, 4–12. [Google Scholar] [CrossRef] [Green Version]
- VandeBerg, J.L.; Williams-Blangero, S.; Hubbard, G.B.; Ley, R.D.; Robinson, E.S. Genetic analysis of ultraviolet radiation-induced skin hyperplasia and neoplasia in a laboratory marsupial model (Monodelphis domestica). Arch. Dermatol. Res. 1994, 286, 12–17. [Google Scholar] [CrossRef]
- Frankenberg, S.R.; de Barros, F.R.; Rossant, J.; Renfree, M.B. The mammalian blastocyst. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 210–232. [Google Scholar] [CrossRef]
- Frankenberg, S.; Shaw, G.; Freyer, C.; Pask, A.J.; Renfree, M.B. Early cell lineage specification in a marsupial: A case for diverse mechanisms among mammals. Development 2013, 140, 965–975. [Google Scholar] [CrossRef] [Green Version]
- Morrison, J.T.; Bantilan, N.S.; Wang, V.N.; Nellett, K.M.; Cruz, Y.P. Expression patterns of Oct4, Cdx2, Tead4, and Yap1 proteins during blastocyst formation in embryos of the marsupial, Monodelphis domestica Wagner. Evol. Dev. 2013, 15, 171–185. [Google Scholar] [CrossRef]
- Frankenberg, S.; Pask, A.; Renfree, M.B. The evolution of class V POU domain transcription factors in vertebrates and their characterisation in a marsupial. Dev. Biol. 2010, 337, 162–170. [Google Scholar] [CrossRef] [Green Version]
- Frankenberg, S.R.; Frank, D.; Harland, R.; Johnson, A.D.; Nichols, J.; Niwa, H.; Scholer, H.R.; Tanaka, E.; Wylie, C.; Brickman, J.M. The POU-er of gene nomenclature. Development 2014, 141, 2921–2923. [Google Scholar] [CrossRef] [Green Version]
- Weeratunga, P.; Shahsavari, A.; Ovchinnikov, D.A.; Wolvetang, E.J.; Whitworth, D.J. Induced pluripotent stem cells from a marsupial, the Tasmanian Devil (Sarcophilus harrisii): Insight into the evolution of mammalian pluripotency. Stem Cells Dev. 2018, 27, 112–122. [Google Scholar] [CrossRef]
- Mahadevaiah, S.K.; Sangrithi, M.N.; Hirota, T.; Turner, J.M.A. A single-cell transcriptome atlas of marsupial embryogenesis and X inactivation. Nature 2020, 586, 612–617. [Google Scholar] [CrossRef]
- Xiong, X.; Samollow, P.B.; Cao, W.; Metz, R.; Zhang, C.; Leandro, A.C.; VandeBerg, J.L.; Wang, X. Genetic and genomic architecture in eight strains of the laboratory opossum Monodelphis domestica. G3 2022, 12, jkab389. [Google Scholar] [CrossRef]
- Okita, K.; Matsumura, Y.; Sato, Y.; Okada, A.; Morizane, A.; Okamoto, S.; Hong, H.; Nakagawa, M.; Tanabe, K.; Tezuka, K.; et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods 2011, 8, 409–412. [Google Scholar] [CrossRef] [Green Version]
- Okita, K.; Yamakawa, T.; Matsumura, Y.; Sato, Y.; Amano, N.; Watanabe, A.; Goshima, N.; Yamanaka, S. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 2013, 31, 458–466. [Google Scholar] [CrossRef]
- Hall, J.; Guo, G.; Wray, J.; Eyres, I.; Nichols, J.; Grotewold, L.; Morfopoulou, S.; Humphreys, P.; Mansfield, W.; Walker, R.; et al. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell 2009, 5, 597–609. [Google Scholar] [CrossRef]
- Lingjaerde, O.C.; Baumbusch, L.O.; Liestol, K.; Glad, I.K.; Borresen-Dale, A.L. CGH-Explorer: A program for analysis of array-CGH data. Bioinformatics 2005, 21, 821–822. [Google Scholar] [CrossRef] [Green Version]
- Weissbein, U.; Schachter, M.; Egli, D.; Benvenisty, N. Analysis of chromosomal aberrations and recombination by allelic bias in RNA-Seq. Nat. Commun. 2016, 7, 12144. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Curran, J.E.; Glahn, D.C.; Blangero, J. Utility of lymphoblastoid cell lines for induced pluripotent stem cell generation. Stem. Cells Int. 2016, 2016, 2349261. [Google Scholar] [CrossRef] [Green Version]
- Franzen, O.; Gan, L.M.; Bjorkegren, J.L.M. PanglaoDB: A web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, 2019, baz046. [Google Scholar] [CrossRef] [Green Version]
- Cardoso-Moreira, M.; Halbert, J.; Valloton, D.; Velten, B.; Chen, C.; Shao, Y.; Liechti, A.; Ascencao, K.; Rummel, C.; Ovchinnikova, S.; et al. Gene expression across mammalian organ development. Nature 2019, 571, 505–509. [Google Scholar] [CrossRef]
