The Origin, Function, Distribution, Quantification, and Research Advances of Extracellular DNA
Abstract
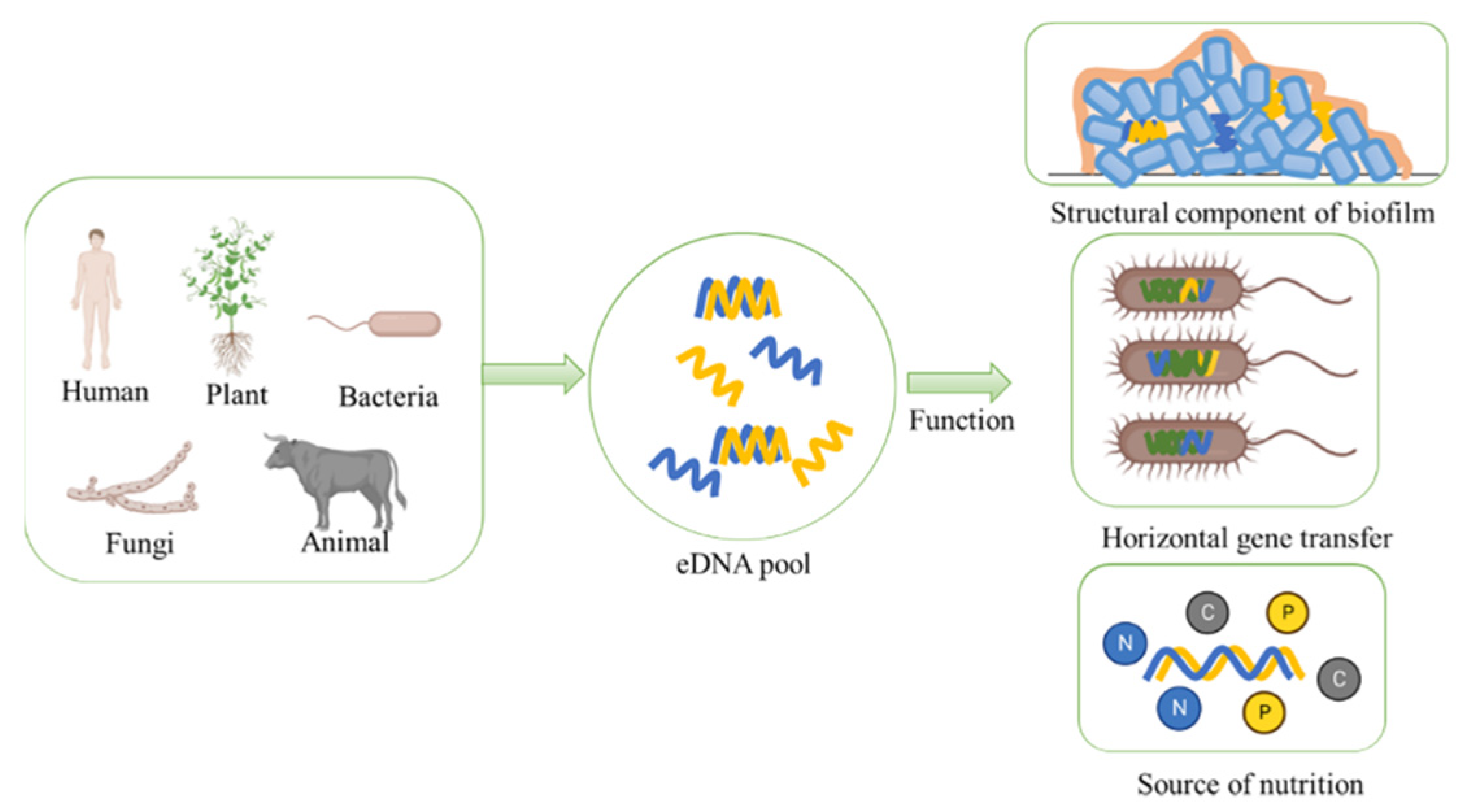
:1. Introduction
2. The Origin of eDNA
2.1. Lysis-Dependent Pathway of eDNA Release
2.2. Lysis-Independent Pathway of eDNA Release
3. Ecological Functions of eDNA
3.1. eDNA Serves as an Essential Structural Component of Biofilm Matrix
3.2. eDNA Acts as Genetic Information Carrier in HGT
3.3. eDNA Serves as a Main Organic Compound in Nutrient Cycle
4. Distribution of eDNA in Different Environments
4.1. eDNA in Soil
4.2. eDNA in Sediments
4.3. eDNA in Feces
4.4. eDNA in Other Ecological Environments
5. Research Advances of eDNA in Model Organisms
5.1. eDNA in Bacillus
5.2. eDNA in Pseudomonas
5.3. eDNA in Escherichia
5.4. eDNA in Other Microorganisms
6. Methods for eDNA Extraction and Quantification
7. Summary and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bairoliya, S.; Xiang, J.K.Z.; Cao, B. Extracellular DNA in Environmental Samples: Occurrence, Extraction, Quantification, and Impact on Microbial Biodiversity Assessment. Appl. Environ. Microbiol. 2022, 88, e01845-21. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, T.A.; Brill, A.; Duerschmied, D.; Schatzberg, D.; Monestier, M.; Myers, D.D.; Wrobleski, S.K.; Wakefield, T.W.; Hartwig, J.H.; Wagner, D.D. Extracellular DNA traps promote thrombosis. Proc. Natl. Acad. Sci. USA 2010, 107, 15880–15885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gartler, S.M. Cellular uptake of deoxyribonucleic acid by human tissue culture cells. Nature 1959, 184 (Suppl. S19), 1505–1506. [Google Scholar] [CrossRef] [PubMed]
- Mandel, P.; Metais, P.; Métais, P. Les acides nucléiques du plasma sanguin chez l’homme. Cr. Acad. Sci. Paris 1948, 142, 241–243. [Google Scholar]
- Torti, A.; Jørgensen, B.B.; Lever, M.A. Preservation of microbial DNA in marine sediments: Insights from extracellular DNA pools. Environ. Microbiol. 2018, 20, 4526–4542. [Google Scholar] [CrossRef]
- Gómez-Brandón, M.; Ascher-Jenull, J.; Bardelli, T.; Fornasier, F.; Sartori, G.; Pietramellara, G.; Arfaioli, P.; Egli, M.; Beylich, A.; Insam, H.; et al. Ground cover and slope exposure effects on micro- and mesobiota in forest soils. Ecol. Indic. 2017, 80, 174–185. [Google Scholar] [CrossRef]
- Gómez-Brandón, M.; Ascher-Jenull, J.; Bardelli, T.; Fornasier, F.; Fravolini, G.; Arfaioli, P.; Ceccherini, M.T.; Pietramellara, G.; Lamorski, K.; Sławiński, C.; et al. Physico-chemical and microbiological evidence of exposure effects on Picea abies—Coarse woody debris at different stages of decay. Forest Ecol. Manag. 2017, 391, 376–389. [Google Scholar] [CrossRef]
- Nagler, M.; Podmirseg, S.M.; Griffith, G.W.; Insam, H.; Ascher-Jenull, J. The use of extracellular DNA as a proxy for specific microbial activity. Appl. Microbiol. Biot. 2018, 102, 2885–2898. [Google Scholar] [CrossRef] [Green Version]
- Nagler, M.; Insam, H.; Pietramellara, G.; Ascher-Jenull, J. Extracellular DNA in natural environments: Features, relevance and applications. Appl. Microbiol. Biot. 2018, 102, 6343–6356. [Google Scholar] [CrossRef] [Green Version]
- Campoccia, D.; Montanaro, L.; Arciola, C.R. Tracing the origins of extracellular DNA in bacterial biofilms: Story of death and predation to community benefit. Biofouling 2021, 37, 1022–1039. [Google Scholar] [CrossRef]
- Pallares, R.M.; Thanh, N.T.K.; Su, X. Tunable plasmonic colorimetric assay with inverse. sensitivity for extracellular DNA quantification. Chem. Commun. 2018, 54, 11260–11263. [Google Scholar] [CrossRef] [PubMed]
- Dominiak, D.M.; Nielsen, J.L.; Nielsen, P.H. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ. Microbiol. 2011, 13, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ. Microbiol. 2010, 12, 1621–1629. [Google Scholar] [CrossRef]
- Nielsen, K.M.; Johnsen, P.J.; Bensasson, D.; Daffonchio, D. Release and persistence of extracellular DNA in the environment. Environ. Biosaf. Res. 2007, 6, 37–53. [Google Scholar] [CrossRef] [Green Version]
- Lappann, M.; Claus, H.; van Alen, T.; Harmsen, M.; Elias, J.; Molin, S.; Vogel, U. A dual role of extracellular DNA during biofilm formation of Neisseria meningitidis. Mol. Microbiol. 2010, 75, 1355–1371. [Google Scholar] [CrossRef]
- Stewart, E.J. Growing Unculturable Bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef] [Green Version]
- Corinaldesi, C.; Danovaro, R.; Dell’Anno, A. Simultaneous Recovery of Extracellular and Intracellular DNA Suitable for Molecular Studies from Marine Sediments. Appl. Environ. Microb. 2005, 71, 46–50. [Google Scholar] [CrossRef] [Green Version]
