Peripheral Ion Channel Genes Screening in Painful Small Fiber Neuropathy
Abstract
:1. Introduction
2. Results
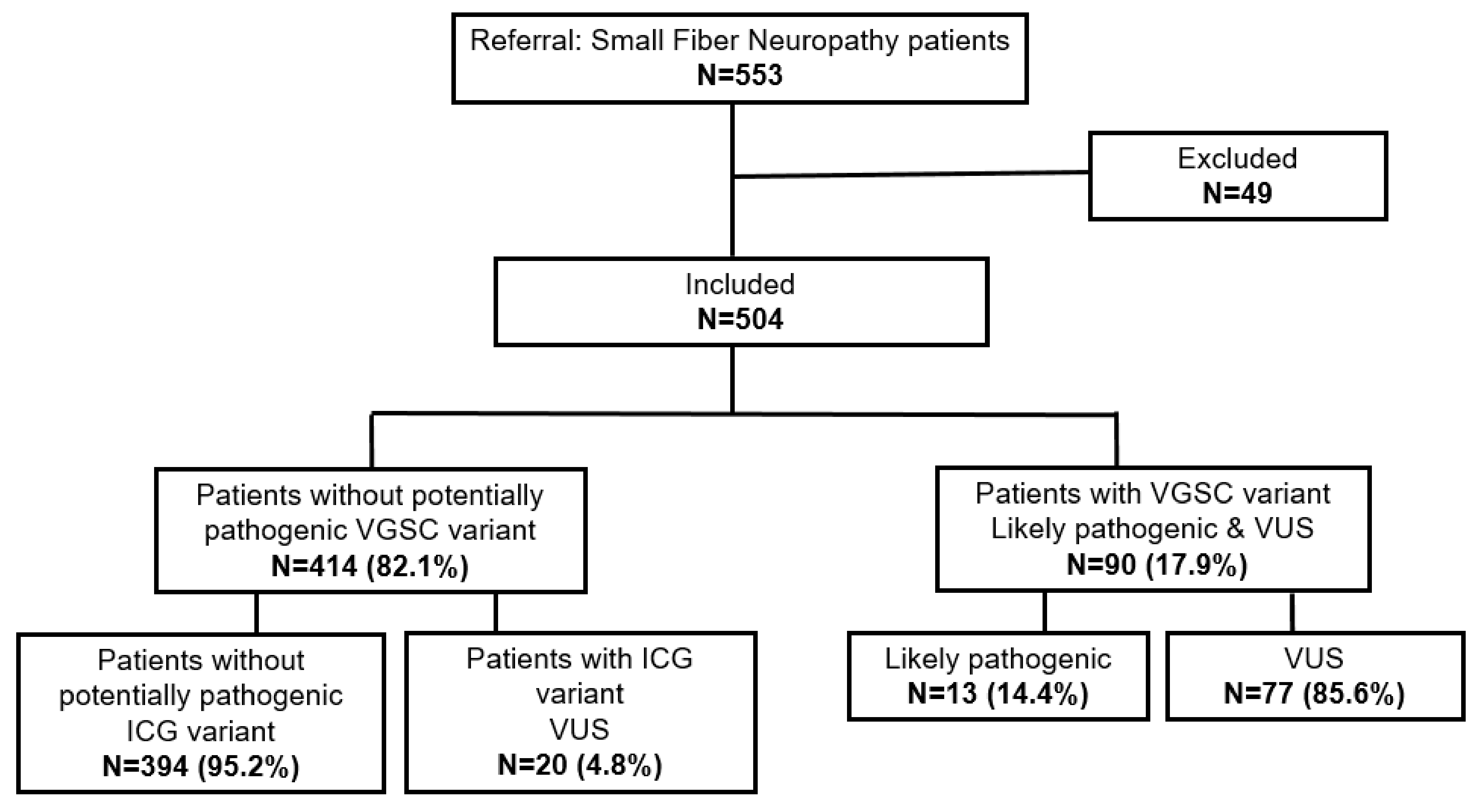
2.1. smMIPs-NGS of Patients with SFN
2.2. Genetic Variants Identified in Ion Channel Genes
2.3. Patients with SFN with an ICG Variant Compared to Patients without ICG/VGSC Variant
3. Discussion
3.1. Summary of ICG Screening in Patients with Small Fiber Neuropathy
3.2. VUS Meaning in Context of Protein Function
3.3. ICG Variants in Relation to Patients’ Clinical Manifestations
3.3.1. Anoctamin 3
3.3.2. Potassium Channels
3.3.3. TRP Channels
3.4. Patients with an ICG Variant Had More Severe Clinical Manifestations and Higher Pain Scores
4. Material and Methods
4.1. Study Population
4.2. DNA Extraction and Storage
4.3. smMIPs-NGS of Peripheral Ion Channels and Data Analysis
4.4. Variant Classification
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Calvo, M.; Davies, A.J.; Hébert, H.L.; Weir, G.A.; Chesler, E.J.; Finnerup, N.B.; Levitt, R.C.; Smith, B.H.; Neely, G.G.; Costigan, M.; et al. The Genetics of Neuropathic Pain from Model Organisms to Clinical Application. Neuron 2019, 104, 637–653. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, E.; Mammana, S.; Nicoletti, F.; Bramanti, P.; Mazzon, E. The neuropathic pain: An overview of the current treatment and future therapeutic approaches. Int. J. Immunopathol. Pharmacol. 2019, 33, 2058738419838383. [Google Scholar] [CrossRef] [Green Version]
- Eijkenboom, I.; Sopacua, M.; Hoeijmakers, J.G.J.; de Greef, B.T.A.; Lindsey, P.; Almomani, R.; Marchi, M.; Vanoevelen, J.; Smeets, H.J.M.; Waxman, S.G.; et al. Yield of peripheral sodium channels gene screening in pure small fibre neuropathy. J. Neurol. Neurosurg. Psychiatry 2019, 90, 342–352. [Google Scholar] [CrossRef]
- Colloca, L.; Ludman, T.; Bouhassira, D.; Baron, R.; Dickenson, A.H.; Yarnitsky, D.; Freeman, R.; Truini, A.; Attal, N.; Finnerup, N.B.; et al. Neuropathic pain. Nat. Rev. Dis. Prim. 2017, 3, 17002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarberg, B.H.; Billington, R. Consequences of neuropathic pain: Quality-of-life issues and associated costs. Am. J. Manag. Care 2006, 12, S263–S268. [Google Scholar] [PubMed]
- Spallone, V.; Greco, C. Painful and painless diabetic neuropathy: One disease or two? Curr. Diabetes Rep. 2013, 13, 533–549. [Google Scholar] [CrossRef]
- Nguyen, P.T.; Yarov-Yarovoy, V. Towards Structure-Guided Development of Pain Therapeutics Targeting Voltage-Gated Sodium Channels. Front. Pharmacol. 2022, 13, 842032. [Google Scholar] [CrossRef]
- Mogil, J.S. Sources of Individual Differences in Pain. Annu. Rev. Neurosci. 2021, 44, 1–25. [Google Scholar] [CrossRef]
