Expression of miRNA-Targeted and Not-Targeted Reporter Genes Shows Mutual Influence and Intercellular Specificity
Abstract
:1. Introduction
2. Results
2.1. miR-21, miR-24 and Let-7 Influence the Expression Not Only of Targeted Renilla Reporter Genes but Also of Non-Targeted Firefly Luciferase Genes
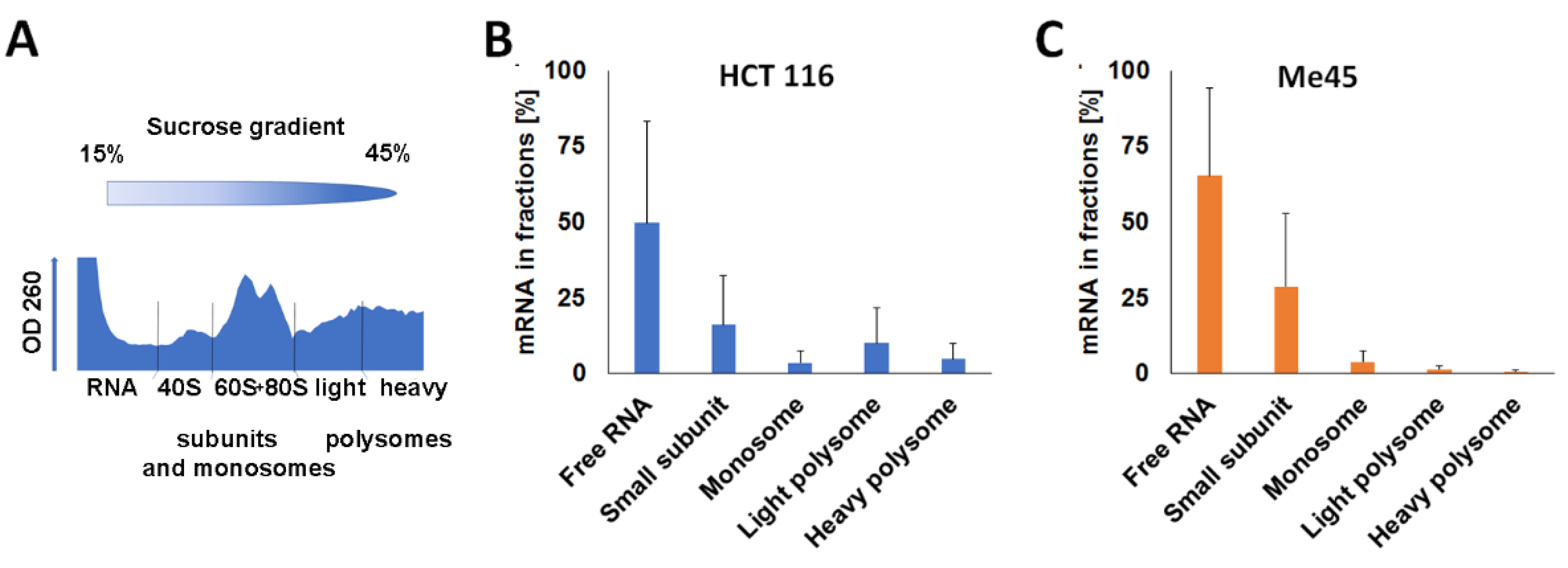
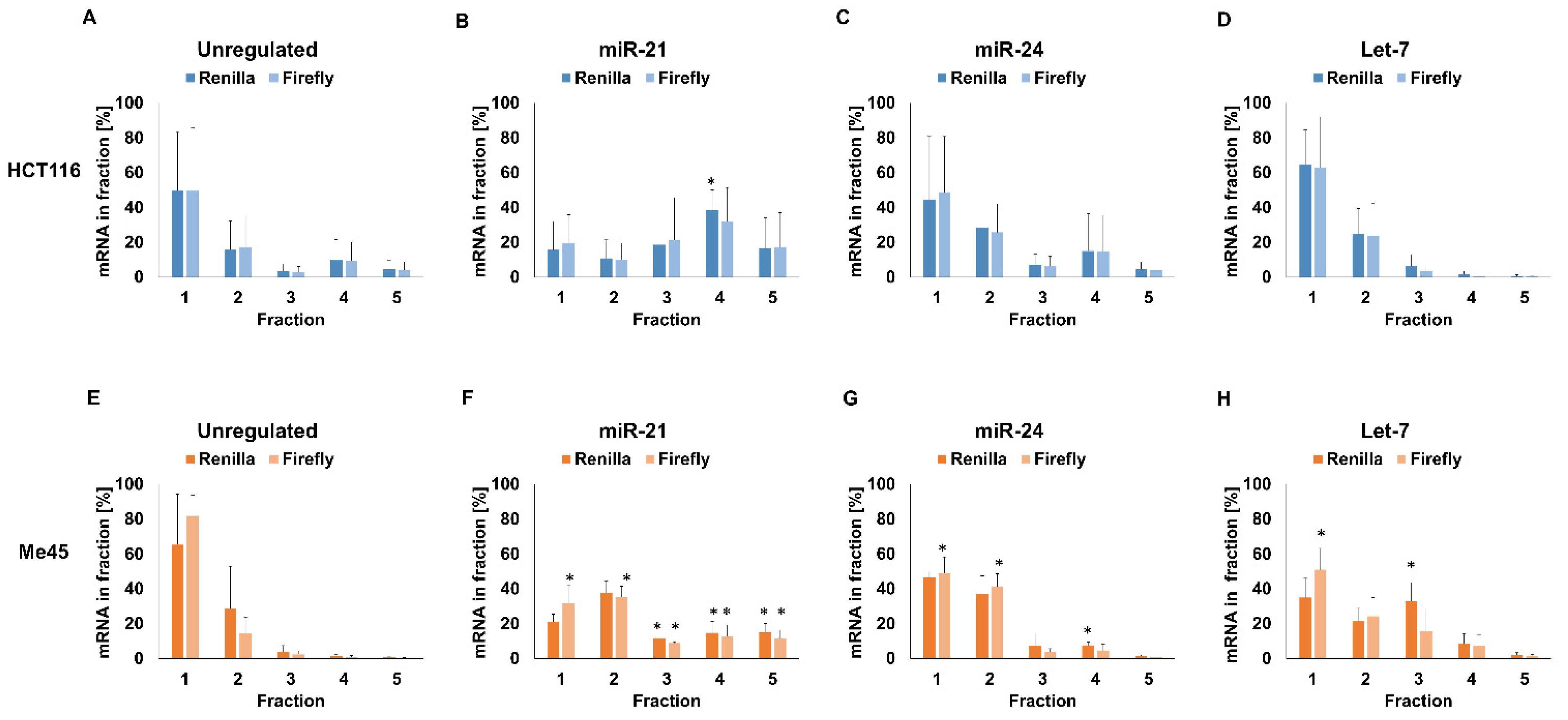
2.2. Sucrose Gradient Centrifugation of Complexes Formed by Firefly and Renilla mRNAs
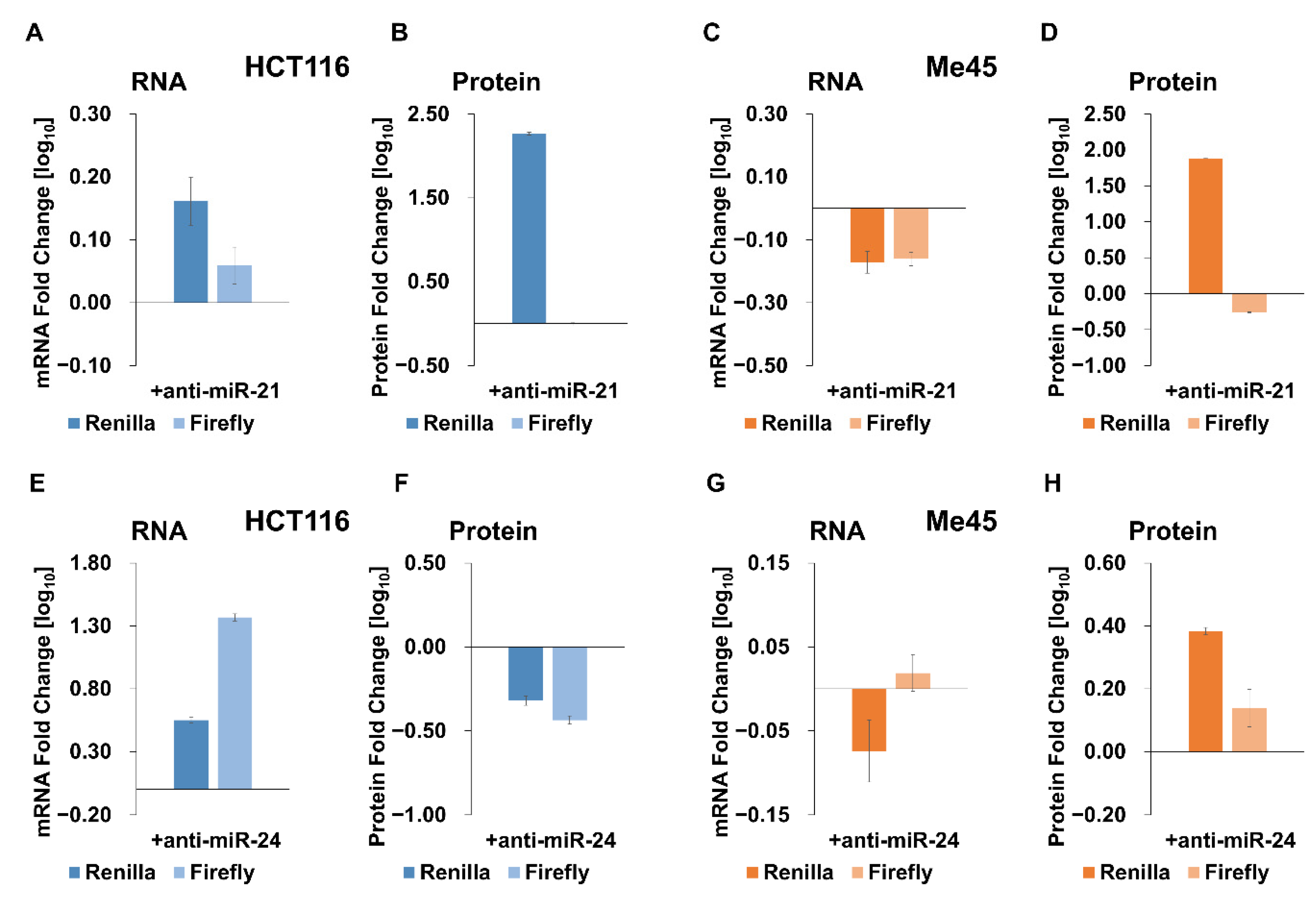
2.3. Influence of Anti-miR-21, Anti-miR-24, and Different Anti-Let-7 Oligonucleotides on Expression of Reporter Luciferases
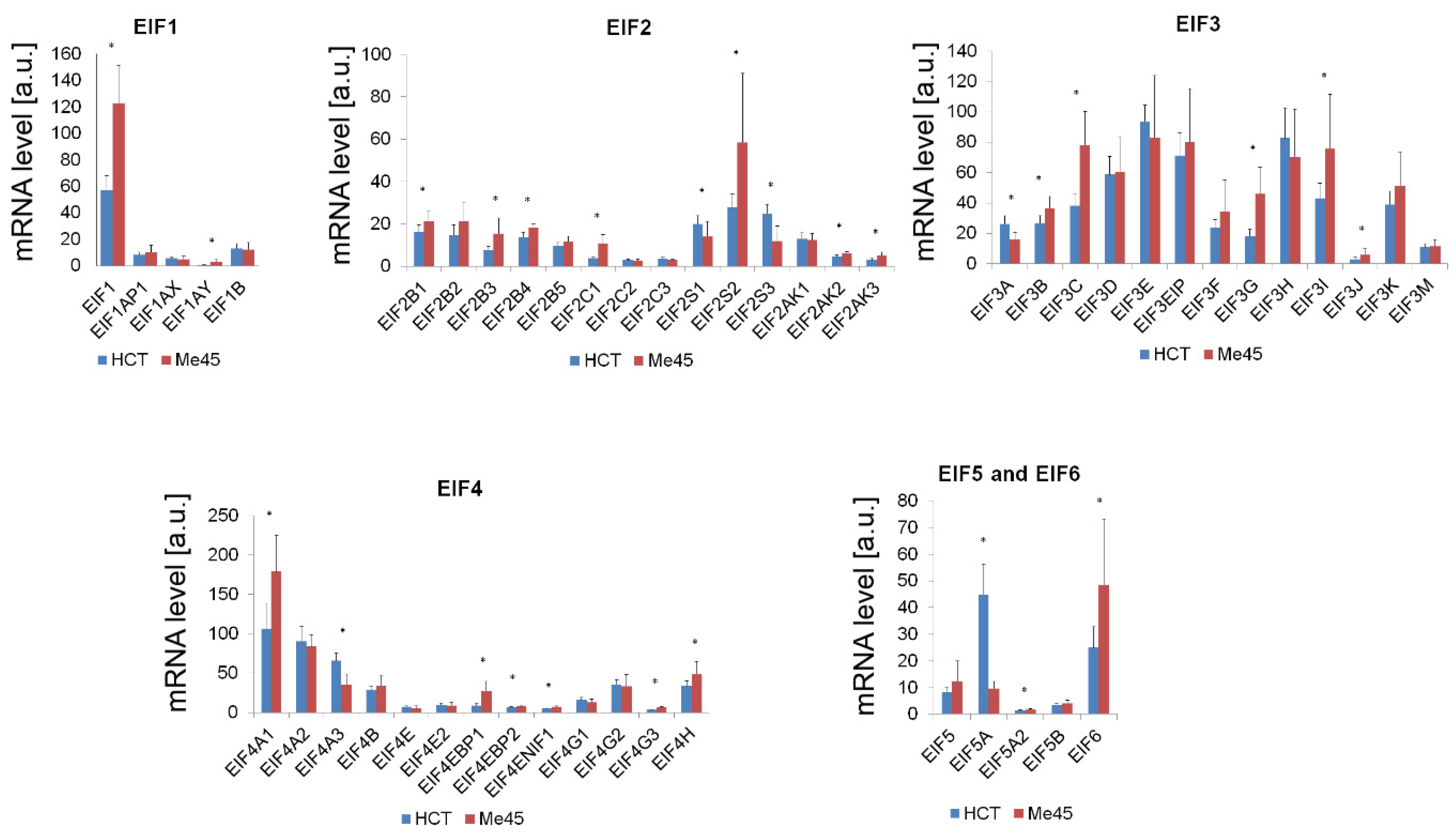
2.4. Proteins Potentially Engaged in Regulation of Translation Are Differently Expressed in Me45 and HCT116 Cells
3. Discussion
3.1. Specific miRNA Effects in Cells of the Same Type and Differences between the Effects of the Same miRNA in Different Cell Types
3.2. Pitfalls in Normalization of Results for miRNA-Targeted Gene Expression to Those for a Non-Targeted Gene
4. Materials and Methods
4.1. Cell Lines
4.2. Plasmids
4.3. Anti-microRNA Oligonucleotides (Anti-miRs)
4.4. Transfection Protocol
4.5. Extraction and Assays of RNA
4.6. Luciferase Assays
4.7. Sucrose Gradient Centrifugation
4.8. Calculation of Cell Numbers Serving for Assays
4.9. Microarray Analyses
4.10. Statistical Tests
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hinnebusch, A.G. Molecular Mechanism of Scanning and Start Codon Selection in Eukaryotes. Microbiol. Mol. Biol. Rev. 2011, 75, 434–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, R.J.; Hellen, C.U.T.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelletier, J.; Sonenberg, N. The Organizing Principles of Eukaryotic Ribosome Recruitment. Annu. Rev. Biochem. 2019, 88, 307–335. [Google Scholar] [CrossRef] [PubMed]
- Shirokikh, N.E.; Preiss, T. Translation initiation by cap-dependent ribosome recruitment: Recent insights and open questions. Wiley Interdiscip. Rev. RNA 2018, 9, e1473. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Höck, J.; Meister, G. The Argonaute protein family. Genome Biol. 2008, 9, 210. [Google Scholar] [CrossRef]
