The Crosstalk between HepG2 and HMC-III Cells: In Vitro Modulation of Gene Expression with Conditioned Media
Abstract
:1. Introduction
2. Results
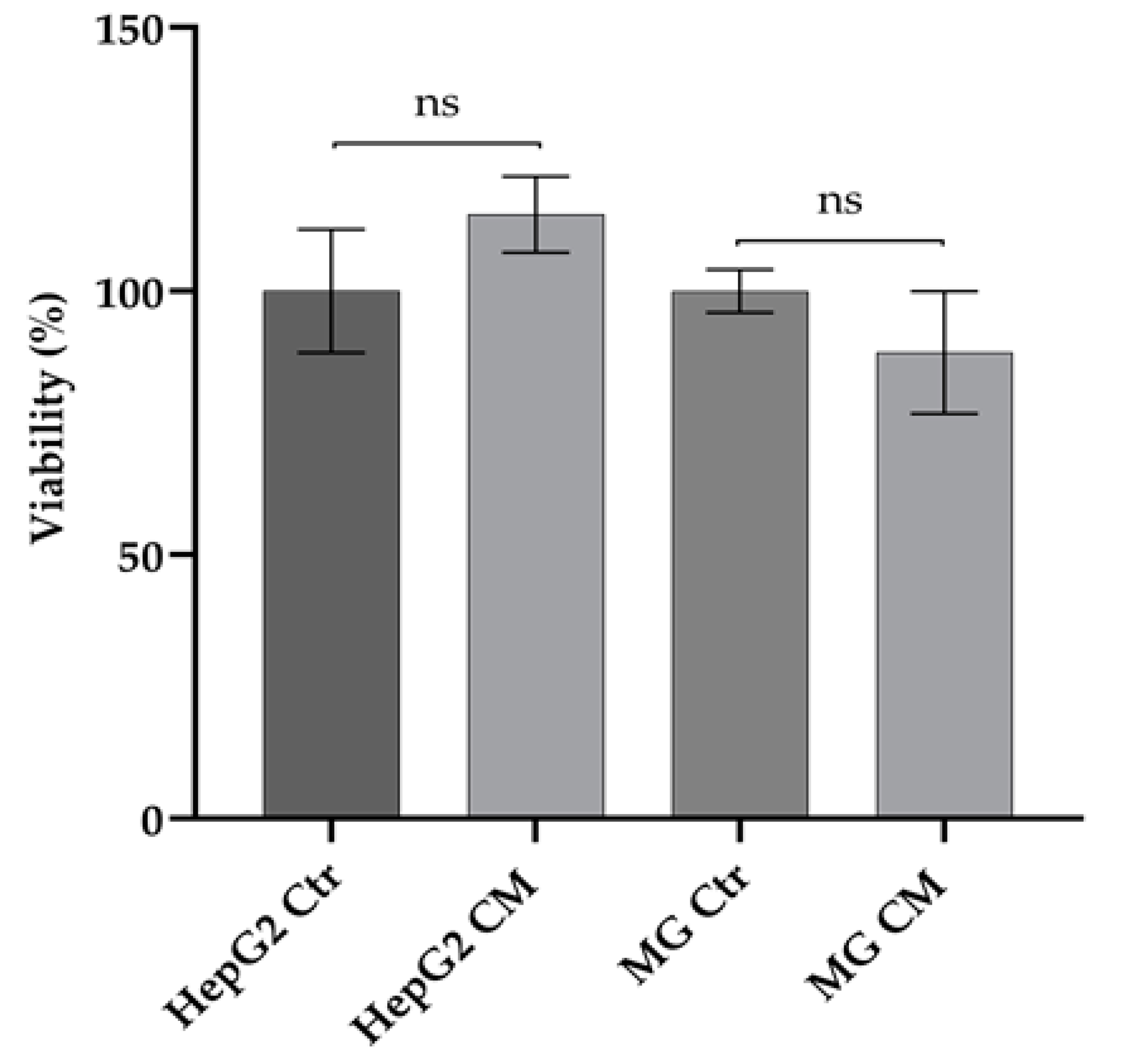
2.1. Effect of Conditioned Media on Viability of HepG2 and HMC-III Cells
2.2. Effect of Conditioned Media on the Expression Level of Endoplasmic Reticulum Stress Markers
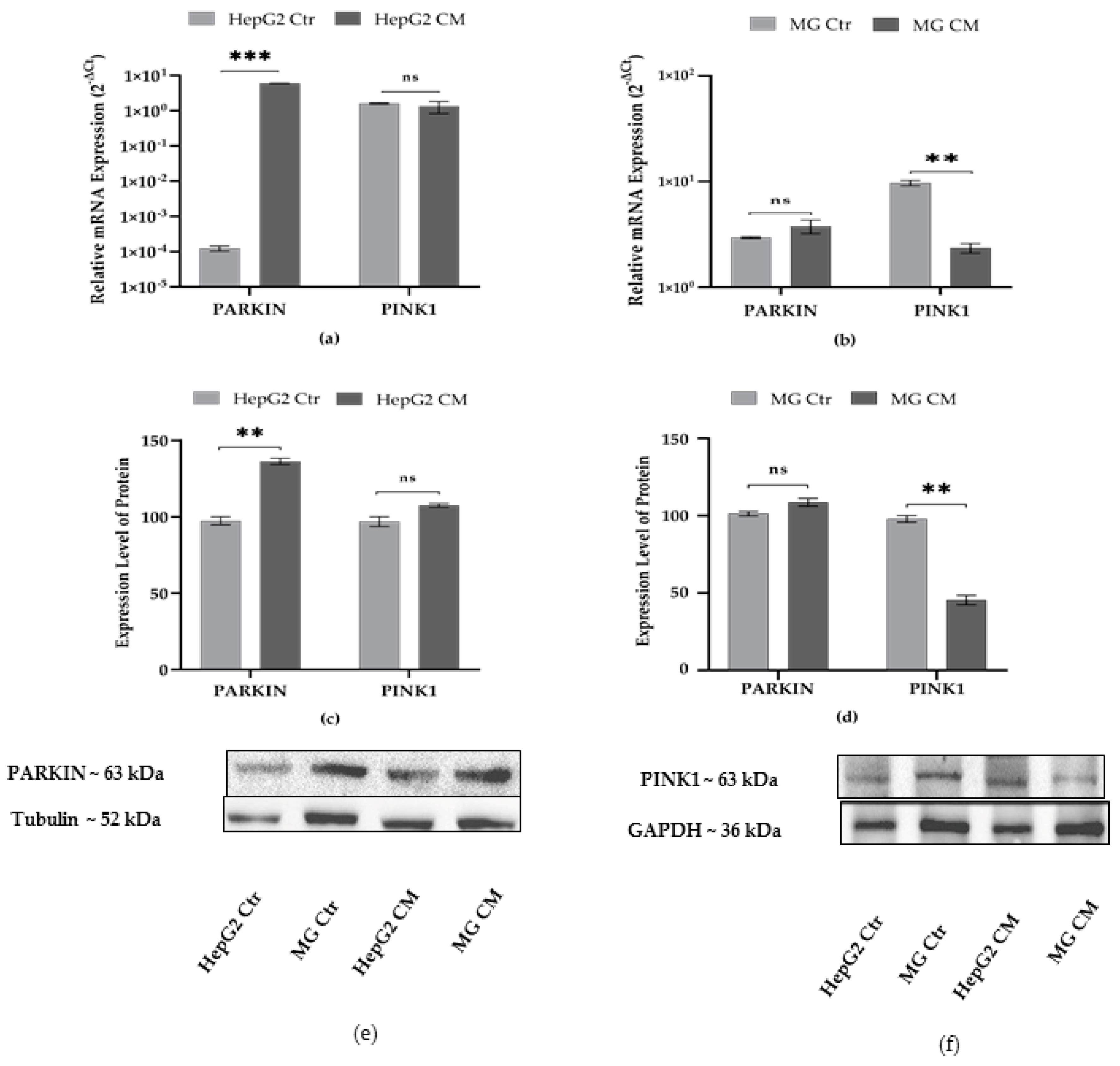
2.3. Effect of Conditioned Media on Mitophagy Markers
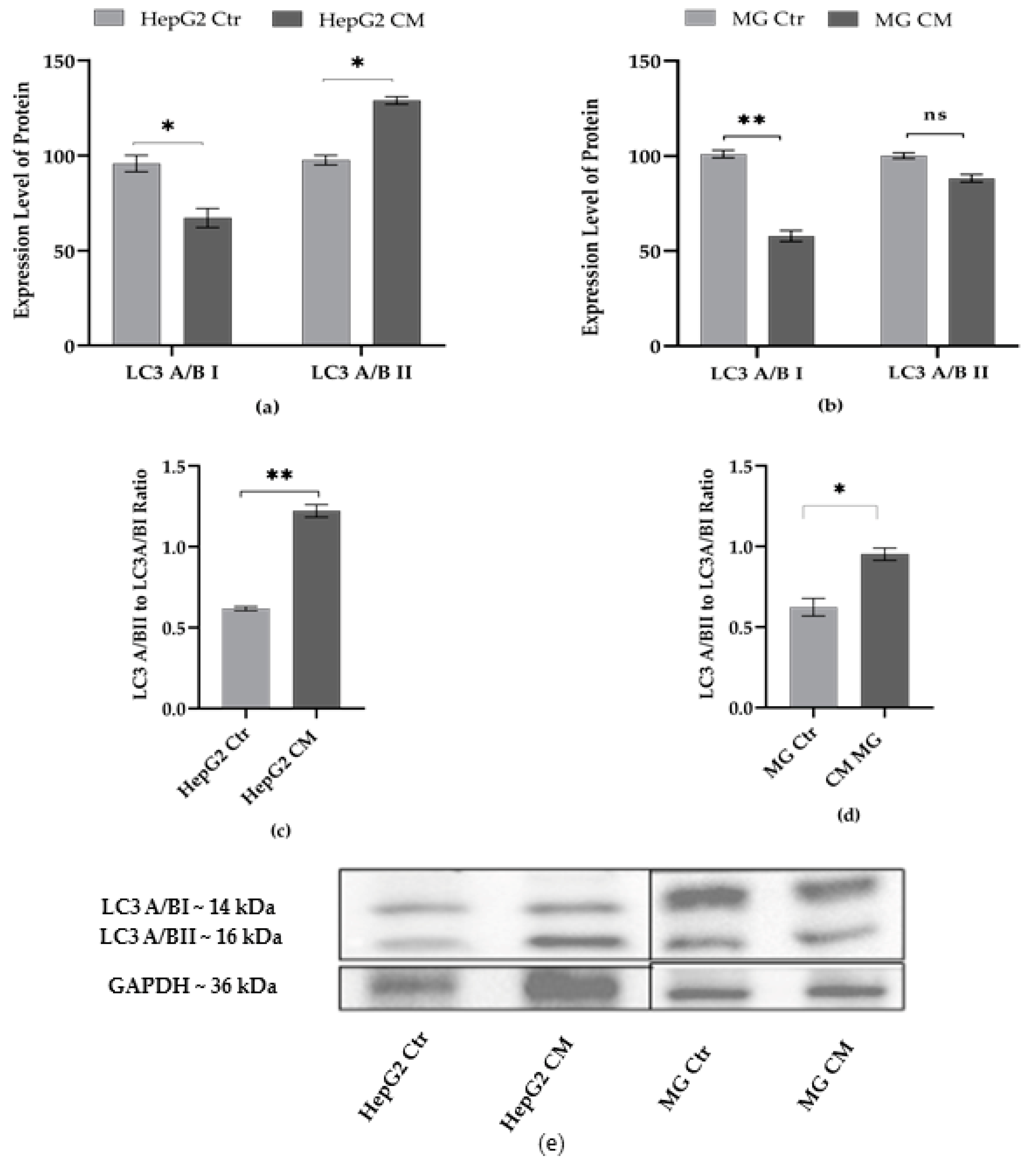
2.4. Effect of Condition Media on Autophagy Markers
2.5. Effect of Conditioned Media on Inflammatory Markers
2.6. Effect of Conditioned Media on Cell Cycle Markers
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Preparation of Conditioned Media
4.2. Viability Assay
4.3. Real-Time PCR
4.4. Western Blot Analysis
4.5. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Houck, A.L.; Seddighi, S.; Driver, J.A. At the crossroads between neurodegeneration and cancer: A review of overlapping biology and its implications. Curr. Aging Sci. 2018, 11, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Driver, J.A. Understanding the link between cancer and neurodegeneration. J. Geriatr. Oncol. 2012, 3, 58–67. [Google Scholar] [CrossRef]
- Seo, J.; Park, M. Molecular crosstalk between cancer and neurodegenerative diseases. Cell Mol. Life Sci. 2020, 77, 2659–2680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plun-Favreau, H.; Lewis, P.A.; Hardy, J.; Martins, L.M.; Wood, N.W. Cancer and neurodegeneration: Between the devil and the deep blue sea. PLoS Genet. 2010, 6, e1001257. [Google Scholar] [CrossRef] [Green Version]
- Bajaj, A.; Driver, J.A.; Schernhammer, E.S. Parkinson’s disease and cancer risk: A systematic review and meta-analysis. Cancer Causes Control 2010, 21, 697–707. [Google Scholar] [CrossRef]
- Ospina-Romero, M.; Glymour, M.M.; Hayes-Larson, E.; Mayeda, E.R.; Graff, R.E.; Brenowitz, W.D.; Ackley, S.F.; Witte, J.S.; Kobayashi, L.C. Association between Alzheimer disease and cancer with evaluation of study biases: A systematic review and meta-analysis. JAMA Netw. Open 2020, 3, e2025515. [Google Scholar] [CrossRef]