- Hynen, A.L.; Lazenby, J.J.; Savva, G.M.; McCaughey, L.C.; Turnbull, L.; Nolan, L.M.; Whitchurch, C.B. Multiple holins contribute to extracellular DNA release in Pseudomonas aeruginosa biofilms. Microbiology 2021, 167, 000990. [Google Scholar] [CrossRef]
- Turnbull, L.; Toyofuku, M.; Hynen, A.L.; Kurosawa, M.; Pessi, G.; Petty, N.K.; Osvath, S.R.; Cárcamo-Oyarce, G.; Gloag, E.S.; Shimoni, R.; et al. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 2016, 7, 11220. [Google Scholar] [CrossRef] [Green Version]
- Das, T.; Sehar, S.; Manefield, M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Env. Microbiol. Rep. 2013, 5, 778–786. [Google Scholar] [CrossRef] [PubMed]
- Thomas, V.C.; Thurlow, L.R.; Boyle, D.; Hancock, L.R. Regulation of Autolysis-Dependent Extracellular DNA Release by Enterococcus faecalis Extracellular Proteases Influences Biofilm Development. J. Bacteriol. 2008, 190, 5690–5698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, C.Y.; Lin, M.H.; Chen, C.C.; Chien, S.C.; Cheng, Y.H.; Su, I.N.; Shu, J.C. Vancomycin promotes the bacterial autolysis, release of extracellular DNA, and biofilm formation in vancomycin-non-susceptible Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 2011, 63, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Manefield, M. Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLoS ONE 2012, 7, e46718. [Google Scholar] [CrossRef] [Green Version]
- Binnenkade, L.; Teichmann, L.; Thormann, K.M. Iron Triggers λSo Prophage Induction and Release of Extracellular DNA in Shewanella oneidensis MR-1 Biofilms. Appl. Environ. Microb. 2014, 80, 5304–5316. [Google Scholar] [CrossRef] [Green Version]
- Okshevsky, M.; Meyer, R.L. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 2015, 41, 341–352. [Google Scholar] [CrossRef]
- Graf, A.C.; Leonard, A.; Schauble, M.; Rieckmann, L.M.; Hoyer, J.; Maass, S.; Lalk, M.; Becher, D.; Pane-Farre, J.; Riedel, K. Virulence Factors Produced by Staphylococcus aureus Biofilms Have a Moonlighting Function Contributing to Biofilm Integrity. Mol. Cell. Proteom. 2019, 18, 1036–1053. [Google Scholar] [CrossRef]
- Svarachorn, A.; Shinmyo, A.; Tsuchido, T.; Takano, M. Autolysis of Bacillus subtilis induced by monovalent cations. Appl. Microbiol. Biot. 1989, 30, 299–304. [Google Scholar] [CrossRef]
- Catlin, B.W. Extracellular deoxyribonucleic acid of bacteria and a deoxyribonuclease inhibitor. Science 1959, 124, 441–442. [Google Scholar] [CrossRef]
- Yin, X.; Stotzky, G. Gene transfer among bacteria in natural environments. Adv. Appl. Microbiol. 1997, 45, 153–212. [Google Scholar]
- Catlin, B.W. Transformation of Neissenia meningitidis by deoxyribonucleates from cells and from culture slime. J. Bacteriol. 1959, 79, 579–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zafra, O.; Lamprecht-Grandio, M.; Gonzalez De Figueras, C.; Eduardo Gonzalez-Pastor, J. Extracellular DNA Release by Undomesticated Bacillus Is Regulated by Early Competence. PLoS ONE 2012, 7, e48716. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, H.L.; Domínguez, N.M.; Schwartz, K.J.; Hackett, K.T.; Dillard, J.P. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion. Mol. Microbiol. 2005, 55, 1704–1721. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, M.G.; Wackernagel, W. Bacterial Gene Transfer by Natural Genetic Transformation in the Environment. Microbiol. Rev. 1994, 58, 563–602. [Google Scholar] [CrossRef]
- Liao, S.; Klein, M.I.; Heim, K.P.; Fan, Y.; Bitoun, J.P.; Ahn, S.; Burne, R.A.; Koo, H.; Brady, L.J.; Wen, Z.T. Streptococcus mutans Extracellular DNA Is Upregulated during Growth in Biofilms, Actively Released via Membrane Vesicles, and Influenced by Components of the Protein Secretion Machinery. J. Bacteriol. 2014, 196, 2355–2366. [Google Scholar] [CrossRef] [Green Version]
- Hawes, M.C.; Curlango-Riverab, G.; Wen, F.; VanEttenb, H.D.; Xiong, Z. Extracellular DNA The tip of root defenses. Plant Sci. 2011, 180, 741–745. [Google Scholar] [CrossRef]
- Ciesluk, M.; Piktel, E.; Watek, M.; Durnas, B.; Wollny, T.; Krol, G.; Bucki, R. Neutrophil extracellular traps as the main source of eDNA. Med. Stud. Stud. Med. 2017, 33, 137–145. [Google Scholar] [CrossRef] [Green Version]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil Extracellular Traps Kill Bacteria. Science 2004, 303, 1532–2535. [Google Scholar] [CrossRef]
- Hawes, M.C.; Wen, F.; Elquza, E. Extracellular DNA-A Bridge to Cancer. Cancer Res. 2015, 75, 4260–4264. [Google Scholar] [CrossRef] [Green Version]
- Lou, H.; Pickering, M.C. Extracellular DNA and autoimmune diseases. Cell. Mol. Immunol. 2018, 15, 746–755. [Google Scholar] [CrossRef] [Green Version]