- Almomani, R.; Sopacua, M.; Marchi, M.; Ślęczkowska, M.; Lindsey, P.; Greef, B.T.d.; Hoeijmakers, J.G.J.; Salvi, E.; Merkies, I.S.J.; Fadavi, H.; et al. Genetic profiling of sodium channels in painful and painless diabetic and idiopathic small fiber neuropathy. 2022; in preparation. [Google Scholar]
- Huang, J.; Han, C.; Estacion, M.; Vasylyev, D.; Hoeijmakers, J.G.; Gerrits, M.M.; Tyrrell, L.; Lauria, G.; Faber, C.G.; Dib-Hajj, S.D.; et al. Gain-of-function mutations in sodium channel Na(v)1.9 in painful neuropathy. Brain J. Neurol. 2014, 137, 1627–1642. [Google Scholar] [CrossRef] [Green Version]
- Faber, C.G.; Lauria, G.; Merkies, I.S.; Cheng, X.; Han, C.; Ahn, H.S.; Persson, A.K.; Hoeijmakers, J.G.; Gerrits, M.M.; Pierro, T.; et al. Gain-of-function Nav1.8 mutations in painful neuropathy. Proc. Natl. Acad. Sci. USA 2012, 109, 19444–19449. [Google Scholar] [CrossRef] [PubMed]
- Faber, C.G.; Hoeijmakers, J.G.; Ahn, H.S.; Cheng, X.; Han, C.; Choi, J.S.; Estacion, M.; Lauria, G.; Vanhoutte, E.K.; Gerrits, M.M.; et al. Gain of function Naν1.7 mutations in idiopathic small fiber neuropathy. Ann. Neurol. 2012, 71, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Alsaloum, M.; Estacion, M.; Almomani, R.; Gerrits, M.M.; Bönhof, G.J.; Ziegler, D.; Malik, R.; Ferdousi, M.; Lauria, G.; Merkies, I.S.; et al. A gain-of-function sodium channel β2-subunit mutation in painful diabetic neuropathy. Mol. Pain 2019, 15, 1744806919849802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoeijmakers, J.G.; Faber, C.G.; Merkies, I.S.; Waxman, S.G. Painful peripheral neuropathy and sodium channel mutations. Neurosci. Lett. 2015, 596, 51–59. [Google Scholar] [CrossRef]
- Dib-Hajj, S.D.; Rush, A.M.; Cummins, T.R.; Hisama, F.M.; Novella, S.; Tyrrell, L.; Marshall, L.; Waxman, S.G. Gain-of-function mutation in Nav1.7 in familial erythromelalgia induces bursting of sensory neurons. Brain J. Neurol. 2005, 128, 1847–1854. [Google Scholar] [CrossRef]
- Basso, L.; Altier, C. Transient Receptor Potential Channels in neuropathic pain. Curr. Opin. Pharmacol. 2017, 32, 9–15. [Google Scholar] [CrossRef]
- Busserolles, J.; Tsantoulas, C.; Eschalier, A.; López García, J.A. Potassium channels in neuropathic pain: Advances, challenges, and emerging ideas. Pain 2016, 157 (Suppl. S1), S7–S14. [Google Scholar] [CrossRef]
- Rivolta, I.; Binda, A.; Masi, A.; DiFrancesco, J.C. Cardiac and neuronal HCN channelopathies. Pflug. Arch. Eur. J. Physiol. 2020, 472, 931–951. [Google Scholar] [CrossRef]
- Cho, H.; Yang, Y.D.; Lee, J.; Lee, B.; Kim, T.; Jang, Y.; Back, S.K.; Na, H.S.; Harfe, B.D.; Wang, F.; et al. The calcium-activated chloride channel anoctamin 1 acts as a heat sensor in nociceptive neurons. Nat. Neurosci. 2012, 15, 1015–1021. [Google Scholar] [CrossRef]