- Hang, Z.; Lei, T.; Zeng, Z.; Cai, S.; Bi, W.; Du, H. Composition of intestinal flora affects the risk relationship between Alzheimer’s disease/Parkinson’s disease and cancer. Biomed. Pharmacother. 2022, 145, 112343. [Google Scholar] [CrossRef]
- Park, J.-H.; Kim, D.-H.; Park, Y.-G.; Kwon, D.-Y.; Choi, M.; Jung, J.-H.; Han, K. Cancer risk in patients with Parkinson’s disease in South Korea: A nationwide, population-based cohort study. Eur. J. Cancer 2019, 117, 5–13. [Google Scholar] [CrossRef]
- Ibáñez, K.; Boullosa, C.; Tabarés-Seisdedos, R.; Baudot, A.; Valencia, A. Molecular evidence for the inverse comorbidity between central nervous system disorders and cancers detected by transcriptomic meta-analyses. PLoS Genet. 2014, 10, e1004173. [Google Scholar] [CrossRef]
- Forés-Martos, J.; Boullosa, C.; Rodrigo-Domínguez, D.; Sánchez-Valle, J.; Suay-García, B.; Climent, J.; Falcó, A.; Valencia, A.; Puig-Butillé, J.-A.; Puig, S.; et al. Transcriptomic and genetic associations between Alzheimer’s disease, Parkinson’s disease, and Cancer. Cancers 2021, 13, 2990. [Google Scholar] [CrossRef]
- Yalçin, M.; Malhan, D.; Basti, A.; Peralta, A.-R.; Ferreira, J.-J.; Relógio, A. A Computational Analysis in a Cohort of Parkinson’s Disease Patients and Clock-Modified Colorectal Cancer Cells Reveals Common Expression Alterations in Clock-Regulated Genes. Cancers 2021, 13, 5978. [Google Scholar] [CrossRef] [PubMed]
- Rudzińska, M.; Parodi, A.; Balakireva, A.V.; Chepikova, O.E.; Venanzi, F.M.; Zamyatnin, A.A., Jr. Cellular aging characteristics and their association with age-related disorders. Antioxidants 2020, 9, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Zhang, Z.; Ren, Y.; Wang, Y.; Fang, J.; Yue, H.; Ma, S.; Guan, F. Aging and age-related diseases: From mechanisms to therapeutic strategies. Biogerontology 2021, 22, 165–187. [Google Scholar] [CrossRef] [PubMed]
- Staropoli, J.F. Tumorigenesis and neurodegeneration: Two sides of the same coin? Bioessays 2008, 30, 719–727. [Google Scholar] [CrossRef]