- Salomon, W.E.; Jolly, S.M.; Moore, M.J.; Zamore, P.D.; Serebrov, V. Single-Molecule Imaging Reveals that Argonaute Reshapes the Binding Properties of Its Nucleic Acid Guides. Cell 2015, 162, 84–95. [Google Scholar] [CrossRef] [Green Version]
- Leung, A.K.L.; Calabrese, J.M.; Sharp, P.A. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. USA 2006, 103, 18125–18130. [Google Scholar] [CrossRef] [Green Version]
- Ørom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a Binds the 5′UTR of Ribosomal Protein mRNAs and Enhances Their Translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef]
- Fehr, C.; Conrad, K.D.; Niepmann, M. Differential stimulation of hepatitis C virus RNA translation by microRNA-122 in different cell cycle phases. Cell Cycle 2012, 11, 277–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, A.P.E.; Lewis, A.P.; Jopling, C.L. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011, 39, 7716–7729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bukhari, S.I.; Truesdell, S.S.; Lee, S.; Kollu, S.; Classon, A.; Boukhali, M.; Jain, E.; Mortensen, R.D.; Yanagiya, A.; Sadreyev, R.I.; et al. A Specialized Mechanism of Translation Mediated by FXR1a-Associated MicroRNP in Cellular Quiescence. Mol. Cell 2016, 61, 760–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truesdell, S.S.; Mortensen, R.D.; Seo, M.; Schroeder, J.C.; Lee, J.H.; LeTonqueze, O.; Vasudevan, S. MicroRNA-mediated mRNA Translation Activation in Quiescent Cells and Oocytes Involves Recruitment of a Nuclear microRNP. Sci. Rep. 2012, 2, 842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Cell cycle control of microRNA-mediated translation regulation. Cell Cycle 2008, 7, 1545–1549. [Google Scholar] [CrossRef] [Green Version]
- Chassé, H.; Boulben, S.; Costache, V.; Cormier, P.; Morales, J. Analysis of translation using polysome profiling. Nucleic Acids Res. 2017, 45, e15. [Google Scholar] [CrossRef] [Green Version]