- Pietramellara, G.; Ascher, J.; Baraniya, D.; Arfaioli, P.; Ceccherini, M.T.; Hawes, M. Relevance of extracellular DNA in rhizosphere. Geophys. Res. Abstr. 2013, 15, EGU2013-8331. [Google Scholar]
- Waheed, H.; Hashmi, I.; Khan, S.J.; Kim, S.R.; Arshad, M.; Nasir, H. Microbial population dynamics and profiling of quorum sensing agents in membrane bioreactor. Int. Biodeter. Biodegr. 2016, 113, 66–73. [Google Scholar] [CrossRef]
- Harmsen, M.; Lappann, M.; Knøchel, S.; Molin, S. Role of Extracellular DNA during Biofilm Formation by Listeria monocytogenes. Appl. Environ. Microb. 2010, 76, 2271–2279. [Google Scholar] [CrossRef]
- Puga, C.H.; Rodríguez-López, P.; Cabo, M.L.; SanJose, C.; Orgaz, B. Enzymatic dispersal of dual-species biofilms carrying Listeria monocytogenes and other associated food industry bacteria. Food Control 2018, 94, 222–228. [Google Scholar] [CrossRef] [Green Version]
- Das, T.; Sharma, P.K.; Busscher, H.J.; van der Mei, H.C.; Krom, B.P. Role of Extracellular DNA in Initial Bacterial Adhesion and Surface Aggregation. Appl. Environ. Microb. 2010, 76, 3405–3408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Liu, X.; Liu, H.; Zhang, L.; Guo, Y.; Yu, S.; Wozniak, D.J.; Ma, L.Z. The exopolysaccharide Psl-eDNA interaction enables the formation of a biofilm skeleton in Pseudomonas aeruginosa. Env. Microbiol. Rep. 2015, 7, 330–340. [Google Scholar] [CrossRef] [Green Version]
- García-Fontana, C.; Narváez-Reinaldo, J.J.; Castillo, F.; González-López, J.; Luque, I.; Manzanera, M. A New Physiological Role for the DNA Molecule as a Protector against Drying Stress in Desiccation-Tolerant Microorganisms. Front. Microbiol. 2016, 7, 2066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and Barriers to, Horizontal Gene Transfer between Bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Mahillon, J.; Chandler, M. Insertion Sequences. Microbiol. Mol. Biol. Rev. 1998, 62, 725–774. [Google Scholar] [CrossRef] [Green Version]
- Dobrindt, U.; Hochhut, B.; Hentschel, U.; Hacker, J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2004, 2, 414–424. [Google Scholar] [CrossRef]
- Hall, R.M.; Collis, C.M. Mobile gene cassettes and integrons: Capture and spread of genes by site-specific recombination. Mol. Microbiol. 1995, 15, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M. Paradigms of plasmid organization. Mol. Microbiol. 2000, 37, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Pietramellara, G.; Ascher, J.; Borgogni, F.; Ceccherini, M.T.; Guerri, G.; Nannipieri, P. Extracellular DNA in soil and sediment: Fate and ecological relevance. Biol. Fert. Soils 2009, 45, 219–235. [Google Scholar] [CrossRef]
- Kaneko, S.; Itaya, M. Designed horizontal transfer of stable giant DNA released from Escherichia coli. J. Biochem. 2010, 147, 819–822. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G.J.; Carlson, C.A.; Ingraham, J.L. Evidence for an active role of donor cells in natural transformation of Pseudomonas stutzeri. J. Bacteriol. 1983, 156, 30–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, E.M.; Prairie, Y.T. Bacterial Metabolism and Growth Efficiency in Lakes: The Importance of Phosphorus Availability. Limnol. Oceanogr. 2004, 49, 137–147. [Google Scholar] [CrossRef]
- Paul, J.H.; Carlson, D.J. Genetic Material in the Marine Environment: Implication for Bacterial DNA. Limnol. Oceanogr. 1984, 29, 1091–1097. [Google Scholar] [CrossRef]
- Dell’Anno, A.; Corinaldesi, C. Degradation and Turnover of Extracellular DNA in Marine Sediments: Ecological and Methodological Considerations. Appl. Environ. Microb. 2004, 70, 4384–4386. [Google Scholar] [CrossRef] [Green Version]
- Pinchuk, G.E.; Ammons, C.; Culley, D.E.; Li, S.W.; McLean, J.S.; Romine, M.F.; Nealson, K.H.; Fredrickson, J.K.; Beliaev, A.S. Utilization of DNA as a Sole Source of Phosphorus, Carbon, and Energy by Shewanella spp.: Ecological and Physiological Implications for Dissimilatory Metal Reduction. Appl. Environ. Microb. 2008, 74, 1198–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palchevskiy, V.; Finkel, S.E. Escherichia coli Competence Gene Homologs Are Essential for Competitive Fitness and the Use of DNA as a Nutrient. J. Bacteriol. 2006, 188, 3902–3910. [Google Scholar] [CrossRef] [Green Version]
- Redfield, R.J. Genes for breakfast: The have-your-cake-and-eat-it-too of bacterial transformation. J. Hered. 1993, 84, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, J.; Gessler, F. Determination of free DNA in soils. J. Plant Nutr. Soil Sc. 2002, 165, 121–124. [Google Scholar] [CrossRef]