- Takayama, Y.; Uta, D.; Furue, H.; Tominaga, M. Pain-enhancing mechanism through interaction between TRPV1 and anoctamin 1 in sensory neurons. Proc. Natl. Acad. Sci. USA 2015, 112, 5213–5218. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.; Wang, X.; Ostertag, E.M.; Nuwal, T.; Huang, B.; Jan, Y.N.; Basbaum, A.I.; Jan, L.Y. TMEM16C facilitates Na+-activated K+ currents in rat sensory neurons and regulates pain processing. Nat. Neurosci. 2013, 16, 1284–1290. [Google Scholar] [CrossRef] [PubMed]
- Ślęczkowska, M.; Almomani, R.; Marchi, M.; de Greef, B.T.A.; Sopacua, M.; Hoeijmakers, J.G.J.; Lindsey, P.; Salvi, E.; Bönhof, G.J.; Ziegler, D.; et al. Peripheral Ion Channel Gene Screening in Painful- and Painless-Diabetic Neuropathy. Int. J. Mol. Sci. 2022, 23, 7190. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, B.A.; Bakkers, M.; Hoeijmakers, J.G.; Faber, C.G.; Merkies, I.S. Improving assessment in small fiber neuropathy. J. Peripher. Nerv. Syst. 2015, 20, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Zech, M.; Gross, N.; Jochim, A.; Castrop, F.; Kaffe, M.; Dresel, C.; Lichtner, P.; Peters, A.; Gieger, C.; Meitinger, T.; et al. Rare sequence variants in ANO3 and GNAL in a primary torsion dystonia series and controls. Mov. Disord. Off. J. Mov. Disord. Soc. 2014, 29, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Perrone, F.; Cacace, R.; van der Zee, J.; Van Broeckhoven, C. Emerging genetic complexity and rare genetic variants in neurodegenerative brain diseases. Genome Med. 2021, 13, 59. [Google Scholar] [CrossRef] [PubMed]
- Federici, G.; Soddu, S. Variants of uncertain significance in the era of high-throughput genome sequencing: A lesson from breast and ovary cancers. J. Exp. Clin. Cancer Res. 2020, 39, 46. [Google Scholar] [CrossRef] [Green Version]
- Grandl, J.; Kim, S.E.; Uzzell, V.; Bursulaya, B.; Petrus, M.; Bandell, M.; Patapoutian, A. Temperature-induced opening of TRPV1 ion channel is stabilized by the pore domain. Nat. Neurosci. 2010, 13, 708–714. [Google Scholar] [CrossRef] [Green Version]
- Papke, D.; Grosman, C. The role of intracellular linkers in gating and desensitization of human pentameric ligand-gated ion channels. J. Neurosci. 2014, 34, 7238–7252. [Google Scholar] [CrossRef] [Green Version]
- Gaudet, R. A primer on ankyrin repeat function in TRP channels and beyond. Mol. Biosyst. 2008, 4, 372–379. [Google Scholar] [CrossRef] [Green Version]
- Mosavi, L.K.; Cammett, T.J.; Desrosiers, D.C.; Peng, Z.Y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. Publ. Protein Soc. 2004, 13, 1435–1448. [Google Scholar] [CrossRef] [Green Version]