- Mathys, H.; Basquin, J.; Ozgur, S.; Czarnocki-Cieciura, M.; Bonneau, F.; Aartse, A.; Dziembowski, A.; Nowotny, M.; Conti, E.; Filipowicz, W. Structural and Biochemical Insights to the Role of the CCR4-NOT Complex and DDX6 ATPase in MicroRNA Repression. Mol. Cell 2014, 54, 751–765. [Google Scholar] [CrossRef] [Green Version]
- Khoshnevis, S.; Gross, T.; Rotte, C.; Baierlein, C.; Ficner, R.; Krebber, H. The iron–sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 2010, 11, 214–219. [Google Scholar] [CrossRef] [Green Version]
- Hondele, M.; Sachdev, R.; Heinrich, S.; Wang, J.; Vallotton, P.; Fontoura, B.M.A.; Weis, K. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 2019, 573, 144–148. [Google Scholar] [CrossRef]
- Yoda, M.; Kawamata, T.; Paroo, Z.; Ye, X.; Iwasaki, S.; Liu, Q.; Tomari, Y. ATP-dependent human RISC assembly pathways. Nat Struct. Mol. Biol. 2010, 17, 17–23. [Google Scholar] [CrossRef]
- Dueck, A.; Ziegler, C.; Eichner, A.; Berezikov, E.; Meister, G. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 2012, 40, 9850–9862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, 6472. [Google Scholar] [CrossRef] [PubMed]
- Landthaler, M.; Gaidatzis, D.; Rothballer, A.; Chen, P.Y.; Soll, S.J.; Dinic, L.; Ojo, T.; Hafner, M.; Zavolan, M.; Tuschl, T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA 2008, 14, 2580–2596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakumani, P.K.; Harvey, L.-M.; Houle, F.; Guitart, T.; Gebauer, F.; Simard, M.J. CSDE1 controls gene expression through the miRNA-mediated decay machinery. Life Sci. Alliance 2020, 3, e201900632. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Landthaler, M.; Peters, L.; Chen, P.Y.; Urlaub, H.; Lührmann, R.; Tuschl, T. Identification of Novel Argonaute-Associated Proteins. Curr. Biol. 2005, 15, 2149–2155. [Google Scholar] [CrossRef] [Green Version]
- Dallaire, A.; Frédérick, P.-M.; Simard, M.J. Somatic and Germline MicroRNAs Form Distinct Silencing Complexes to Regulate Their Target mRNAs Differently. Dev. Cell 2018, 47, 239–247. [Google Scholar] [CrossRef] [Green Version]