- Borneman, J.; Hartin, R.J. PCR Primers That Amplify Fungal rRNA Genes from Environmental Samples. Appl. Environ. Microb. 2000, 66, 4356–4360. [Google Scholar] [CrossRef]
- Pathan, S.I.; Arfaioli, P.; Ceccherini, M.T.; Ascher-Jenull, J.; Pietramellara, G. Preliminary evidences of the presence of extracellular DNA single stranded forms in soil. PLoS ONE 2020, 15, e0227296. [Google Scholar] [CrossRef]
- Poté, J.; Ceccherini, M.T.; Van, V.T.; Rosselli, W.; Wildi, W.; Simonet, P.; Vogel, T.M. Fate and transport of antibiotic resistance genes in saturated soil columns. Eur. J. Soil Biol. 2003, 39, 65–71. [Google Scholar] [CrossRef]
- Levy-Booth, D.J.; Campbell, R.G.; Gulden, R.H.; Hart, M.M.; Powell, J.R.; Klironomos, J.N.; Pauls, K.P.; Swanton, C.J.; Trevors, J.T.; Dunfield, K.E. Cycling of extracellular DNA in the soil environment. Soil Biol. Biochem. 2007, 39, 2977–2991. [Google Scholar] [CrossRef]
- Paungfoo-Lonhienne, C.; Lonhienne, T.G.A.; Schmidt, S. DNA uptake by Arabidopsis induces changes in the expression of CLE peptides which control root morphology. Plant Signal. Behav. 2010, 153, 799–805. [Google Scholar] [CrossRef] [Green Version]
- Mazzoleni, S.; Bonanomi, G.; Incerti, G.; Chiusano, M.L.; Termolino, P.; Mingo, A.; Senatore, M.; Giannino, F.; Cartenì, F.; Rietkerk, M.; et al. Inhibitory and toxic effects of extracellular self-DNA in litter: A mechanism for negative plant–soil feedbacks? New Phytol. 2015, 205, 1195–1210. [Google Scholar] [CrossRef] [Green Version]
- Hawes, M.; McLain, J.; Ramirez-Andreotta, M.; Curlango-Rivera, G.; Flores-Lara, Y.; Brigham, L. Extracellular Trapping of Soil Contaminants by Root Border Cells: New Insights into Plant Defense. Agronomy 2016, 6, 5. [Google Scholar] [CrossRef]
- Dell’Anno, A.; Danovaro, R. Extracellular DNA Plays a Key Role in Deep-Sea Ecosystem Functioning. Science 2005, 309, 2179. [Google Scholar] [CrossRef]
- Mao, D.; Luo, Y.; Mathieu, J.; Wang, Q.; Feng, L.; Mu, Q.; Feng, C.; Alvarez, P.J.J. Persistence of Extracellular DNA in River Sediment Facilitates Antibiotic Resistance Gene Propagation. Environ. Sci. Technol. 2014, 48, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Vuillemin, A.; Horn, F.; Alawi, M.; Henny, C.; Wagner, D.; Crowe, S.A.; Kallmeyer, J. Preservation and Significance of Extracellular DNA in Ferruginous Sediments from Lake Towuti, Indonesia. Front. Microbiol. 2017, 8, 1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dell’Anno, A.; Bompadre, S.; Danovaro, R. Quantification, base composition, and fate of extracellular DNA in marine sediments. Limnol. Oceanogr. 2002, 47, 899–905. [Google Scholar] [CrossRef]
- Dell’Anno, A.; Corinaldesi, C.; Stavrakakis, S.; Lykousis, V.; Danovaro, R. Pelagic-Benthic Coupling and Diagenesis of Nucleic Acids in a Deep-Sea Continental Margin and an Open-Slope System of the Eastern Mediterranean. Appl. Environ. Microb. 2005, 71, 6070–6076. [Google Scholar] [CrossRef] [Green Version]
- Ellegaard, M.; Clokie, M.R.J.; Czypionka, T.; Frisch, D.; Godhe, A.; Kremp, A.; Letarov, A.; McGenity, T.J.; Ribeiro, S.; John Anderson, N. Dead or alive: Sediment DNA archives as tools for tracking aquatic evolution and adaptation. Commun. Biol. 2020, 3, 169. [Google Scholar] [CrossRef] [Green Version]
- Chroňáková, A.; Ascher, J.; Jirout, J.; Ceccherini, M.T.; Elhottová, D.; Pietramellara, G.; Šimek, M. Cattle impact on composition of archaeal, bacterial, and fungal communities by comparative fingerprinting of total and extracellular DNA. Biol. Fert. Soils 2013, 49, 351–361. [Google Scholar] [CrossRef]
- Dong, P.; Wang, H.; Fang, T.; Wang, Y.; Ye, Q. Assessment of extracellular antibiotic resistance genes (eARGs) in typical environmental samples and the transforming ability of eARG. Environ. Int. 2019, 125, 90–96. [Google Scholar] [CrossRef]
- Xu, Y.; Li, H.; Shi, R.; Lv, J.; Li, B.; Yang, F.; Zheng, X.; Xu, J. Antibiotic resistance genes in different animal manures and their derived organic fertilizer. Environ. Sci. Eur. 2020, 32, 102. [Google Scholar] [CrossRef]
- Castro-Mejia, J.L.; Deng, L.; Vogensen, F.K.; Reyes, A.; Nielsen, D.S. Extraction and Purification of Viruses from Fecal Samples for Metagenome and Morphology Analyses. Methods Mol. Biol. 2018, 1838, 49–57. [Google Scholar] [CrossRef]
- Wu, H.; Tremaroli, V.; Bäckhed, F. Linking Microbiota to Human Diseases: A Systems Biology Perspective. Trends Endocrinol. Metab. 2015, 26, 758–770. [Google Scholar] [CrossRef]
- Liang, Z.; Keeley, A. Filtration Recovery of Extracellular DNA from Environmental Water Samples. Environ. Sci. Technol. 2013, 47, 9324–9331. [Google Scholar] [CrossRef] [PubMed]