- Strumpf, D.; Mao, C.A.; Yamanaka, Y.; Ralston, A.; Chawengsaksophak, K.; Beck, F.; Rossant, J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 2005, 132, 2093–2102. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Gentile, L.; Fuchikami, T.; Sutter, J.; Psathaki, K.; Esteves, T.C.; Arauzo-Bravo, M.J.; Ortmeier, C.; Verberk, G.; Abe, K.; et al. Initiation of trophectoderm lineage specification in mouse embryos is independent of Cdx2. Development 2010, 137, 4159–4169. [Google Scholar] [CrossRef]
- Yagi, R.; Kohn, M.J.; Karavanova, I.; Kaneko, K.J.; Vullhorst, D.; DePamphilis, M.L.; Buonanno, A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 2007, 134, 3827–3836. [Google Scholar] [CrossRef] [Green Version]
- Grant, J.; Mahadevaiah, S.K.; Khil, P.; Sangrithi, M.N.; Royo, H.; Duckworth, J.; McCarrey, J.R.; VandeBerg, J.L.; Renfree, M.B.; Taylor, W.; et al. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 2012, 487, 254–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lachmann, A.; Xu, H.; Krishnan, J.; Berger, S.I.; Mazloom, A.R.; Ma’ayan, A. ChEA: Transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 2010, 26, 2438–2444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duchene, D.A.; Bragg, J.G.; Duchene, S.; Neaves, L.E.; Potter, S.; Moritz, C.; Johnson, R.N.; Ho, S.Y.W.; Eldridge, M.D.B. Analysis of phylogenomic tree space resolves relationships among marsupial families. Syst. Biol. 2018, 67, 400–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsson, M.A.; Arnason, U.; Spencer, P.B.; Janke, A. Marsupial relationships and a timeline for marsupial radiation in South Gondwana. Gene 2004, 340, 189–196. [Google Scholar] [CrossRef]
- Chan, J.; Sharkey, F.E.; Kushwaha, R.S.; VandeBerg, J.F.; VandeBerg, J.L. Steatohepatitis in laboratory opossums exhibiting a high lipemic response to dietary cholesterol and fat. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G12–G19. [Google Scholar] [CrossRef] [Green Version]
- Hansen, V.L.; Faber, L.S.; Salehpoor, A.A.; Miller, R.D. A pronounced uterine pro-inflammatory response at parturition is an ancient feature in mammals. Proc. Biol. Sci. 2017, 284, 20171694. [Google Scholar] [CrossRef] [Green Version]
- Farrell, G.C.; van Rooyen, D. Liver cholesterol: Is it playing possum in NASH? Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G9–G11. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Wei, Y.; Duan, J.; Schmitz, D.A.; Sakurai, M.; Wang, L.; Wang, K.; Zhao, S.; Hon, G.C.; Wu, J. Blastocyst-like structures generated from human pluripotent stem cells. Nature 2021, 591, 620–626. [Google Scholar] [CrossRef]
- Dooley, T.P.; Mattern, V.L.; Moore, C.M.; Porter, P.A.; Robinson, E.S.; VandeBerg, J.L. Cell lines derived from ultraviolet radiation-induced benign melanocytic nevi in Monodelphis domestica exhibit cytogenetic aneuploidy. Cancer Genet. Cytogenet. 1993, 71, 55–66. [Google Scholar] [CrossRef]
- Robinson, E.S.; Dooley, T.P.; Williams, K.L. UV-induced melanoma cell lines and their potential for proteome analysis: A review. J. Exp. Zool. 1998, 282, 48–53. [Google Scholar] [CrossRef]
- Ma, X.; Dighe, A.; Maziarz, J.; Neumann, E.; Erkenbrack, E.; Hei, Y.Y.; Liu, Y.; Suhail, Y.; Kshitiz; Pak, I.; et al. Evolution of higher mesenchymal CD44 expression in the human lineage: A gene linked to cancer malignancy. Evol. Med. Public Health. 2022, 10, 447–462. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, S.; De Leon, E.M.; Granados, J.; Whitworth, D.J.; VandeBerg, J.L. Monodelphis domestica Induced Pluripotent Stem Cells Reveal Metatherian Pluripotency Architecture. Int. J. Mol. Sci. 2022, 23, 12623. https://doi.org/10.3390/ijms232012623
Kumar S, De Leon EM, Granados J, Whitworth DJ, VandeBerg JL. Monodelphis domestica Induced Pluripotent Stem Cells Reveal Metatherian Pluripotency Architecture. International Journal of Molecular Sciences. 2022; 23(20):12623. https://doi.org/10.3390/ijms232012623
Chicago/Turabian StyleKumar, Satish, Erica M. De Leon, Jose Granados, Deanne J. Whitworth, and John L. VandeBerg. 2022. "Monodelphis domestica Induced Pluripotent Stem Cells Reveal Metatherian Pluripotency Architecture" International Journal of Molecular Sciences 23, no. 20: 12623. https://doi.org/10.3390/ijms232012623