- Munakata, F.; Suzawa, M.; Ui-Tei, K. Identification of Phosphorylated Amino Acids in Human TNRC6A C-Terminal Region and Their Effects on the Interaction with the CCR4-NOT Complex. Genes 2021, 12, 271. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef] [Green Version]
- Darty, K.; Denise, A.; Ponty, Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 2009, 25, 1974–1975. [Google Scholar] [CrossRef] [Green Version]
- Morisaki, T.; Lyon, K.; DeLuca, K.F.; DeLuca, J.G.; English, B.P.; Zhang, Z.; Lavis, L.D.; Grimm, J.B.; Viswanathan, S.; Looger, L.L.; et al. Real-time quantification of single RNA translation dynamics in living cells. Science 2016, 352, 1425–1429. [Google Scholar] [CrossRef]
- Protter, D.S.W.; Parker, R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016, 26, 668–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banani, S.F.; Rice, A.M.; Peeples, W.B.; Lin, Y.; Jain, S.; Parker, R.; Rosen, M.K. Compositional Control of Phase-Separated Cellular Bodies. Cell 2016, 166, 651–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kedersha, N.; Panas, M.D.; Achorn, C.A.; Lyons, S.; Tisdale, S.; Hickman, T.; Thomas, M.; Lieberman, J.; McInerney, G.M.; Ivanov, P.; et al. G3BP–Caprin1–USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 2016, 212, 845–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panas, M.D.; Ivanov, P.; Anderson, P. Mechanistic insights into mammalian stress granule dynamics. J. Cell Biol. 2016, 215, 313–323. [Google Scholar] [CrossRef] [Green Version]
- van Treeck, B.; Protter, D.S.W.; Matheny, T.; Khong, A.; Link, C.D.; Parker, R. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. USA 2018, 115, 2734–2739. [Google Scholar] [CrossRef] [Green Version]
- Mugler, C.F.; Hondele, M.; Heinrich, S.; Sachdev, R.; Vallotton, P.; Koek, A.Y.; Chan, L.Y.; Weis, K. ATPase activity of the DEAD-box protein Dhh1 controls processing body formation. eLife 2016, 5, e18746. [Google Scholar] [CrossRef]
- Khong, A.; Matheny, T.; Jain, S.; Mitchell, S.F.; Wheeler, J.R.; Parker, R. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol. Cell 2017, 68, 808–820. [Google Scholar] [CrossRef]