- Waldemar Siuda, H.G. Determination of dissolved deoxyribonucleic acid concentration in lake water. Aquat. Microb. Ecol. 1996, 11, 193–202. [Google Scholar] [CrossRef]
- Takahashi, I. Genetic transformation of Bacillus subtilis by extracellular DNA. Biochem. Bioph. Res. Co. 1962, 7, 467–470. [Google Scholar] [CrossRef]
- Garchitorena, M.R. Evolution of Extracellular DNA (eDNA) Secretion in Bacillus subtilis. Sr. Proj. Spring 2017, 2017, 127. [Google Scholar]
- Kaneko, S.; Fukushima, H.; Nakahama, M.; Asano, S.; Miyazaki, Y.; Aizawa, Y.; Itaya, M. DNA synthesis by fragment assembly using extra-cellular DNA delivered by artificial controlled horizontal transfer. J. Biochem. 2018, 163, 305–312. [Google Scholar] [CrossRef]
- Kampf, J.; Stülke, J. Cyclic-di-GMP signalling meets extracellular polysaccharide synthesis in Bacillus subtilis. Env. Microbiol. Rep. 2017, 9, 182–185. [Google Scholar] [CrossRef]
- Vilain, S.; Pretorius, J.M.; Theron, J.; Broözel, V.S. DNA as an Adhesin: Bacillus cereus Requires Extracellular DNA To Form Biofilms. Appl. Environ. Microb. 2009, 75, 2861–2868. [Google Scholar] [CrossRef] [Green Version]
- Peng, N.; Cai, P.; Mortimer, M.; Wu, Y.; Gao, C.; Huang, Q. The exopolysaccharide–eDNA interaction modulates 3D architecture of Bacillus subtilis biofilm. BMC Microbiol. 2020, 20, 115. [Google Scholar] [CrossRef]
- Randrianjatovo Gbalou, I.; Rouquette, P.; Lefebvre, D.; Girbal Neuhauser, E.; Marcato Romain, C.E. In situ analysis of Bacillus licheniformis biofilms: Amyloid-like polymers and eDNA are involved in the adherence and aggregation of the extracellular matrix. J. Appl. Microbiol. 2017, 122, 1262–1274. [Google Scholar] [CrossRef]
- Yu, R.; Hou, C.; Liu, A.; Peng, T.; Xia, M.; Wu, X.; Shen, L.; Liu, Y.; Li, J.; Yang, F.; et al. Extracellular DNA enhances the adsorption of Sulfobacillus thermosulfidooxidans strain ST on chalcopyrite surface. Hydrometallurgy 2018, 176, 97–103. [Google Scholar] [CrossRef]
- Peix, A.; Ramirez-Bahena, M.H.; Velazquez, E. Historical evolution and current status of the taxonomy of genus Pseudomonas. Infect. Genet. Evol. 2009, 9, 1132–1147. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T.; Jensen, P.Ø.; Fiandaca, M.J.; Pedersen, J.; Hansen, C.R.; Andersen, C.B.; Pressler, T.; Givskov, M.; Høiby, N. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulm. 2009, 44, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Ciofu, O.; Tolker-Nielsen, T. Tolerance and Resistance of Pseudomonas aeruginosa Biofilms to Antimicrobial Agents—How P. aeruginosa Can Escape Antibiotics. Front. Microbiol. 2019, 10, 913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allesen-Holm, M.; Barken, K.B.; Yang, L.; Klausen, M.; Webb, J.S.; Kjelleberg, S.; Molin, S.; Givskov, M.; Tolker-Nielsen, T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 2006, 59, 1114–1128. [Google Scholar] [CrossRef]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitchurch, C.B.; Tolker-Nielsen, T.; Ragas, P.C.; Mattick, J.S. Extracellular DNA required for bacterial biofilm formation. Science 2002, 295, 1487. [Google Scholar] [CrossRef] [PubMed]
- Wilton, M.; Charron-Mazenod, L.; Moore, R.; Lewenza, S. Extracellular DNA Acidifies Biofilms and Induces Aminoglycoside Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Ch. 2016, 60, 544–553. [Google Scholar] [CrossRef] [Green Version]
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008, 4, e1000213. [Google Scholar] [CrossRef] [Green Version]
- Bass, J.I.F.; Russo, D.M.; Gabelloni, M.L.; Geffner, J.R.; Giordano, M.; Catalano, M.; Trevani, A.S. Extracellular DNA: A Major Proinflammatory Component of Pseudomonas aeruginosa Biofilms. J. Immunol. 2010, 184, 6386–6395. [Google Scholar] [CrossRef] [Green Version]
- Priester, J.H.; Olson, S.G.; Webb, S.M.; Neu, M.P.; Hersman, L.E.; Holden, P.A. Enhanced Exopolymer Production and Chromium Stabilization in Pseudomonas putida Unsaturated Biofilms. Appl. Environ. Microb. 2006, 72, 1988–1996. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Wang, C.; Zhao, H.; Chen, G.; Chen, X. Interaction between copper and extracellular nucleic acids in the EPS of unsaturated Pseudomonas putida CZ1 biofilm. Environ. Sci. Pollut. R. 2018, 25, 24172–24180. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S. Release mechanisms and molecular interactions of Pseudomonas aeruginosa extracellular DNA. Appl. Microbiol. Biot. 2020, 104, 6549–6564. [Google Scholar] [CrossRef]