- Varela, L.; Garcia-Rendueles, M.E. Oncogenic Pathways in Neurodegenerative Diseases. Int. J. Mol. Sci. 2022, 23, 3223. [Google Scholar] [CrossRef]
- Zhao, Y.; Seluanov, A.; Gorbunova, V. Revelations about aging and disease from unconventional vertebrate model organisms. Annu. Rev. Genet. 2021, 55, 135–159. [Google Scholar] [CrossRef]
- Kennedy, S.R.; Loeb, L.A.; Herr, A.J. Somatic mutations in aging, cancer and neurodegeneration. Mechanisms of ageing and development. Mech. Ageing Dev. 2012, 133, 118–126. [Google Scholar] [CrossRef] [Green Version]
- Lagoumtzi, S.M.; Chondrogianni, N. Senolytics and senomorphics: Natural and synthetic therapeutics in the treatment of aging and chronic diseases. Free Radic. Biol. Med. 2021, 171, 169–190. [Google Scholar] [CrossRef]
- Si, H.; Liu, D. Dietary antiaging phytochemicals and mechanisms associated with prolonged survival. J. Nutr. Biochem. 2014, 25, 581–591. [Google Scholar] [CrossRef] [Green Version]
- Niccoli, T.; Partridge, L. Ageing as a risk factor for disease. Curr. Boil. 2012, 22, R741–R752. [Google Scholar] [CrossRef]
- Rosenberg, G.A. Neurological diseases in relation to the blood–brain barrier. J. Cereb. Blood Flow Metab. 2012, 32, 1139–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, M.; Xiao, Z.J.; Yang, B.; Lan, Z.; Fang, F. Blood-brain barrier: More contributor to disruption of central nervous system homeostasis than victim in neurological disorders. Front. Neurosci. 2020, 14, 764. [Google Scholar] [CrossRef] [PubMed]
- Takata, F.; Nakagawa, S.; Matsumoto, J.; Dohgu, S. Blood-brain barrier dysfunction amplifies the development of neuroinflammation: Understanding of cellular events in brain microvascular endothelial cells for prevention and treatment of BBB dysfunction. Front. Cell. Neurosci. 2021, 15, 344. [Google Scholar] [CrossRef] [PubMed]
- Sade, D.; Shaham-Niv, S.; Arnon, Z.A.; Tavassoly, O.; Gazit, E. Seeding of proteins into amyloid structures by metabolite assemblies may clarify certain unexplained epidemiological associations. Open Biol. 2018, 8, 170229. [Google Scholar] [CrossRef] [Green Version]