- Krishna, M.M.; Englander, S.W. The N-terminal to C-terminal motif in protein folding and function. Proc. Natl. Acad. Sci. USA 2005, 102, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Brewster, M.S.J.; Gaudet, R. How the TRPA1 receptor transmits painful stimuli: Inner workings revealed by electron cryomicroscopy. Bioessays 2015, 37, 1184–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.F.A.; Chen, C.; Huang, F.; Feng, S.J.; Tien, J.S.; Braz, J.M.; Basbaum, A.I.; Jan, Y.N.; Jan, L.Y. TMEM16C is involved in thermoregulation and protects rodent pups from febrile seizures. Proc. Natl. Acad. Sci. USA 2021, 118, e2023342118. [Google Scholar] [CrossRef] [PubMed]
- Tsantoulas, C. Emerging potassium channel targets for the treatment of pain. Curr. Opin. Support Palliat. Care 2015, 9, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yang, C.X.; Zhong, J.Y.; Wang, H.B. Intrathecal TRESK gene recombinant adenovirus attenuates spared nerve injury-induced neuropathic pain in rats. Neuroreport 2013, 24, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chen, H.T.; Yang, C.X.; Zhong, J.Y.; He, W.Y.; Xiong, Q.M. Reversal of TRESK Downregulation Alleviates Neuropathic Pain by Inhibiting Activation of Gliocytes in the Spinal Cord. Neurochem. Res. 2017, 42, 1288–1298. [Google Scholar] [CrossRef]
- Yuan, J.H.; Estacion, M.; Mis, M.A.; Tanaka, B.S.; Schulman, B.R.; Chen, L.; Liu, S.; Dib-Hajj, F.B.; Dib-Hajj, S.D.; Waxman, S.G. KCNQ variants and pain modulation: A missense variant in Kv7.3 contributes to pain resilience. Brain Commun. 2021, 3, fcab212. [Google Scholar] [CrossRef]
- Gonzalez-Ramirez, R.; Chen, Y.; Liedtke, W.B.; Morales-Lazaro, S.L. TRP Channels and Pain. In Neurobiology of TRP Channels; Emir, T.L.R., Ed.; CRC Press: Boca Raton, FL, USA, 2017; pp. 125–147. [Google Scholar] [CrossRef]
- Obata, K.; Katsura, H.; Mizushima, T.; Yamanaka, H.; Kobayashi, K.; Dai, Y.; Fukuoka, T.; Tokunaga, A.; Tominaga, M.; Noguchi, K. TRPA1 induced in sensory neurons contributes to cold hyperalgesia after inflammation and nerve injury. J. Clin. Investig. 2005, 115, 2393–2401. [Google Scholar] [CrossRef] [Green Version]
- Dai, Y.; Wang, S.; Tominaga, M.; Yamamoto, S.; Fukuoka, T.; Higashi, T.; Kobayashi, K.; Obata, K.; Yamanaka, H.; Noguchi, K. Sensitization of TRPA1 by PAR2 contributes to the sensation of inflammatory pain. J. Clin. Investig. 2007, 117, 1979–1987. [Google Scholar] [CrossRef] [Green Version]
- McNamara, C.R.; Mandel-Brehm, J.; Bautista, D.M.; Siemens, J.; Deranian, K.L.; Zhao, M.; Hayward, N.J.; Chong, J.A.; Julius, D.; Moran, M.M.; et al. TRPA1 mediates formalin-induced pain. Proc. Natl. Acad. Sci. USA 2007, 104, 13525–13530. [Google Scholar] [CrossRef]
- Kremeyer, B.; Lopera, F.; Cox, J.J.; Momin, A.; Rugiero, F.; Marsh, S.; Woods, C.G.; Jones, N.G.; Paterson, K.J.; Fricker, F.R.; et al. A gain-of-function mutation in TRPA1 causes familial episodic pain syndrome. Neuron 2010, 66, 671–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nirenberg, M.J.; Chaouni, R.; Biller, T.M.; Gilbert, R.M.; Paisán-Ruiz, C. A novel TRPA1 variant is associated with carbamazepine-responsive cramp-fasciculation syndrome. Clin. Genet. 