- Zeng, B.; Wang, C.; Zhang, P.; Guo, Z.; Chen, L.; Duan, K. Heat Shock Protein DnaJ in Pseudomonas aeruginosa Affects Biofilm Formation via Pyocyanin Production. Microorganisms 2020, 8, 395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meto, A.; Colombari, B.; Meto, A.; Boaretto, G.; Pinetti, D.; Marchetti, L.; Benvenuti, S.; Pellati, F.; Blasi, E. Propolis Affects Pseudomonas aeruginosa Growth, Biofilm Formation, eDNA Release and Phenazine Production: Potential Involvement of Polyphenols. Microorganisms 2020, 8, 243. [Google Scholar] [CrossRef] [PubMed]
- Thomann, A.; Brengel, C.; Börger, C.; Kail, D.; Steinbach, A.; Empting, M.; Hartmann, R.W. Structure-Activity Relationships of 2-Sufonylpyrimidines as Quorum-Sensing Inhibitors to Tackle Biofilm Formation and eDNA Release of Pseudomonas aeruginosa. ChemMedChem 2016, 11, 2522–2533. [Google Scholar] [CrossRef]
- Bhongir, R.K.V.; Kasetty, G.; Papareddy, P.; Mörgelin, M.; Herwald, H.; Egesten, A. DNA-fragmentation is a source of bactericidal activity against Pseudomonas aeruginosa. Biochem. J. 2017, 474, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Burgess, N.R.H.; McDermott, S.N.; Whiting, J. Aerobic bacteria occurring in the hind-gut of the cockroach, Blatta orientalis. J. Hyg. 1973, 71, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Kaper, J.B.; Nataro, J.P. Diarrheagenic Escherichia coli. Am. Soc. Microbiol. Clin. Microbiol. Rev. 1998, 11, 142–201. [Google Scholar] [CrossRef] [Green Version]
- Huys, G.; Cnockaert, M.; Janda, J.M.; Swings, J. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Int. J. Syst. Evol. Micr. 2003, 53, 807–810. [Google Scholar] [CrossRef] [Green Version]
- Farmer, J.R.; Fanning, G.R.; Davis, B.R.; O’Hara, C.M.; Riddle, C.; Hickman-Brenner, F.W.; Asbury, M.A.; Lowery, V.R.; Brenner, D.J. Escherichia fergusonii and Enterobacter taylorae, two new species of Enterobacteriaceae isolated from clinical specimens. J. Clin. Microbiol. 1985, 21, 77–81. [Google Scholar] [CrossRef] [Green Version]
- Brenner, D.J.; McWhorter, A.C.; Knutson, J.K.; Steigerwalt, A.G. Escherichia vulneris: A new species of Enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 1982, 15, 1133–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, D.; Banting, G.; Neumann, N.F. A review of the taxonomy, genetics, and biology of the genus Escherichia and the type species Escherichia coli. Can. J. Microbiol. 2021, 67, 553–571. [Google Scholar] [CrossRef] [PubMed]
- Clements, A.; Young, J.C.; Constantinou, N.; Frankel, G. Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes 2012, 3, 71–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reisner, A.; Maierl, M.; Jorger, M.; Krause, R.; Berger, D.; Haid, A.; Tesic, D.; Zechner, E.L. Type 1 Fimbriae Contribute to Catheter-Associated Urinary Tract Infections Caused by Escherichia coli. J. Bacteriol. 2014, 196, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Taniuchi, A.; May, T.; Kawata, K.; Okabe, S. Increased Antibiotic Resistance of Escherichia coli in Mature Biofilms. Appl. Environ. Microb. 2009, 75, 4093–4100. [Google Scholar] [CrossRef] [Green Version]
- Matsui, K.; Ishii, N.; Kawabata, Z. Release of Extracellular Transformable Plasmid DNA from Escherichia coli Cocultivated with Algae. Appl. Environ. Microb. 2003, 69, 2399–2404. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Yang, Y.; Shen, X.; Zhang, Z.; Shen, P.; Xie, Z. Role of DNA in Bacterial Aggregation. Curr. Microbiol. 2008, 57, 139–144. [Google Scholar] [CrossRef]
- Pachucki, R.J.; Corradetti, C.; Kohler, L.; Ghadiali, J.; Gallo, P.M.; Nicastro, L.; Tursi, S.A.; Gallucci, S.; Tükel, Ç.; Caricchio, R. Persistent Bacteriuria and Antibodies Recognizing Curli/eDNA Complexes From Escherichia coli Are Linked to Flares in Systemic Lupus Erythematosus. Arthritis Rheumatol. 2020, 72, 1872–1881. [Google Scholar] [CrossRef]
- Qiu, C.C.; Caricchio, R.; Gallucci, S. Triggers of Autoimmunity: The Role of Bacterial Infections in the Extracellular Exposure of Lupus Nuclear Autoantigens. Front. Immunol. 2019, 10, 2608. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Torres, V.; Maeda, T.; Wood, T.K. Global regulator H-NS and lipoprotein NlpI influence production of extracellular DNA in Escherichia coli. Biochem. Bioph. Res. Co. 2010, 401, 197–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Wang, Q.; Li, M.; Heijstra, B.D.; Wang, S.; Liang, Q.; Qi, Q. Escherichia coli toxin gene hipA affects biofilm formation and DNA release. Microbiology+ 2013, 159, 633–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castan, A.; Enfors, S. Formate accumulation due to DNA release in aerobic cultivations of Escherichia coli. Biotechnol. Bioeng. 2002, 77, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Yang, Q.; Fang, F.; Li, Y. The camelliagenin from defatted seeds of Camellia oleifera as antibiotic substitute to treat chicken against infection of Escherichia coli and Staphylococcus aureus. BMC Vet. Res. 2015, 11, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zweig, M.; Schork, S.; Koerdt, A.; Siewering, K.; Sternberg, C.; Thormann, K.; Albers, S.; Molin, S.; van der Does, C. Secreted single-stranded DNA is involved in the initial phase of biofilm formation by Neisseria gonorrhoeae. Environ. Microbiol. 2014, 16, 1040–1052. [Google Scholar] [CrossRef]