- Bartman, C.R.; TeSlaa, T.; Rabinowitz, J.D. Quantitative flux analysis in mammals. Nat. Metab. 2021, 3, 896–908. [Google Scholar] [CrossRef]
- Ou, G.-Y.; Lin, W.-W.; Zhao, W.-J. Neuregulins in neurodegenerative diseases. Front. Aging Neurosci. 2021, 13, 170. [Google Scholar] [CrossRef]
- Di Santo, S.; Widmer, H.R. Paracrine factors for neurodegenerative disorders: Special emphasis on Parkinson’s disease. Neural Regen. Res. 2016, 11, 570. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, K.; Raz, A. Autocrine motility factor and its receptor expression in musculoskeletal tumors. J. Bone Oncol. 2020, 24, 100318. [Google Scholar] [CrossRef]
- Thomas, S.K.; Lee, J.; Beatty, G.L. Paracrine and cell autonomous signalling in pancreatic cancer progression and metastasis. EBioMedicine 2020, 53, 102662. [Google Scholar] [CrossRef]
- Dowling, P.; Clynes, M. Conditioned media from cell lines: A complementary model to clinical specimens for the discovery of disease-specific biomarkers. Proteomics 2011, 11, 794–804. [Google Scholar] [CrossRef]
- Song, P.; Kwon, Y.; Joo, J.-Y.; Kim, D.-G.; Yoon, J.-H. Secretomics to discover regulators in diseases. Int. J. Mol. Sci. 2019, 20, 3893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuda, T.; Sankowski, R.; Staszewski, O.; Prinz, M. Microglia heterogeneity in the single-cell era. Cell Rep. 2020, 30, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Colonna, M.; Butovsky, O. Microglia function in the central nervous system during health and neurodegeneration. Annu. Rev. Immunol. 2017, 35, 441. [Google Scholar] [CrossRef] [PubMed]
- Hickman, S.; Izzy, S.; Sen, P.; Morsett, L.; El Khoury, J. Microglia in neurodegeneration. Nat. Neurosci. 2018, 21, 1359–1369. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Jin, M.-Z.; Yang, Z.-Y.; Jin, W.-L. Microglia in neurodegenerative diseases. Neural Regen. Res. 2021, 16, 270. [Google Scholar] [CrossRef]