2018, 93, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.T.; Wang, G.X.; Wei, N.N.; Wang, K. A pivotal role for the activation of TRPV3 channel in itch sensations induced by the natural skin sensitizer carvacrol. Acta Pharmacol. Sin. 2018, 39, 331–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, J.H.; Schulman, B.R.; Effraim, P.R.; Sulayman, D.H.; Jacobs, D.S.; Waxman, S.G. Genomic analysis of 21 patients with corneal neuralgia after refractive surgery. Pain Rep. 2020, 5, e826. [Google Scholar] [CrossRef]
- Estacion, M.; Vohra, B.P.S.; Liu, S.; Hoeijmakers, J.; Faber, C.G.; Merkies, I.S.J.; Lauria, G.; Black, J.A.; Waxman, S.G. Ca2+ toxicity due to reverse Na+/Ca2+ exchange contributes to degeneration of neurites of DRG neurons induced by a neuropathy-associated Nav1.7 mutation. J. Neurophysiol. 2015, 114, 1554–1564. [Google Scholar] [CrossRef] [Green Version]
- De Greef, B.T.A.; Hoeijmakers, J.G.J.; Gorissen-Brouwers, C.M.L.; Geerts, M.; Faber, C.G.; Merkies, I.S.J. Associated conditions in small fiber neuropathy—A large cohort study and review of the literature. Eur. J. Neurol. 2018, 25, 348–355. [Google Scholar] [CrossRef] [Green Version]
- Delgado, D.A.; Lambert, B.S.; Boutris, N.; McCulloch, P.C.; Robbins, A.B.; Moreno, M.R.; Harris, J.D. Validation of Digital Visual Analog Scale Pain Scoring With a Traditional Paper-based Visual Analog Scale in Adults. J. Am. Acad. Orthop. Surgeons. Glob. Res. Rev. 2018, 2, e088. [Google Scholar] [CrossRef]
- Williamson, A.; Hoggart, B. Pain: A review of three commonly used pain rating scales. J. Clin. Nurs. 2005, 14, 798–804. [Google Scholar] [CrossRef]
- Almomani, R.; Marchi, M.; Sopacua, M.; Lindsey, P.; Salvi, E.; Koning, B.; Santoro, S.; Magri, S.; Smeets, H.J.M.; Martinelli Boneschi, F.; et al. Evaluation of molecular inversion probe versus TruSeq® custom methods for targeted next-generation sequencing. PLoS ONE 2020, 15, e0238467. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015, 17, 405–424. [Google Scholar] [CrossRef]
| Gene | c.Position & | p.Position | Number of Patients | Classification According to Richards et.al 2015 | Location | MAF gnomAD (%) |
|---|---|---|---|---|---|---|
| ANO3 | c.2656A>T | p.(Ile886Phe) | 1 | VUS | Linker between transmembrane domain VII and VIII | 0 |
| c.3100G>C | p.(Gly1034Arg) | 1 | VUS | C-terminus | 0.011 | |
| KCNK18 | c.1107del | p.(Met370Cysfs*?) | 1 | VUS | Frame shift starting at codon Met370 | 0.0016 |
| KCNQ3 | c.1885G>A | p.(Val629Ile) | 1 | VUS | C-terminus | 0.052 |
| c.1706A>G | p.(Asp569Gly) | 1 | VUS | C-terminus | 0.0018 | |
| TRPA1 | c.932C>A | p.(Thr311Asn) | 1 | VUS | Ankyrin repeat VIII-containing domain | 0.044 |
| c.980A>G | p.(Tyr327Cys) | 1 | VUS | Ankyrin repeat VIII-containing domain | 0 | |