- Heijstra, B.D.; Pichler, F.B.; Liang, Q.; Blaza, R.G.; Turner, S.J. Extracellular DNA and Type IV pili mediate surface attachment by Acidovorax temperans. Antonie Leeuwenhoek 2009, 95, 343–349. [Google Scholar] [CrossRef]
- Peterson, B.W.; van der Mei, H.C.; Sjollema, J.; Busscher, H.J.; Sharma, P.K. A distinguishable role of eDNA in the viscoelastic relaxation of biofilms. mBio 2013, 4, e413–e497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berne, C.; Kysela, D.T.; Brun, Y.V. A bacterial extracellular DNA inhibits settling of motile progeny cells within a biofilm. Mol. Microbiol. 2010, 77, 815–829. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Huang, Y.; Wu, S.; Li, Y.; Ye, Y.; Zheng, Y.; Huang, R. Extracellular DNA Inhibits Salmonella enterica Serovar Typhimurium and S. enterica Serovar Typhi Biofilm Development on Abiotic Surfaces. Curr. Microbiol. 2014, 68, 262–268. [Google Scholar] [CrossRef]
- Roose-Amsaleg, C.L.; Garnier-Sillam, E.; Harry, M. Extraction and purification of microbial DNA from soil and sediment samples. Appl. Soil Ecol. 1987, 18, 47–60. [Google Scholar] [CrossRef]
- Agnelli, A.; Ascher, J.; Corti, G.; Ceccherini, M.T.; Nannipieri, P.; Pietramellara, G. Distribution of microbial communities in a forest soil profile investigated by microbial biomass, soil respiration and DGGE of total and extracellular DNA. Soil Biol. Biochem. 2004, 36, 859–868. [Google Scholar] [CrossRef]
- Alawi, M.; Schneider, B.; Kallmeyer, J. A procedure for separate recovery of extra- and intracellular DNA from a single marine sediment sample. J. Microbiol. Meth. 2014, 104, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Snow, D.D.; Parker, D.; Zhou, Z.; Li, X. Intracellular and Extracellular Antimicrobial Resistance Genes in the Sludge of Livestock Waste Management Structures. Environ. Sci. Technol. 2013, 47, 10206–10213. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Hata, A.; Katayama, H.; Kasuga, I. Consecutive ultrafiltration and silica adsorption for recovery of extracellular antibiotic resistance genes from an urban river. Environ. Pollut. 2020, 260, 114062. [Google Scholar] [CrossRef]
- Lever, M.A.; Torti, A.; Eickenbusch, P.; Michaud, A.B.; Santl-Temkiv, T.; Jorgensen, B.B. A modular method for the extraction of DNA and RNA, and the separation of DNA pools from diverse environmental sample types. Front. Microbiol. 2015, 6, 476. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Yang, Y.; Lu, D.; Niu, Z.; Feng, J.; Chen, Y.; Tou, F.; Garner, E.; Xu, J.; Liu, M.; et al. Biofilms as a sink for antibiotic resistance genes (ARGs) in the Yangtze Estuary. Water Res. 2018, 129, 277–286. [Google Scholar] [CrossRef]
- Maruyama, A.; Oda, M.; Higashihara, T. Abundance of Virus-Sized Non-DNase-Digestible DNA (Coated DNA) in Eutrophic Seawater. Appl. Environ. Microb. 1993, 59, 712–717. [Google Scholar] [CrossRef] [Green Version]
- Pillal, T.N.V.; Ganguly, A.K. Nucleic acids in the dissolved constituents of sea-water. Curr. Sci. 1970, 39, 501–504. [Google Scholar]
- Zhang, Y.; Li, A.; Dai, T.; Li, F.; Xie, H.; Chen, L.; Wen, D. Cell-free DNA: A Neglected Source for Antibiotic Resistance Genes Spreading from WWTPs. Environ. Sci. Technol. 2018, 52, 248–257. [Google Scholar] [CrossRef]
- Metcalf, D.; Weese, J.S. Evaluation of commercial kits for extraction of DNA and RNA from Clostridium difficile. Anaerobe 2012, 18, 608–613. [Google Scholar] [CrossRef]
- Wang, D.; Liu, L.; Qiu, Z.; Shen, Z.; Guo, X.; Yang, D.; Li, J.; Liu, W.; Jin, M.; Li, J. A new adsorption-elution technique for the concentration of aquatic extracellular antibiotic resistance genes from large volumes of water. Water Res. 2016, 92, 188–198. [Google Scholar] [CrossRef]
- O’Malley, K.; McDonald, W.; McNamara, P. An Extraction Method to Quantify the Fraction of Extracellular and Intracellular Antibiotic Resistance Genes in Aquatic Environments. J. Environ. Eng. 2022, 148. [Google Scholar] [CrossRef]
- Yuan, Q.; Huang, Y.; Wu, W.; Zuo, P.; Hu, N.; Zhou, Y.; Alvarez, P.J.J. Redistribution of intracellular and extracellular free & adsorbed antibiotic resistance genes through a wastewater treatment plant by an enhanced extracellular DNA extraction method with magnetic beads. Environ. Int. 2019, 131, 104986. [Google Scholar] [CrossRef] [PubMed]
