The Impact of Nanobody Density on the Targeting Efficiency of PEGylated Liposomes
Abstract
:1. Introduction
2. Results and Discussion
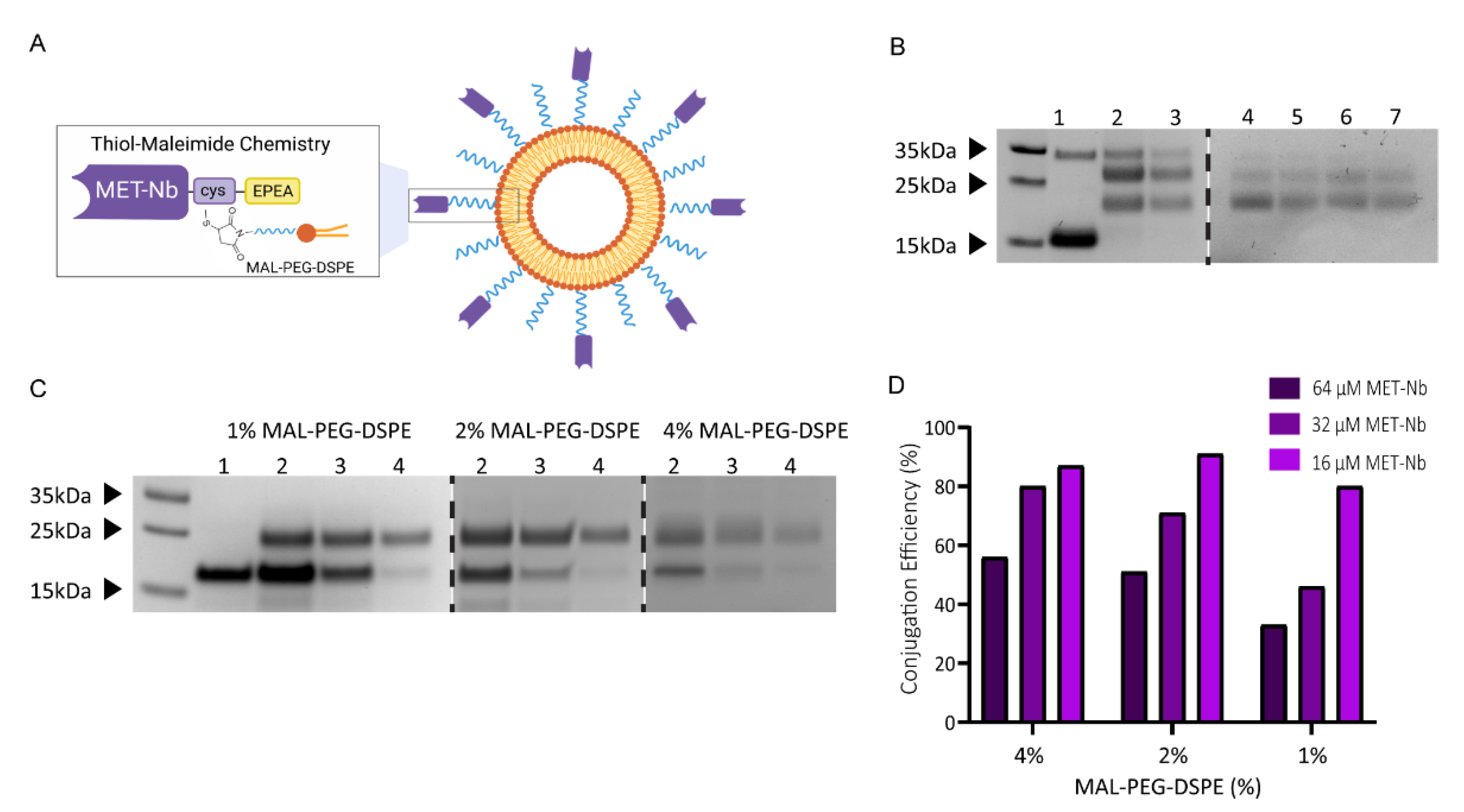
2.1. Characterization of Non-Targeted and MET-Targeted Liposomes
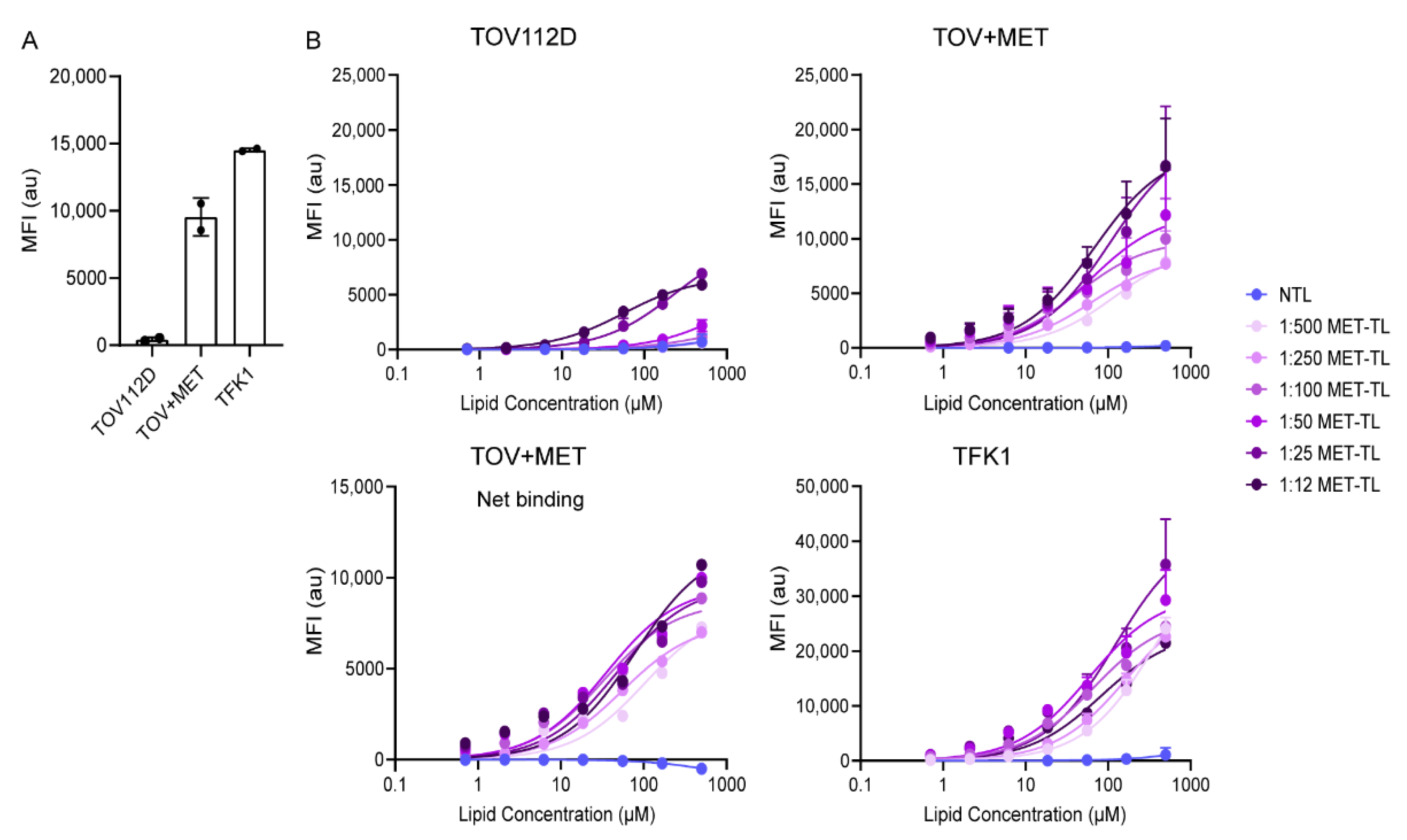
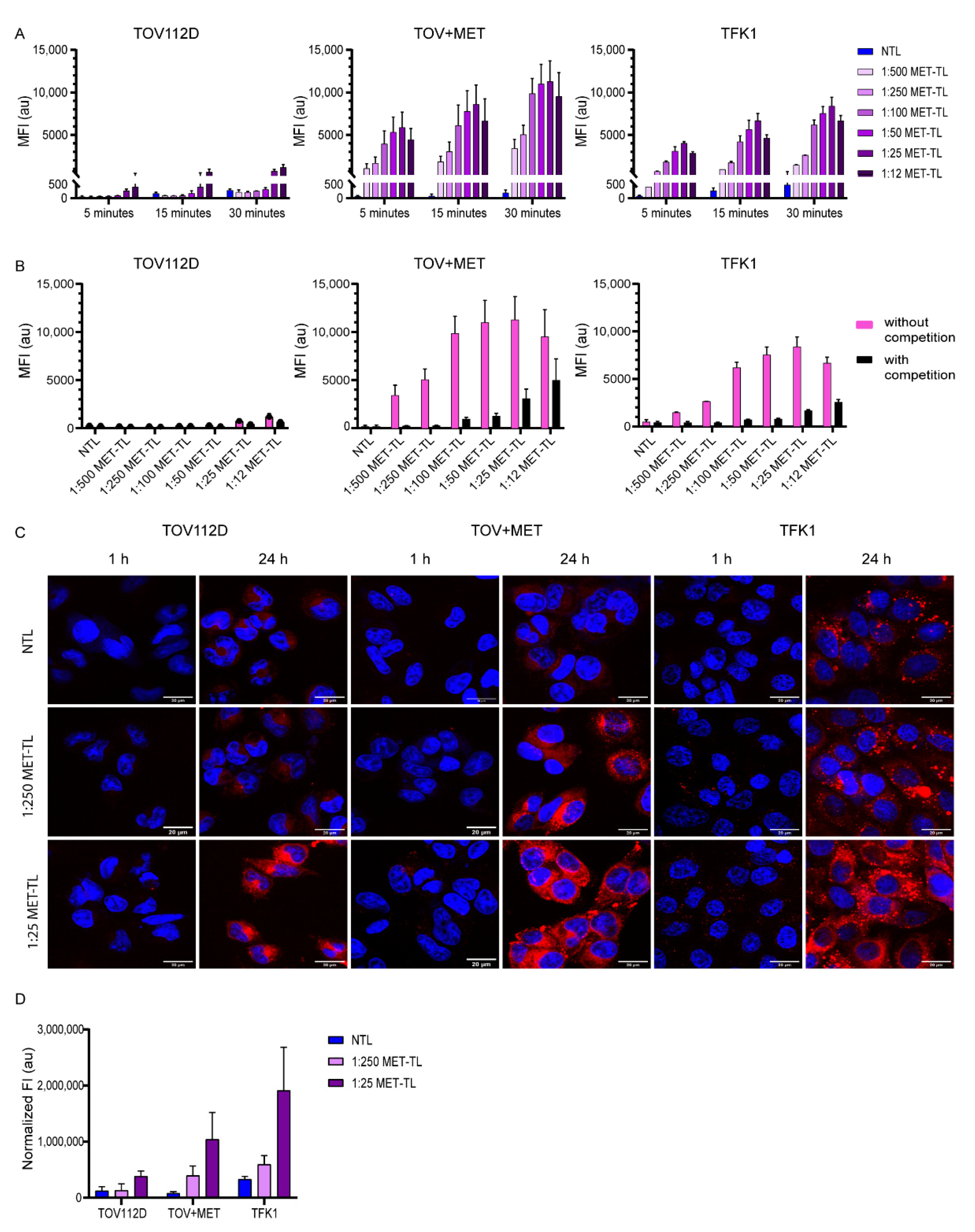
2.2. MET-Targeted Liposome Binding to and Association with Cells
2.3. Targeting Efficiency of MET-Targeting Liposomes in the Presence of Excess Proteins
2.4. Interaction of Non-Targeted and MET-Targeted Liposomes with Human Blood Cells
3. Materials and Methods
3.1. Materials
3.2. Production and Purification of Nanobodies
3.3. Cell Culture
3.4. MET Expression Assessment by Flow Cytometry
3.5. Preparation of (Fluorescent) Liposomes and Conjugation of MET-Nanobodies
3.6. Characterization of Non-Targeted Liposomes and MET-Targeted Liposomes
3.7. Nanobody Surface Density
3.8. Binding of MET-Targeted Liposomes to Cells at 4 °C
3.9. Cell Association with and Uptake of MET-Targeted Liposomes
3.9.1. Flow Cytometry
3.9.2. Confocal Microscopy
3.10. Cell Association with and Uptake of MET-Targeted Liposomes in the Presence of Excess Proteins
3.11. Interaction of Non-Targeted Liposomes and MET-Targeted Liposomes with Whole Blood
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van der Meel, R.; Vehmeijer, L.J.; Kok, R.J.; Storm, G.; van Gaal, E.V. Ligand-targeted particulate nanomedicines undergoing clinical evaluation: Current status. Intracell. Deliv. III 2016, 163–200. [Google Scholar] [CrossRef]
- Pearce, A.K.; O’Reilly, R.K. Insights into active targeting of nanoparticles in drug delivery: Advances in clinical studies and design considerations for cancer nanomedicine. Bioconjug. Chem. 2019, 30, 2300–2311. [Google Scholar] [CrossRef] [PubMed]
- Srinivasarao, M.; Galliford, C.V.; Low, P.S. Principles in the design of ligand-targeted cancer therapeutics and imaging agents. Nat. Rev. Drug Discov. 2015, 14, 203–219. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H. Polymer therapeutics and the EPR effect. J. Drug Target. 2017, 25, 781–785. [Google Scholar] [CrossRef]
- Matsumura, Y.; Maeda, H. A new concept for macromolecular therapeutics in cancer chemotherapy: Mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986, 46, 6387–6392. [Google Scholar]