| c.1177C>T | p.(Arg393*) | 1 | VUS | Stop codon in Ankyrin repeat X–containing domain | 0.018 | |
| c.1954C>T | p.(Arg652*) | 1 | VUS | Cytoplasmic domain between ANK repeats and transmembrane domain I | 0.015 | |
| c.2065A>G | p.(Met689Val) | 1 | VUS | Cytoplasmic domain between ANK repeats and transmembrane domain I | 0.0068 | |
| c.3136A>G | p.(Lys1046Glu) | 2 * | VUS | inositol-phosphate binding site in C-terminus | 0.0008 | |
| TRPM8 | c.665A>G | p.(Asn222Ser) | 1 | VUS | N-terminus | 0.0004 |
| c.1102C>T | p.(Arg368Trp) | 1 | VUS | N-terminus | 0.0016 | |
| c.2945C>T | p.(Thr982Met) | 1 | VUS | C-terminus | 0.005 | |
| TRPV1 | c.914T>G | p.(Phe305Cys) | 1 | VUS | Ankyrin repeat V-containing domain | 0.00054 |
| c.1348A>G | p.(Thr450Ala) | 1 | VUS | Transmembrane domain I | 0 | |
| c.1735C>T | p.(Arg579Cys) | 1 | VUS | Transmembrane domain V | 0.0017 | |
| TRPV3 | c.1242+1G>A | p.? ^ | 1 | VUS | Donor splice site of intron 9 | 0.0029 |
| c.2006T>C | p.(Leu669Pro) | 1 | VUS | Transmembrane domain VI | 0.0019 |
| Patients with SFN and ICG Variant N = 20 | Patients with SFN without ICG/VGSC Variant N = 394 | |
|---|---|---|
| Mean age at recruitment [years ± SD] | 52.2 (±13.3) | 54.2 (±13.9) |
| Females (n, %) | 11 (55.0) | 240 (±60.9) |
| Males (n, %) | 9 (45.0) | 154 (±39.1) |
| Mean age of onset neuropathy [years ± SD] | 46.2 (±12.2) | 47.1 (±13.4) |
| Duration of neuropathy [years ± SD] | 4.4 (±5.0) | 8.1 (±9.3) |
| Positive family history for neuropathy (n, %) | 5 (38.5) | 50 (22.2) |
| Negative family history for neuropathy (n, %) | 8 (61.5) | 175 (77.8) |
| Normal skin biopsy (n, %) | 4 (22.2) | 178 (58) |
| Abnormal skin biopsy (n, %) | 14 (77.8) | 129 (42) |
| Normal TTT (n, %) | 0 (0) | 25 (8.2) |
| Abnormal TTT (n, %) | 9 (100) | 280 (91.8) |
| Patient with ANO3 Variant (n = 1) | Patients with KCNQ3 Variant (n = 2) | Patient with TRPA1 Variant (n = 4) | Patients with ICG Variant (n = 7) | Patients without ICG Variant and without VGSC Variant (n = 204) | |
|---|---|---|---|---|---|
| Maximal pain [VAS] [±SD] | 9.7 | 8.6 (±0.14) | 9.65 (±0.7) | 9.36 (±0.72) | 7.47 (±2.37) |
| Gene | Variant | Gender | Onset Complaints | NCS | TTT | IENFD | Itch | Muscle Cramps | Warmth Influence | Cold Influence | Exercise Influence | Rest Influence | Temperature Sensation | Pain Sensation | Allodynia | Sleep Pattern |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANO3 | (Gly1034Arg) | M | 18 | n | a/n | n | N | N | - | - | N | N | n | ↑ | Y | n |
| KCNQ3 | (Val629Ile) | M | 49 | a/n | a/n | a/n | N | N | - | Y ↑ | Y ↑ | Y ↑ | n | - | Y | n |
| KCNQ3 | (Asp569Gly) | M | 46 | n | a/n | n | N | Y | - | - | - | Y ↑ | - | - | Y | - |
| TRPA1 | (Arg393*) | M | 50 | a/n | a/n | a/n | N | Y | N | N | Y ↑ | - | n | - | Y | a/n |