- Lull, M.E.; Block, M.L. Microglial activation and chronic neurodegeneration. Neurotherapeutics 2010, 7, 354–365. [Google Scholar] [CrossRef] [Green Version]
- Šešelja, K.; Bazina, I.; Welss, J.; Schicht, M.; Paulsen, F.; Bijelić, N.; Rođak, E.; Horvatić, A.; Gelemanović, A.; Mihalj, M.; et al. Effect of Tff3 deficiency and ER stress in the liver. Int. J. Mol. Sci. 2019, 20, 4389. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.; Kim, B. Anti-cancer natural products and their bioactive compounds inducing ER stress-mediated apoptosis: A review. Nutrients 2018, 10, 1021. [Google Scholar] [CrossRef] [Green Version]
- Sprenkle, N.-T.; Sims, S.-G.; Sánchez, C.-L.; Meares, G.-P. Endoplasmic reticulum stress and inflammation in the central nervous system. Mol. Neurodegener. 2017, 12, 42. [Google Scholar] [CrossRef] [Green Version]
- Mao, L.; Liu, H.; Zhang, R.; Deng, Y.; Hao, Y.; Liao, W.; Yuan, M.; Sun, S. PINK1/Parkin-mediated mitophagy inhibits warangalone-induced mitochondrial apoptosis in breast cancer cells. Aging 2021, 13, 12955. [Google Scholar] [CrossRef]
- Denisenko, T.V.; Gogvadze, V.; Zhivotovsky, B. Mitophagy in carcinogenesis and cancer treatment. Discov. Oncol. 2021, 12, 42. [Google Scholar] [CrossRef] [PubMed]
- Quinn, P.M.; Moreira, P.I.; Ambrósio, A.F.; Alves, C.H. PINK1/PARKIN signalling in neurodegeneration and neuroinflammation. Acta Neuropathol. Commun. 2020, 8, 189. [Google Scholar] [CrossRef] [PubMed]
- Ge, P.; Dawson, V.L.; Dawson, T.M. PINK1 and Parkin mitochondrial quality control: A source of regional vulnerability in Parkinson’s disease. Mol. Neurodegener. 2020, 15, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, J.; Shen, H.-M.; Lim, L.-H.-K. The role of autophagy in liver cancer: Crosstalk in signaling pathways and potential therapeutic targets. Pharmaceuticals 2020, 13, 432. [Google Scholar] [CrossRef]
- Nah, J.; Yuan, J.; Jung, Y.-K. Autophagy in neurodegenerative diseases: From mechanism to therapeutic approach. Mol. Cells 2015, 38, 381. [Google Scholar] [CrossRef] [Green Version]