- Ahn, J.; Miura, Y.; Yamada, N.; Chida, T.; Liu, X.; Kim, A.; Sato, R.; Tsumura, R.; Koga, R.; Yasunaga, M.; et al. Antibody fragment-conjugated polymeric micelles incorporating platinum drugs for targeted therapy of pancreatic cancer. Biomaterials 2015, 39, 23–30. [Google Scholar] [CrossRef]
- Kim, B.; Shin, J.; Wu, J.; Omstead, D.T.; Kiziltepe, T.; Littlepage, L.E.; Bilgicer, B. Engineering peptide-targeted liposomal nanoparticles optimized for improved selectivity for HER2-positive breast cancer cells to achieve enhanced in vivo efficacy. J. Control. Release 2020, 322, 530–541. [Google Scholar] [CrossRef]
- Palanikumar, L.; Al-Hosani, S.; Kalmouni, M.; Nguyen, V.P.; Ali, L.; Pasricha, R.; Barrera, F.N.; Magzoub, M. pH-responsive high stability polymeric nanoparticles for targeted delivery of anticancer therapeutics. Commun. Biol. 2020, 3, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhou, J.; Qiu, L.; Wang, X.; Chen, L.; Liu, T.; Di, W. Cisplatin–alginate conjugate liposomes for targeted delivery to EGFR-positive ovarian cancer cells. Biomaterials 2014, 35, 4297–4309. [Google Scholar] [CrossRef]
- Goswami, S.; Wang, W.; Arakawa, T.; Ohtake, S. Developments and challenges for mAb-based therapeutics. Antibodies 2013, 2, 452–500. [Google Scholar] [CrossRef] [Green Version]
- Vaisman-Mentesh, A.; Gutierrez-Gonzalez, M.; DeKosky, B.J.; Wine, Y. The molecular mechanisms that underlie the immune biology of anti-drug antibody formation following treatment with monoclonal antibodies. Front. Immunol. 2020, 11, 1951. [Google Scholar] [CrossRef] [PubMed]
- Bates, A.; Power, C.A. David vs. Goliath: The structure, function, and clinical prospects of antibody fragments. Antibodies 2019, 8, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hammers, C.; Songa, E.B.; Bendahman, N.; Hammers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altintas, I.; Heukers, R.; van der Meel, R.; Lacombe, M.; Amidi, M.; van Bergen en Henegouwen, P.M.P.; Hennink, W.E.; Schiffelers, R.M.; Kok, R.J. Nanobody-albumin nanoparticles (NANAPs) for the delivery of a multikinase inhibitor 17864 to EGFR overexpressing tumor cells. J. Control. Release 2013, 165, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Broekgaarden, M.; van Vught, R.; Oliveira, S.; Roovers, R.C.; Henegouwen, P.M.P.V.B.E.; Pieters, R.J.; Van Gulik, T.M.; Breukink, E.; Heger, M. Site-specific conjugation of single domain antibodies to liposomes enhances photosensitizer uptake and photodynamic therapy efficacy. Nanoscale 2016, 8, 6490–6494. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Scrivano, L.; Peterson, J.D.; Fens, M.H.A.M.; Hernández, I.B.; Mesquita, B.; Toraño, J.S.; Hennink, W.E.; Van Nostrum, C.F.; Oliveira, S. EGFR-targeted nanobody functionalized polymeric micelles loaded with mTHPC for selective photodynamic therapy. Mol. Pharm. 2020, 17, 1276–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Jothar, L.; Beztsinna, N.; Van Nostrum, C.F.; Hennink, W.E.; Oliveira, S. Selective cytotoxicity to HER2 positive breast cancer cells by saporin-loaded nanobody-targeted polymeric nanoparticles in combination with photochemical internalization. Mol. Pharm. 2019, 16, 1633–1647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Meel, R.; Oliveira, S.; Altintas, I.; Haselberg, R.; van der Veeken, J.; Roovers, R.C.; Henegouwen, P.M.V.B.E.; Storm, G.; Hennink, W.E.; Schiffelers, R.M.; et al. Tumor-targeted Nanobullets: Anti-EGFR nanobody-liposomes loaded with anti-IGF-1R kinase inhibitor for cancer treatment. J. Control. Release 2012, 159, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Talelli, M.; Oliveira, S.; Rijcken, C.J.; Pieters, E.H.; Etrych, T.; Ulbrich, K.; van Nostrum, R.C.; Storm, G.; Hennink, W.E.; Lammers, T. Intrinsically active nanobody-modified polymeric micelles for tumor-targeted combination therapy. Biomaterials 2013, 34, 1255–1260. [Google Scholar] [CrossRef]
- Nogueira, J.C.; Greene, M.K.; Richards, D.A.; Furby, A.O.; Steven, J.; Porter, A.; Barelle, C.; Scott, C.J.; Chudasama, V. Oriented attachment of V NAR proteins, via site-selective modification, on PLGA–PEG nanoparticles enhances nanoconjugate performance. Chem. Commun. 2019, 55, 7671–7674. [Google Scholar] [CrossRef] [PubMed]
- Greene, M.K.; Nogueira, J.C.F.; Tracey, S.R.; Richards, D.A.; McDaid, W.J.; Burrows, J.F.; Campbell, K.; Longley, D.B.; Chudasama, V.; Scott, C.J. Refined construction of antibody-targeted nanoparticles leads to superior antigen binding and enhanced delivery of an entrapped payload to pancreatic cancer cells. Nanoscale 2020, 12, 11647–11658. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Kwon, I. Expansion of bioorthogonal chemistries towards site-specific polymer–protein conjugation. Polym. Chem. 2016, 7, 4584–4598. [Google Scholar] [CrossRef]