| TRPA1 | (Arg652*) | F | 17 | n | a/n | n | N | N | Y ↑ | Y ↑ | Y ↑ | N | ↓ | ↓ | Y | a/n |
| TRPA1 | (Met689Val) | F | 34 | n | a/n | a/n | Y | N | Y ↑ | - | Y ↑ | Y ↑ | - | ↑ | Y | a/n |
| TRPA1 | (Tyr327Cys) | M | 45 | n | - | a/n | N | Y | - | - | - | - | - | - | - | - |
| TRPM8 | (Arg368Trp) | M | 45 | n | a/n | a/n | N | N | - | - | - | - | ↓ | n | - | - |
| TRPV1 | (Arg579Cys) | M | 43 | n | a/n | n | Y | Y | N | N | - | Y ↑ | ↓ | n | Y | a/n |
| Feature | NCS N = 308 | TTT N = 305 | IENFD N = 307 | Temperature Sensation N = 204 | Pain Sensation N = 145 | Sleep Pattern N = 130 | |
|---|---|---|---|---|---|---|---|
| Normal | 297, 96.4% | 25, 8.2% | 178, 58% | 120, 58.8% | 73, 50.3% | 35, 26.9% | |
| Abnormal | 11, 3.6% | 280, 91.8% | 129, 42% | 84, 41.2% (1, 0.5% ↑, 83, 40.7% ↓) | 72, 49.7% (33, 22.8% ↑, 39, 26.9% ↓) | 95, 73.1% | |
| Feature | Itch N = 308 | Muscle Cramps N = 308 | Warmth Influence N = 172 | Cold Influence N = 169 | Exercise Influence N = 195 | Rest Influence N = 123 | Allodynia N = 211 |
| yes | 25, 8.1% | 56, 18.2% | 84, 48.8% (63, 36.6% complains ↑, 21, 12.2% complains ↓) | 82, 48.5% (54, 32% complains ↑, 28, 16.6% complains ↓) | 161, 82.6% (144,. 73.8% complains ↑, 17, 8.7% complains ↓) | 54, 68.3% (45, 36.6% complains ↑, 39, 31.7% complains ↓) | 193, 91.5% |
| no | 283, 91.9% | 252, 81.8% | 88, 51.2% | 87, 51.5% | 34, 17.4% | 39, 31.7% | 18, 8.5% |
| Gene | Variant | Sweating Change | Diarrhea | Constipation | Micturition Problem | Dry Eyes | Dry Mouth | Dizziness on Standing | Palpitations | Hot Flashes | Hypersensitivity of Leg’s Skin | Burning Feet | Sheet Intolerance | Restless Leg |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANO3 | (Gly1034Arg) | 4 | 1 | 1 | 3 | 3 | 4 | 2 | - | 1 | 4 | 4 | 4 | 4 |
| KCNQ3 | (Val629Ile) | 2 | 2 | 2 | 2 | 1 | 2 | 1 | 1 | 2 | 1 | 3 | 2 | 3 |
| KCNQ3 | (Asp569Gly) | 3 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 2 | 3 | 4 | 3 | 4 |
| TRPA1 | (Arg393*) | - | 2 | 3 | 3 | 1 | 3 | 2 | 1 | 2 | 4 | 4 | 4 | 4 |
| TRPA1 | (Arg652*) | 3 | 2 | 3 | 2 | 1 | 3 | 3 | 2 | 2 | 3 | 3 | 3 | 3 |
| TRPA1 | (Met689Val) | 3 | 3 | 3 | 2 | 4 | 4 | 3 | 3 | 3 | 4 | 3 | 4 | 4 |
| Frequency of Complaint | Sweating Change N = 268 | Diarrhea N = 272 | Constipation N = 275 | Micturition Problem N = 273 | Dry Eyes N = 273 | Dry Mouth N = 273 | Orthostatic Dizziness N = 271 | Palpitations N = 268 | Hot Flashes N = 270 | Hypersensitivity of Leg’s Skin N = 273 | Burning Feet n = 269 | Sheet Intolerance N = 272 | Restless Leg N = 272 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| never | 56, 20.9% | 114, 41.9% | 115, 41.8% | 91, 33.3% | 95, 34.8% | 54, 19.8% | 73, 26.9% | 105, 39.2% | 86, 31.9% | 52, 19.0% | 21, 7.8% | 70, 25.7% | 36, 13.2% |
| sometimes | 94, 35.1% | 110, 40.4% | 86, 31.3% | 75, 27.5% | 87, 31.9% | 97, 35.5% | 130, 48.0% | 108, 40.3% | 94, 34.8% | 75, 27.5% | 35, 13.0% | 76, 27.9% | 80, 29.4% |