- Vannini, F.; Kashfi, K.; Nath, N. The dual role of iNOS in cancer. Redox Biol. 2015, 6, 334–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, Y.; Shen, S.; Verma, I.-M. NF-κB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaghloul, N.; Kurepa, D.; Bader, M.-Y.; Nagy, N.; Ahmed, M.-N. Prophylactic inhibition of NF-κB expression in microglia leads to attenuation of hypoxic ischemic injury of the immature brain. J. Neuroinflamm. 2020, 17, 365. [Google Scholar] [CrossRef]
- Saha, R.-N.; Pahan, K. Regulation of inducible nitric oxide synthase gene in glial cells. Antioxid Redox Signal. 2006, 8, 929–947. [Google Scholar] [CrossRef]
- Ahmed, S.M.U.; Luo, L.; Namani, A.; Wang, X.J.; Tang, X. Nrf2 signaling pathway: Pivotal roles in inflammation. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 585–597. [Google Scholar] [CrossRef]
- Esteras, N.; Dinkova-Kostova, A.T.; Abramov, A.Y. Nrf2 activation in the treatment of neurodegenerative diseases: A focus on its role in mitochondrial bioenergetics and function. Biol. Chem. 2016, 397, 383–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Islam, M.-S.; Hu, Y.; Chen, X. TNFR2: Role in cancer immunology and immunotherapy. Immunotargets Ther. 2021, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Zhang, Y.; Ding, B.; Li, H. Targeting TNFR2 in cancer: All roads lead to Rome. Front. Immunol. 2022, 13, 460. [Google Scholar] [CrossRef] [PubMed]
- Veroni, C.; Gabriele, L.; Canini, I.; Castiello, L.; Coccia, E.; Remoli, M.-E.; Columba-Cabezas, S.; Aricò, E.; Aloisi, F.; Agresti, C. Activation of TNF receptor 2 in microglia promotes induction of anti-inflammatory pathways. Mol. Cell. Neurosci. 2010, 45, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Papazian, I.; Tsoukala, E.; Boutou, A.; Karamita, M.; Kambas, K.; Iliopoulou, L.; Fischer, R.; Kontermann, R.E.; Denis, M.C.; Kollias, G.; et al. Fundamentally different roles of neuronal TNF receptors in CNS pathology: TNFR1 and IKKβ promote microglial responses and tissue injury in demyelination while TNFR2 protects against excitotoxicity in mice. J. Neuroinflamm. 2021, 18, 222. [Google Scholar] [CrossRef]