- Byzova, N.A.; Safenkova, I.V.; Slutskaya, E.S.; Zherdev, A.V.; Dzantiev, B.B. Less is more: A comparison of antibody–gold nanoparticle conjugates of different ratios. Bioconjug. Chem. 2017, 28, 2737–2746. [Google Scholar] [CrossRef] [PubMed]
- Sivaram, A.J.; Wardiana, A.; Alcantara, S.; Sonderegger, S.E.; Fletcher, N.L.; Houston, Z.H.; Howard, C.B.; Mahler, S.M.; Alexander, C.; Kent, S.J.; et al. Controlling the biological fate of micellar nanoparticles: Balancing stealth and targeting. ACS Nano 2020, 14, 13739–13753. [Google Scholar] [CrossRef] [PubMed]
- Kunjachan, S.; Pola, R.; Gremse, F.; Theek, B.; Ehling, J.; Moeckel, D.; Hermanns-Sachweh, B.; Pechar, M.; Ulbrich, K.; Hennink, W.E.; et al. Passive versus active tumor targeting using RGD-and NGR-modified polymeric nanomedicines. Nano Lett. 2014, 14, 972–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNeeley, K.M.; Annapragada, A.; Bellamkonda, R.V. Decreased circulation time offsets increased efficacy of PEGylated nanocarriers targeting folate receptors of glioma. Nanotechnology 2007, 18, 385101. [Google Scholar] [CrossRef]
- Einav, T.; Gentles, L.E.; Bloom, J.D. SnapShot: Influenza by the numbers. Cell 2020, 182, 532. [Google Scholar] [CrossRef]
- Klein, J.S.; Bjorkman, P.J. Few and far between: How HIV may be evading antibody avidity. PLoS Pathog. 2010, 6, e1000908. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Sun, Z.; Pryce, R.; Parsy, M.L.; Fehling, S.K.; Schlie, K.; Sierbert, C.A.; Garten, W.; Bowden, T.A.; Strecker, T.; et al. Acidic pH-induced conformations and LAMP1 binding of the Lassa virus glycoprotein spike. PLoS Pathog. 2016, 12, e1005418. [Google Scholar] [CrossRef] [Green Version]
- Alkilany, A.M.; Zhu, L.; Weller, H.; Mews, A.; Parak, W.J.; Barz, M.; Feliu, N. Ligand density on nanoparticles: A parameter with critical impact on nanomedicine. Adv. Drug Deliv. Rev. 2019, 143, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Chen, C. The crown and the scepter: Roles of the protein corona in nanomedicine. Adv. Mater. 2019, 31, 1805740. [Google Scholar] [CrossRef] [PubMed]
- Salvati, A.; Pitek, A.; Monopoli, M.P.; Prapainop, K.; Bombelli, F.B.; Hristov, D.; Kelly, P.; Åberg, C.; Mahon, E.; Dawson, K.A. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 2013, 8, 137–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heukers, R.; Altintas, I.; Raghoenath, S.; De Zan, E.; Pepermans, R.; Roovers, R.C.; Haselberg, R.; Hennink, W.E.; Schiffelers, R.M.; Kok, R.J.; et al. Targeting hepatocyte growth factor receptor (Met) positive tumor cells using internalizing nanobody-decorated albumin nanoparticles. Biomaterials 2014, 35, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Heukers, R.; Mashayekhi, V.; Ramírez-Escudero, M.; De Haard, H.; Verrips, T.C.; van Bergen en Henegouwen, P.M.P.; Oliveira, S. VHH-photosensitizer conjugates for targeted photodynamic therapy of met-overexpressing tumor cells. Antibodies 2019, 8, 26. [Google Scholar] [CrossRef] [Green Version]
- Raghav, K.P.; Wang, W.; Liu, S.; Chavez-MacGregor, M.; Meng, X.; Hortobagyi, G.N.; Mills, G.B.; Meric-Bernstam, F.; Blumenschein, G.R.; Gonzalez-Angulo, A.M. cMET and phospho-cMET protein levels in breast cancers and survival outcomes. Clin. Cancer Res. 2012, 18, 2269–2277. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, M.; Ojima, H.; Iwasaki, M.; Shimizu, H.; Kokubu, A.; Hiraoka, N.; Kosuge, T.; Yoshikawa, D.; Kono, T.; Furukawa, H.; et al. Prognostic significance of overexpression of c-Met oncoprotein in cholangiocarcinoma. Br. J. Cancer 2011, 105, 131–138. [Google Scholar] [CrossRef]
- Kong, D.S.; Song, S.-Y.; Kim, D.-H.; Joo, K.M.; Yoo, J.-S.; Koh, J.S.; Dong, S.M.; Suh, Y.-L.; Lee, J.-I.; Park, K.; et al. Prognostic significance of c-Met expression in glioblastomas. Cancer 2009, 115, 140–148. [Google Scholar] [CrossRef]
- Kim, K.-H.; Kim, H. Progress of antibody-based inhibitors of the HGF–cMET axis in cancer therapy. Exp. Mol. Med. 2017, 49, e307. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Anderson, M.G.; Oleksijew, A.; Vaidya, K.S.; Boghaert, E.R.; Tucker, L.; Zhang, Q.; Han, E.K.; Palma, J.P.; Naumovski, L.; et al. ABBV-399, a c-Met antibody–Drug conjugate that targets both MET–Amplified and c-Met–Overexpressing tumors, irrespective of MET pathway dependencea tumor-targeting c-Met ADC. Clin. Cancer Res. 2017, 23, 992–1000. [Google Scholar] [CrossRef] [Green Version]
- Camidge, D.R.; Morgensztern, D.; Heist, R.S.; Barve, M.; Vokes, E.; Goldman, J.W.; Hong, D.S.; Bauer, T.M.; Strickler, J.H.; Angevin, E.; et al. Phase I study of 2-or 3-week dosing of telisotuzumab vedotin, an antibody–drug conjugate targeting c-Met, monotherapy in patients with advanced non–small cell lung carcinoma. Clin. Cancer Res. 2021, 27, 5781–5792. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, S.; Schiffelers, R.M.; van der Veeken, J.; van der Meel, R.; Vongpromek, R.; van Bergen En Henegouwen, P.M.P.; Storm, G.; Roovers, R.C. Downregulation of EGFR by a novel multivalent nanobody-liposome platform. J. Control. Release 2010, 145, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Dézsi, L.; Fülöp, T.; Mészáros, T.; Szénási, G.; Urbanics, R.; Vázsonyi, C.; Őrfi, E.; Rosivall, L.; Nemes, R.; Kok, R.J.; et al. Features of complement activation-related pseudoallergy to liposomes with different surface charge and PEGylation: Comparison of the porcine and rat responses. J. Control. Release 2014, 195, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, N.; Kok, R.J.; Bos, C.; Zandvliet, M.; Geerts, W.J.; Storm, G.; Moonen, C.T.W.; Lammers, T.; Deckers, R. Hyperthermia-triggered release of hypoxic cell radiosensitizers from temperature-sensitive liposomes improves radiotherapy efficacy in vitro. Nanotechnology 2019, 30, 264001. [Google Scholar] [CrossRef]
- Bhattacharjee, S. DLS and zeta potential–what they are and what they are not? J. Control. Release 2016, 235, 337–351. [Google Scholar] [CrossRef]
- Stetefeld, J.; McKenna, S.A.; Patel, T.R. Dynamic light scattering: A practical guide and applications in biomedical sciences. Biophys. Rev. 2016, 8, 409–427. [Google Scholar] [CrossRef] [Green Version]
- Kijanka, M.; Dorresteijn, B.; Oliveira, S.; van Bergen en Henegouwen, P.M.P. Nanobody-based cancer therapy of solid tumors. Nanomedicine 2015, 10, 161–174. [Google Scholar] [CrossRef]
- Yong, K.W.; Yuen, D.; Chen, M.Z.; Johnston, A.P.R. Engineering the orientation, density, and flexibility of single-domain antibodies on nanoparticles to improve cell targeting. ACS Appl. Mater. Interfaces 2020, 12, 5593–5600. [Google Scholar] [CrossRef]
- Woythe, L.; Madhikar, P.; Feiner-Gracia, N.; Storm, C.; Albertazzi, L. A single-molecule view at nanoparticle targeting selectivity: Correlating ligand functionality and cell receptor density. ACS Nano 2022, 16, 3785–3796. [Google Scholar] [CrossRef]
- Rodallec, A.; Franco, C.; Robert, S.; Sicard, G.; Giacometti, S.; Lacarelle, B.; Bouquet, F.; Savina, A.; Lacroix, R.; Dignat-George, F.; et al. Prototyping trastuzumab docetaxel immunoliposomes with a new FCM-based method to quantify optimal antibody density on nanoparticles. Sci. Rep. 2020, 10, 4147. [Google Scholar] [CrossRef] [Green Version]