- Jakymiw, A.; Lian, S.; Eystathioy, T.; Li, S.; Satoh, M.; Hamel, J.C.; Fritzler, M.J.; Chan, E. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005, 7, 1267–1274. [Google Scholar] [CrossRef]
- Eulalio, A.; Behm-Ansmant, I.; Schweizer, D.; Izaurralde, E. P-Body Formation Is a Consequence, Not the Cause, of RNA-Mediated Gene Silencing. Mol. Cell Biol. 2007, 27, 3970–3981. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Jove Navarro, M.; Kashida, S.; Chouaib, R.; Souquere, S.; Pierron, G.; Weil, D.; Gueroui, Z. RNA is a critical element for the sizing and the composition of phase-separated RNA–protein condensates. Nat. Commun. 2019, 10, 3230. [Google Scholar] [CrossRef]
- Sachdev, R.; Hondele, M.; Linsenmeier, M.; Vallotton, P.; Mugler, C.F.; Arosio, P.; Weis, K. Pat1 promotes processing body assembly by enhancing the phase separation of the DEAD-box ATPase Dhh1 and RNA. eLife 2019, 8, e41415. [Google Scholar] [CrossRef] [PubMed]
- Vicens, Q.; Kieft, J.S.; Rissland, O.S. Revisiting the Closed-Loop Model and the Nature of mRNA 5′–3′ Communication. Mol. Cell 2018, 72, 805–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, S.E.; Hillner, P.E.; Vale, R.D.; Sachs, A.B. Circularization of mRNA by Eukaryotic Translation Initiation Factors. Mol. Cell 1998, 2, 135–140. [Google Scholar] [CrossRef] [Green Version]
- Alekhina, O.; Terenin, I.; Dmitriev, S.; Vassilenko, K. Functional Cyclization of Eukaryotic mRNAs. Int. J. Mol. Sci. 2020, 21, 1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallie, D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991, 5, 2108–2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Pillai, R.S.; Bhattacharyya, S.N.; Filipowicz, W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007, 17, 118–126. [Google Scholar] [CrossRef]
- Mathonnet, G.; Fabian, M.R.; Svitkin, Y.V.; Parsyan, A.; Huck, L.; Murata, T.; Biffo, S.; Merrick, W.C.; Darzynkiewicz, E.; Pillai, R.S.; et al. MicroRNA Inhibition of Translation Initiation in Vitro by Targeting the Cap-Binding Complex eIF4F. Science 2007, 317, 1764–1767. [Google Scholar] [CrossRef] [Green Version]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef] [Green Version]