- Jebelli, J.; Hooper, C.; Pocock, J.-M. Microglial p53 activation is detrimental to neuronal synapses during activation-induced inflammation: Implications for neurodegeneration. Neurosci. Lett. 2014, 583, 92–97. [Google Scholar] [CrossRef]
- Aloi, M.-S.; Su, W.; Garden, G.-A. The p53 transcriptional network influences microglia behavior and neuroinflammation. Crit. Rev. Immunol. 2015, 35, 401–415. [Google Scholar] [CrossRef] [Green Version]
- Corazzari, M.; Gagliardi, M.; Fimia, G.M.; Piacentini, M. Endoplasmicreticulum stress, unfoldedproteinresponse, and cancercell fate. Front. Oncol. 2017, 7, 78. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Ackerman, S.L. Endoplasmic reticulum stress in health and disease. Curr. Opin. Cell Biol. 2006, 18, 444–452. [Google Scholar] [CrossRef]
- Riha, R.; Gupta-Saraf, P.; Bhanja, P.; Badkul, S.; Saha, S. Stressed out–therapeutic implications of ER stress related cancer research. Oncomedicine 2017, 2, 156. [Google Scholar] [CrossRef]
- Yadav, R.K.; Chae, S.-W.; Kim, H.-R.; Chae, H.J. Endoplasmic reticulum stress and cancer. J. Cancer Prev. 2014, 19, 75. [Google Scholar] [CrossRef] [PubMed]
- Lindholm, D.; Wootz, H.; Korhonen, L. ER stress and neurodegenerative diseases. Cell Death Differ. 2006, 13, 385–392. [Google Scholar] [CrossRef] [Green Version]
- Senft, D.; Ze’ev, A.R. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem. Sci. 2015, 40, 141–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panigrahi, D.P.; Praharaj, P.P.; Bhol, C.S.; Mahapatra, K.K.; Patra, S.; Behera, B.P.; Mishra, S.R. The emerging, multifaceted role of mitophagy in cancer and cancer therapeutics. Semin. Cancer Biol. 2019, 66, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Lin, M.; Wu, R.; Wang, X.; Yang, B.; Levine, A.J.; Hu, W.; Feng, Z. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc. Natl. Acad. Sci. USA 2011, 108, 16259–16264. [Google Scholar] [CrossRef] [Green Version]