- Kirpotin, D.; Park, J.W.; Hong, K.; Zalipsky, S.; Li, W.-L.; Carter, P.; Benz, A.C.C.; Papahadjopoulos, D. Sterically stabilized anti-HER2 immunoliposomes: Design and targeting to human breast cancer cells in vitro. Biochemistry 1997, 36, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Lara, S.; Alnasser, F.; Polo, E.; Garry, D.; Giudice, M.C.L.; Hristov, D.R.; Rocks, L.; Salvati, A.; Yan, Y.; Dawson, K.A. Identification of receptor binding to the biomolecular corona of nanoparticles. ACS Nano 2017, 11, 1884–1893. [Google Scholar] [CrossRef] [PubMed]
- Böhmert, L.; Voß, L.; Stock, V.; Braeuning, A.; Lampen, A.; Sieg, H. Isolation methods for particle protein corona complexes from protein-rich matrices. Nanoscale Adv. 2020, 2, 563–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Mesquita, B.; Horvatovich, P.; Salvati, A. Tuning liposome composition to modulate corona formation in human serum and cellular uptake. Acta Biomater. 2020, 106, 314–327. [Google Scholar] [CrossRef]
- Dai, Q.; Yan, Y.; Ang, C.-S.; Kempe, K.; Kamphuis, M.M.J.; Dodds, S.J.; Caruso, F. Monoclonal antibody-functionalized multilayered particles: Targeting cancer cells in the presence of protein coronas. ACS Nano 2015, 9, 2876–2885. [Google Scholar] [CrossRef] [Green Version]
- Hadjidemetriou, M.; Al-Ahmady, Z.; Mazza, M.; Collins, R.F.; Dawson, K.; Kostarelos, K. In vivo biomolecule corona around blood-circulating, clinically used and antibody-targeted lipid bilayer nanoscale vesicles. ACS Nano 2015, 9, 8142–8156. [Google Scholar] [CrossRef]
- Zarschler, K.; Prapainop, K.; Mahon, E.; Rocks, L.; Bramini, M.; Kelly, P.M.; Stephan, H.; Dawson, K.A. Diagnostic nanoparticle targeting of the EGF-receptor in complex biological conditions using single-domain antibodies. Nanoscale 2014, 6, 6046–6056. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Wu, T.; Yu, W.; Ruan, S.; He, Q.; Gao, H. Ligand size and conformation affect the behavior of nanoparticles coated with in vitro and in vivo protein corona. ACS Appl. Mater. Interfaces 2018, 10, 9094–9103. [Google Scholar] [CrossRef]
- Lundqvist, M.; Stigler, J.; Elia, G.; Lynch, I.; Cedervall, T.; Dawson, K.A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. USA 2008, 105, 14265–14270. [Google Scholar] [CrossRef] [Green Version]