- Archer, S.K.; Shirokikh, N.E.; Hallwirth, C.V.; Beilharz, T.H.; Preiss, T. Probing the closed-loop model of mRNA translation in living cells. RNA Biol. 2015, 12, 248–254. [Google Scholar] [CrossRef]
- Ziv, O.; Gabryelska, M.M.; Lun, A.T.L.; Gebert, L.F.R.; Sheu-Gruttadauria, J.; Meredith, L.W.; Liu, Z.Y.; Kwok, C.K.; Qin, C.F.; MacRae, I.J.; et al. COMRADES determines in vivo RNA structures and interactions. Nat. Methods. 2018, 15, 785–788. [Google Scholar] [CrossRef] [PubMed]
- Cialek, C.A.; Galindo, G.; Morisaki, T.; Zhao, N.; Montgomery, T.A.; Stasevich, T.J. Imaging translational control by Argonaute with single-molecule resolution in live cells. Nat. Commun. 2022, 13, 3345. [Google Scholar] [CrossRef] [PubMed]
- Kramer-Marek, G.; Serpa, C.; Szurko, A.; Widel, M.; Sochanik, A.; Snietura, M.; Kus, P.; Nunes, R.M.; Arnaut, L.G.; Ratuszna, A. Spectroscopic properties and photodynamic effects of new lipophilic porphyrin derivatives: Efficacy, localisation and cell death pathways. J. Photochem. Photobiol. B 2006, 84, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herok, R.; Konopacka, M.; Polanska, J.; Swierniak, A.; Rogolinski, J.; Jaksik, R.; Hancock, R.; Rzeszowska-Wolny, J. Bystander Effects Induced by Medium From Irradiated Cells: Similar Transcriptome Responses in Irradiated and Bystander K562 Cells. Int. J. Radiat. Oncol. Biol. Phys. 2010, 77, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Rzeszowska-Wolny, J.; Herok, R.; Widel, M.; Hancock, R. X-irradiation and bystander effects induce similar changes of transcript profiles in most functional pathways in human melanoma cells. DNA Repair 2009, 8, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Athar, A.; Füllgrabe, A.; George, N.; Iqbal, H.; Huerta, L.; Ali, A.; Snow, C.; Fonseca, N.A.; Petryszak, R.; Papatheodorou, I.; et al. ArrayExpress update—From bulk to single-cell expression data. Nucleic Acids Res. 2019, 47, D711–D715. [Google Scholar] [CrossRef]