| often | 85, 31.7% | 44, 16.2% | 55, 20.0% | 80, 29.3% | 65, 23.8% | 91, 33.3% | 59, 21.8% | 54, 20.1% | 86, 31.9% | 70, 25.6% | 101, 37.5% | 68, 25.0% | 91, 33.5% |
| always | 33, 12.3% | 4, 1.5% | 19, 6.9% | 27, 9.9% | 26, 9.5% | 31, 11.4% | 9, 3.3% | 1, 0.4% | 4, 1.5% | 76, 27.8% | 112, 41.6% | 58, 21.3% | 65, 23.9% |
| Ion Channel Family | Gene | OMIM Number | Full Gene Name |
|---|---|---|---|
| Anoctamins | ANO1 | 610108 | Anoctamin 1, calcium activated chloride channel |
| ANO3 | 610110 | Anoctamin 3 | |
| Non-selective cation channels | HCN1 | 602780 | Hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| Potassium channels | KCNA2 | 176262 | Potassium voltage-gated channel, shaker-related subfamily, member 2 |
| KCNA4 | 176266 | Potassium voltage-gated channel, shaker-related subfamily, member 4 | |
| KCNK18 | 613655 | Potassium channel, subfamily K, member 18 | |
| KCNN1 | 602982 | Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 | |
| KCNQ3 | 602232 | Potassium voltage-gated channel, KQT-like subfamily, member 3 | |
| KCNQ5 | 607357 | Potassium voltage-gated channel, KQT-like subfamily, member 5 | |
| KCNS1 | 602905 | Potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 | |
| Transient receptors | TRPA1 | 604775 | Transient receptor potential cation channel, subfamily A, member 1 |
| TRPM8 | 606678 | Transient receptor potential cation channel, subfamily M, member 8 | |
| TRPV1 | 602076 | Transient receptor potential cation channel, subfamily V, member 1 | |
| TRPV3 | 607066 | Transient receptor potential cation channel, subfamily V, member 3 | |
| TRPV4 | 605427 | Transient receptor potential cation channel, subfamily V, member 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ślęczkowska, M.; Almomani, R.; Marchi, M.; Salvi, E.; de Greef, B.T.A.; Sopacua, M.; Hoeijmakers, J.G.J.; Lindsey, P.; Waxman, S.G.; Lauria, G.; et al. Peripheral Ion Channel Genes Screening in Painful Small Fiber Neuropathy. Int. J. Mol. Sci. 2022, 23, 14095. https://doi.org/10.3390/ijms232214095
Ślęczkowska M, Almomani R, Marchi M, Salvi E, de Greef BTA, Sopacua M, Hoeijmakers JGJ, Lindsey P, Waxman SG, Lauria G, et al. Peripheral Ion Channel Genes Screening in Painful Small Fiber Neuropathy. International Journal of Molecular Sciences. 2022; 23(22):14095. https://doi.org/10.3390/ijms232214095
Chicago/Turabian StyleŚlęczkowska, Milena, Rowida Almomani, Margherita Marchi, Erika Salvi, Bianca T A de Greef, Maurice Sopacua, Janneke G J Hoeijmakers, Patrick Lindsey, Stephen G Waxman, Giuseppe Lauria, and et al. 2022. "Peripheral Ion Channel Genes Screening in Painful Small Fiber Neuropathy" International Journal of Molecular Sciences 23, no. 22: 14095. https://doi.org/10.3390/ijms232214095