- Lundqvist, M.; Augustsson, C.; Lilja, M.; Lundkvist, K.; Dahlbäck, B.; Linse, S.; Cedervall, T. The nanoparticle protein corona formed in human blood or human blood fractions. PLoS ONE 2017, 12, e0175871. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, B.; Poon, W.; Zhang, Y.-N.; Lin, Z.P.; Kingston, B.R.; Tavares, A.J.; Zhang, Y.; Chen, J.; Valic, M.S.; Syed, A.M.; et al. The dose threshold for nanoparticle tumour delivery. Nat. Mater. 2020, 19, 1362–1371. [Google Scholar] [CrossRef] [PubMed]
- Lammers, T. Just dose it. Nat. Mater. 2020, 19, 1257–1258. [Google Scholar] [CrossRef] [PubMed]
- Leiske, M.N.; Lai, M.; Amarasena, T.; Davis, T.P.; Thurecht, K.J.; Kent, S.J.; Kempe, K. Interactions of core cross-linked poly (2-oxazoline) and poly (2-oxazine) micelles with immune cells in human blood. Biomaterials 2021, 274, 120843. [Google Scholar] [CrossRef]
- Ong, Y.R.; de Rose, R.; Johnston, A.P. In vivo quantification of nanoparticle association with immune cell subsets in blood. Adv. Healthc. Mater. 2021, 10, 2002160. [Google Scholar] [CrossRef] [PubMed]
- Xenaki, K.T.; Oliveira, S.; Henegouwen, P.M.v.E. Antibody or antibody fragments: Implications for molecular imaging and targeted therapy of solid tumors. Front. Immunol. 2017, 8, 1287. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H. Thin-film hydration followed by extrusion method for liposome preparation. In Liposomes: Methods and Protocols; D’Souza, G.G.M., Ed.; Springer: New York, NY, USA, 2017; pp. 17–22. [Google Scholar]
- Rouser, G.; Fleischer, S.; Yamamoto, A. Two dimensional thin layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids 1970, 5, 494–496. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mesquita, B.S.; Fens, M.H.A.M.; Di Maggio, A.; Bosman, E.D.C.; Hennink, W.E.; Heger, M.; Oliveira, S. The Impact of Nanobody Density on the Targeting Efficiency of PEGylated Liposomes. Int. J. Mol. Sci. 2022, 23, 14974. https://doi.org/10.3390/ijms232314974
Mesquita BS, Fens MHAM, Di Maggio A, Bosman EDC, Hennink WE, Heger M, Oliveira S. The Impact of Nanobody Density on the Targeting Efficiency of PEGylated Liposomes. International Journal of Molecular Sciences. 2022; 23(23):14974. https://doi.org/10.3390/ijms232314974
Chicago/Turabian StyleMesquita, Bárbara S., Marcel H. A. M. Fens, Alessia Di Maggio, Esmeralda D. C. Bosman, Wim E. Hennink, Michal Heger, and Sabrina Oliveira. 2022. "The Impact of Nanobody Density on the Targeting Efficiency of PEGylated Liposomes" International Journal of Molecular Sciences 23, no. 23: 14974. https://doi.org/10.3390/ijms232314974