- Wardyn, J.D.; Ponsford, A.H.; Sanderson, C.M. Dissecting molecular cross-talk between Nrf2 and NF-κB response pathways. Biochem. Soc. Trans. 2015, 43, 621–626. [Google Scholar] [CrossRef] [Green Version]
- Abruzzese, V.; Matera, I.; Martinelli, F.; Carmosino, M.; Koshal, P.; Milella, L.; Bisaccia, F.; Ostuni, A. Effect of Quercetin on ABCC6 Transporter: Implication in HepG2 Migration. Int. J. Mol. Sci. 2021, 22, 3437. [Google Scholar] [CrossRef]



| Genes | Forward Primer | Reverse Primer |
|---|---|---|
| β-Actina | 5′-CCTGGCACCCAGCACAAT-3′ | 5′-GCCGATCCACACGGAGTACT-3′ |
| CHOP | 5′-GTACCTATGTTTCACCTCCTG-3′ | 5′-TCTCCTTCATGCGCTGCTTTC-3′ |
| IRE1α | 5′-AAGAGCCTGCCTTTCTCCCA-3′ | 5′-ATCTGAACTTCGGCATGGGG-3′ |
| PARKIN | 5′-ACTGTGCAGAATTGTGACCT-3′ | 5′-TTCTGGGGTCGTCGCCTCC-3′ |
| PINK1 | 5’-CAGTCACTTACAGAAAATCCA-3’ | 5’-CCTGCCGAGATGTTCCACA-3’ |
| iNOS | 5′-TCCAGAAGCAGAATGTGACC-3′ | 5′-AGCCAAATCCAGTCTGCCG-3′ |
| NF-kB | 5′-GATGAAGCATGGAACCATGG-3′ | 5′-CCCAAAGAGGTTTACAGTGT-3′ |
| TNFR2 | 5′-GGACTGATTGTGGGTGTGAC-3′ | 5′-CTTATCGGCAGGCAAGTGAG-3′ |
| p53 | 5′-TGAATGAGGCCTTGGAACTC-3′ | 5′-ACTTCAGGTGGCTGGAGTG-3′ |
| p21 | 5′-CTGTCTTGTACCCTTGTGCCT-3′ | 5′-CGTTTGGAGTGGTAGAAATCTGTC-3′ |
| Nrf2 | 5′-AACTACTCCCAGGTTGCCCA-3′ | 5′CATTGTCATCTACAAACGGGAA-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koshal, P.; Matera, I.; Abruzzese, V.; Ostuni, A.; Bisaccia, F. The Crosstalk between HepG2 and HMC-III Cells: In Vitro Modulation of Gene Expression with Conditioned Media. Int. J. Mol. Sci. 2022, 23, 14443. https://doi.org/10.3390/ijms232214443
Koshal P, Matera I, Abruzzese V, Ostuni A, Bisaccia F. The Crosstalk between HepG2 and HMC-III Cells: In Vitro Modulation of Gene Expression with Conditioned Media. International Journal of Molecular Sciences. 2022; 23(22):14443. https://doi.org/10.3390/ijms232214443
Chicago/Turabian StyleKoshal, Prashant, Ilenia Matera, Vittorio Abruzzese, Angela Ostuni, and Faustino Bisaccia. 2022. "The Crosstalk between HepG2 and HMC-III Cells: In Vitro Modulation of Gene Expression with Conditioned Media" International Journal of Molecular Sciences 23, no. 22: 14443. https://doi.org/10.3390/ijms232214443