- Dahllöf, I. Molecular community analysis of microbial diversity. Curr. Opin. Biotech. 2002, 13, 213–217. [Google Scholar] [CrossRef]
- Stach, J.E.M.; Bathe, S.; Clapp, J.P.; Burns, R.G. PCR-SSCP comparison of 16S rDNA sequence diversity in soil DNA obtained using di¡erent isolation and puri¢cation methods. FEMS Microbiol. Ecol. 2001, 36, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Haque, K.A.; Pfeiffer, R.M.; Beerman, M.B.; Struewing, J.P.; Chanock, S.J.; Bergen, A.W. Performance of high-throughput DNA quantification methods. BMC Biotechnol. 2003, 3, 20. [Google Scholar] [CrossRef]
- Ren, P.; Chen, T.; Liu, N.; Sun, W.; Hu, G.; Yu, Y.; Yu, B.; Ouyang, P.; Liu, D.; Chen, Y. Efficient Biofilm-Based Fermentation Strategies by eDNA Formation for L-Proline Production with Corynebacterium glutamicum. ACS Omega 2020, 5, 33314–33322. [Google Scholar] [CrossRef]
- Ahn, S.J.; Costa, J.; Emanuel, J.R. PicoGreen quantitation of DNA: Effective evaluation of samples pre- or post-PCR. Nucleic Acids Res. 1996, 24, 2623–2625. [Google Scholar] [CrossRef]
- Borneman, J.; Skroch, P.W.; O’Sullivan, K.M.; Palus, J.A.; Rumjanek, N.G.; Jansen, J.L.; Nienhuis, J.; Triplett, E.W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl. Environ. Microbiol. 1996, 62, 1935–1943. [Google Scholar] [CrossRef] [Green Version]
- Becker, S.; Boger, P.; Oehlmann, R.; Ernst, A. PCR Bias in Ecological Analysis: A Case Study for Quantitative Taq Nuclease Assays in Analyses of Microbial Communities. Appl. Environ. Microb. 2000, 66, 4945–4953. [Google Scholar] [CrossRef] [Green Version]
- Chandler, D.P.; Fredrickson, J.K.; Brockman, F.J. Effect of PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol. Ecol. 2003, 6, 475–482. [Google Scholar] [CrossRef]
- Davies, C.E.; Wilson, M.J.; Hill, K.E.; Stephens, P.; Hill, C.M.; Harding, K.G.; Thomas, D.W. Use of molecular techniques to study microbial diversity in the skin: Chronic wounds reevaluated. Wound Repair Regen. 2001, 9, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Carini, P.; Marsden, P.J.; Leff, J.W.; Morgan, E.E.; Strickland, M.S.; Fierer, N. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2017, 2, 16242. [Google Scholar] [CrossRef] [PubMed]
- Manu, S.; Umapathy, G. A Novel Metagenomic Workflow for Biomonitoring across the Tree of Life using PCR-free Ultra-deep Sequencing of Extracellular eDNA. Authorea Prepr. 2021, 4, e64864. [Google Scholar] [CrossRef]
- Jensen, M.R.; Sigsgaard, E.E.; Liu, S.; Manica, A.; Bach, S.S.; Hansen, M.M.; Møller, P.R.; Thomsen, P.F. Genome-scale target capture of mitochondrial and nuclear environmental DNA from water samples. Mol. Ecol. Resour. 2021, 21, 690–702. [Google Scholar] [CrossRef] [PubMed]
- Horn, S. Target Enrichment via DNA Hybridization Capture. Anc. DNA 2012, 840, 177–188. [Google Scholar]
- Rajendhran, J.; Gunasekaran, P. Strategies for accessing soil metagenome for desired applications. Biotechnol. Adv. 2008, 26, 576–590. [Google Scholar] [CrossRef]
- Nelson, M.T.; Pope, C.E.; Marsh, R.L.; Wolter, D.J.; Weiss, E.J.; Hager, K.R.; Vo, A.T.; Brittnacher, M.J.; Radey, M.C.; Hayden, H.S.; et al. Human and Extracellular DNA Depletion for Metagenomic Analysis of Complex Clinical Infection Samples Yields Optimized Viable Microbiome Profiles. Cell Rep. 2019, 26, 2227–2240. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; Zhu, Y.; Yan, Y.; Wang, W.; Wang, Y. Deciphering extracellular antibiotic resistance genes (eARGs) in activated sludge by metagenome. Water Res. 2019, 161, 610–620. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, K.; Wang, L.; Cao, X.; Gu, Z.; Zhao, G.; Ran, M.; Yan, Y.; Yan, J.; Xu, L.; Gao, C.; et al. The Origin, Function, Distribution, Quantification, and Research Advances of Extracellular DNA. Int. J. Mol. Sci. 2022, 23, 13690. https://doi.org/10.3390/ijms232213690
Yang K, Wang L, Cao X, Gu Z, Zhao G, Ran M, Yan Y, Yan J, Xu L, Gao C, et al. The Origin, Function, Distribution, Quantification, and Research Advances of Extracellular DNA. International Journal of Molecular Sciences. 2022; 23(22):13690. https://doi.org/10.3390/ijms232213690
Chicago/Turabian StyleYang, Kaixin, Lishuang Wang, Xinghong Cao, Zhaorui Gu, Guowei Zhao, Mengqu Ran, Yunjun Yan, Jinyong Yan, Li Xu, Chunhui Gao, and et al. 2022. "The Origin, Function, Distribution, Quantification, and Research Advances of Extracellular DNA" International Journal of Molecular Sciences 23, no. 22: 13690. https://doi.org/10.3390/ijms232213690