- Wilson, C.L.; Miller, C.J. Simpleaffy: A BioConductor package for Affymetrix Quality Control and data analysis. Bioinformatics 2005, 21, 3683–3685. [Google Scholar] [CrossRef] [Green Version]
- Dai, M. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005, 33, e175. [Google Scholar] [CrossRef] [Green Version]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef] [Green Version]
- Camps, C.; Saini, H.K.; Mole, D.R.; Choudhry, H.; Reczko, M.; A Guerra-Assunção, J.; Tian, Y.-M.; Buffa, F.M.; Harris, A.L.; Hatzigeorgiou, A.G.; et al. Integrated analysis of microRNA and mRNA expression and association with HIF binding reveals the complexity of microRNA expression regulation under hypoxia. Mol. Cancer 2014, 13, 28. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Belinda, P.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [PubMed]

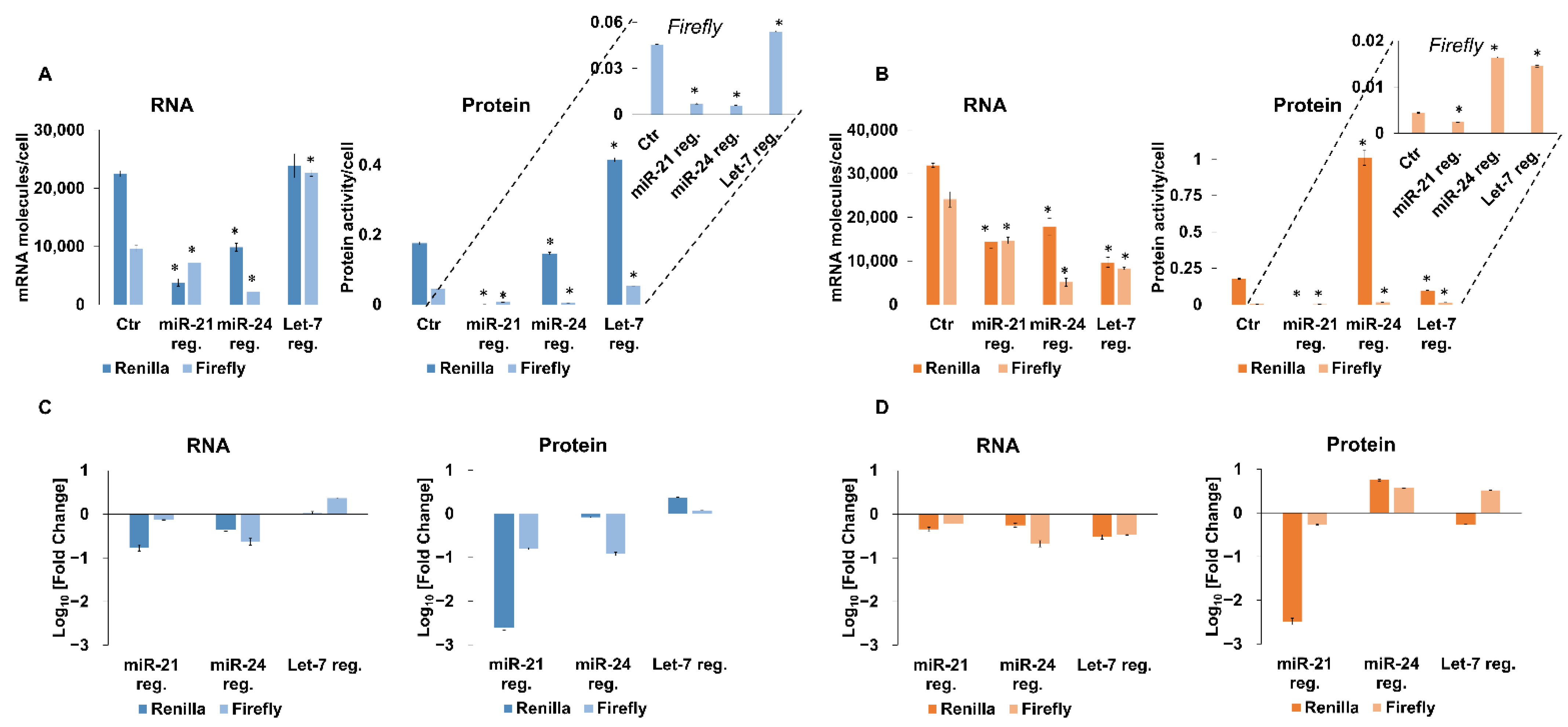

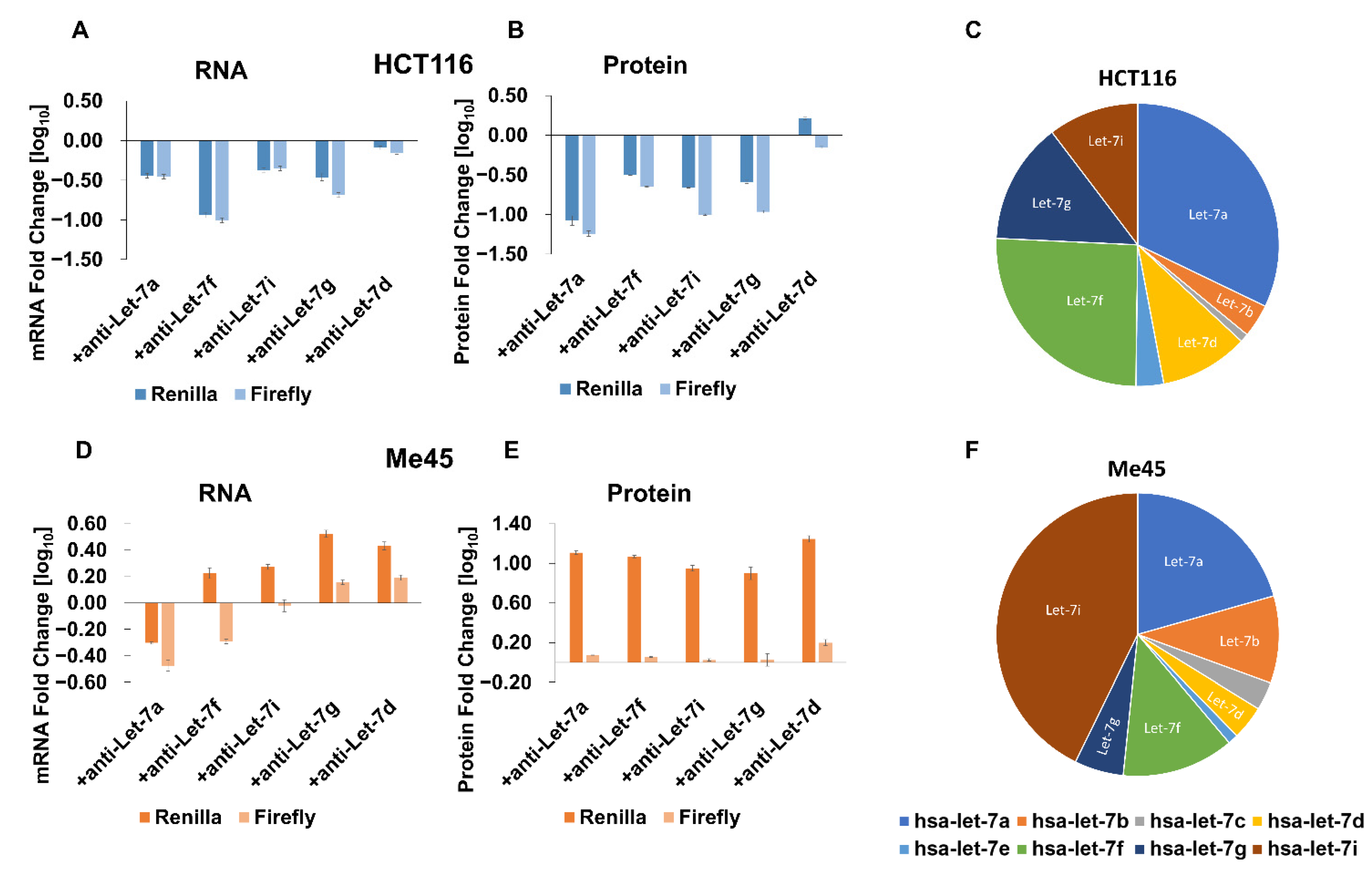
| No Target | miR-21 | miR-24 | Let-7 | ||
|---|---|---|---|---|---|
| Me45 cells | Renilla | 5.56 ± 0.10 | 0.04 ± 0.01 | 56.82 ± 5.62 | 10.15 ± 1.15 |
| firefly | 0.18 ± 0.01 | 0.16 ± 0.01 | 3.23 ± 0.46 | 1.75 ± 0.06 | |
| HCT116 cells | Renilla | 7.82 ± 0.21 | 0.12 ± 0.02 | 14.88 ± 0.94 | 17.45 ± 1.40 |
| firefly | 4.71 ± 0.25 | 1.01 ± 0.05 | 2.46 ± 0.42 | 2.39 ± 0.05 |
| Gene | HCT116 Cells | Me45 Cells |
|---|---|---|
| ABCE1 | 18.68 ± 3.05 | 11.79 ± 6.36 |
| CNOT6/CCR4 | 5.66 ± 0.78 | 4.43 ± 2.35 |
| CNOT7/CAF1 | 34.72 ± 6.09 | 17.75 ± 7.97 |
| CNOT8/CAF2 | 6.19 ± 0.81 | 4.99 ± 1.84 |
| CSDE1 | 19.50 ± 2.20 | 19.16 ± 4.97 |
| DDX17 | 4.05 ± 1.74 | 4.49 ± 1.45 |
| DDX3X | 12.05 ± 1.73 | 10.60 ± 4.78 |
| DDX3Y | 0.85 ± 0.80 | 6.23 ± 2.39 |
| DDX5 | 62.49 ± 15.75 | 84.26 ± 21.25 |
| DICER1 | 1.67 ± 0.41 | 2.16 ± 0.38 |
| EEF2 | 59.93 ± 10.37 | 104.99 ± 36.10 |
| EIF2C1/AGO1 | 3.81 ± 0.74 | 10.81 ± 4.05 |
| EIF2C2/AGO2 | 2.87 ± 0.52 | 2.71 ± 0.71 |
| HNRNPH1 | 22.61 ± 5.34 | 21.80 ± 2.97 |
| HNRNPL | 20.70 ± 1.39 | 8.18 ± 2.19 |
| HNRNPM | 26.78 ± 5.77 | 20.08 ± 11.52 |
| HNRNPU | 32.20 ± 6.20 | 25.66 ± 7.63 |
| HSP90AA1 | 149.67 ± 14.05 | 112.88 ± 20.58 |
| HSP90AB1 | 186.57 ± 26.71 | 145.69 ± 33.39 |
| HSPA1A | 20.82 ± 10.19 | 10.42 ± 4.24 |
| HSPA5 | 89.45 ± 23.53 | 114.94 ± 28.48 |
| HSPA8 | 156.33 ± 28.72 | 149.77 ± 81.01 |
| IGF2BP2 | 8.73 ± 0.93 | 17.40 ± 3.16 |
| IGF2BP3 | 9.22 ± 1.37 | 8.03 ± 4.71 |
| MKNK2 | 10.94 ± 2.08 | 7.51 ± 2.32 |
| PABPC1 | 127.80 ± 22.45 | 144.75 ± 20.27 |
| PABPC3 | 42.33 ± 7.50 | 31.14 ± 5.15 |
| PABPC4 | 15.94 ± 3.45 | 51.53 ± 8.44 |
| RBM14 | 6.77 ± 1.31 | 13.66 ± 3.41 |
| SRP14 | 52.32 ± 12.92 | 78.13 ± 19.86 |
| SYNCRIP | 16.92 ± 2.65 | 8.68 ± 3.94 |
| TNRC6B | 4.38 ± 0.85 | 5.49 ± 0.89 |
| Gene Name | Forward Primer | Reverse Primer |
|---|---|---|
| Renilla Luciferase | ACAAGTACCTCACCGCTTGG | GACACTCTCAGCATGGACGA |
| Firefly Luciferase | GCTAAGAGCACCCTGATCG | CCTCTGGGGTAATCAGAATGG |
| GAPDH | TTTGGCTACAGCAACAGGGTG | TTCCTCTTGTGCTCTTGCTGG |
| RPL41 | TCCTGCGTTGGGATTCCGTG | ACGGTGCAACAAGCTAGCGG |
| hsa-Let-7i-5p | GTGAGGTAGTAGTTTGTGCTGTT | Universal from miRNA 1st-Strand cDNA Synthesis kit |
| hsa-miR-24-3p | TGGCTCAGTTCAGCAGGAACA | Universal from miRNA 1st-Strand cDNA Synthesis kit |
| U75 | AGCCTGTGATGCTTTAAGAGTAG | Universal from miRNA 1st-Strand cDNA Synthesis kit |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hudy, D.; Rzeszowska-Wolny, J. Expression of miRNA-Targeted and Not-Targeted Reporter Genes Shows Mutual Influence and Intercellular Specificity. Int. J. Mol. Sci. 2022, 23, 15059. https://doi.org/10.3390/ijms232315059
Hudy D, Rzeszowska-Wolny J. Expression of miRNA-Targeted and Not-Targeted Reporter Genes Shows Mutual Influence and Intercellular Specificity. International Journal of Molecular Sciences. 2022; 23(23):15059. https://doi.org/10.3390/ijms232315059
Chicago/Turabian StyleHudy, Dorota, and Joanna Rzeszowska-Wolny. 2022. "Expression of miRNA-Targeted and Not-Targeted Reporter Genes Shows Mutual Influence and Intercellular Specificity" International Journal of Molecular Sciences 23, no. 23: 15059. https://doi.org/10.3390/ijms232315059






