Structural Change from Nonparallel to Parallel G-Quadruplex Structures in Live Cancer Cells Detected in the Lysosomes Using Fluorescence Lifetime Imaging Microscopy
Abstract
:1. Introduction
2. Results
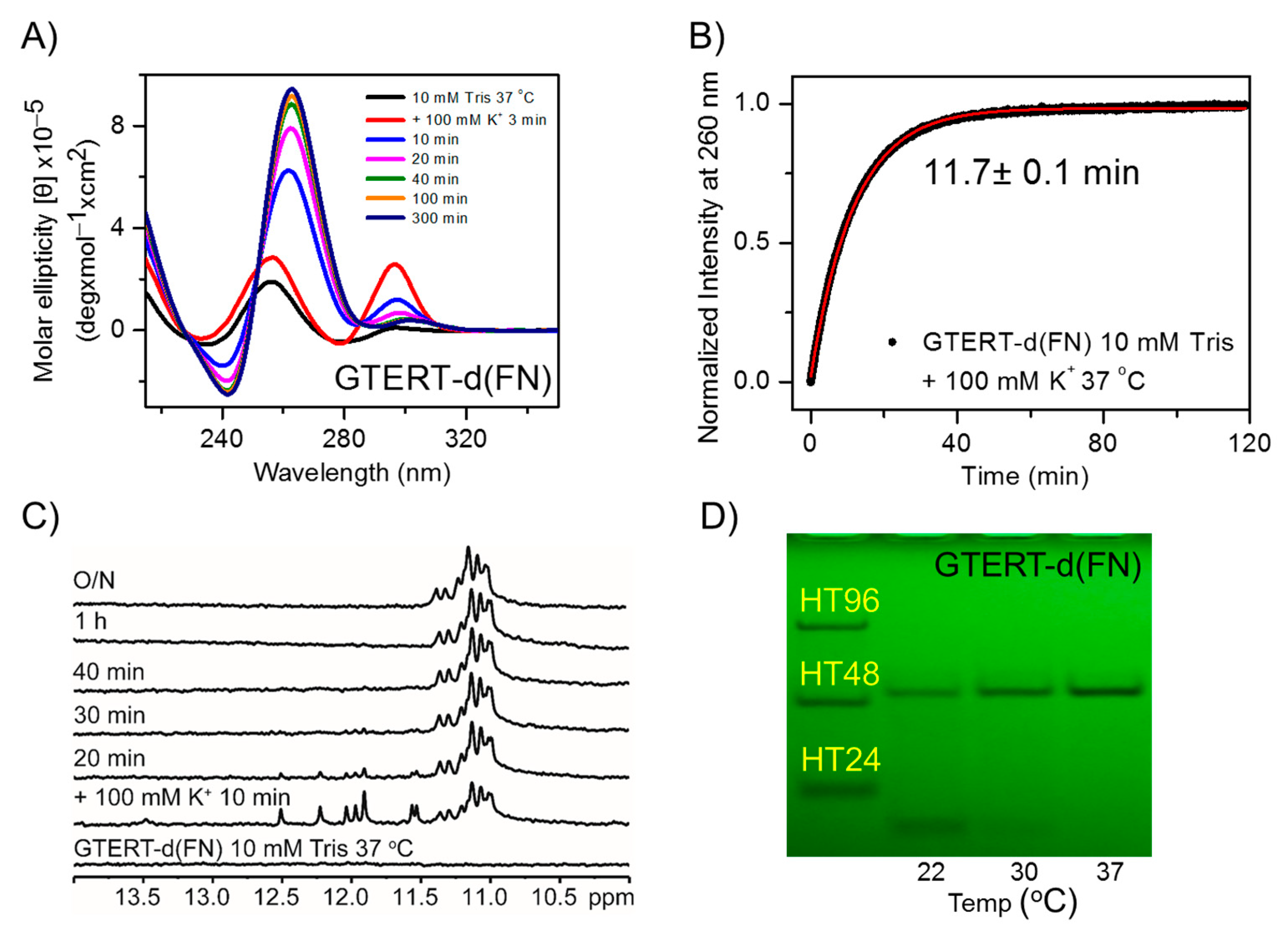
2.1. Temperature Effect on the Structural Conversion of GTERT-d(FN) G4s in Solution
2.2. Time-Gated FLIM Images of the Exogenous GTERT-d(FN) G4s in CL1-0 Live Cells
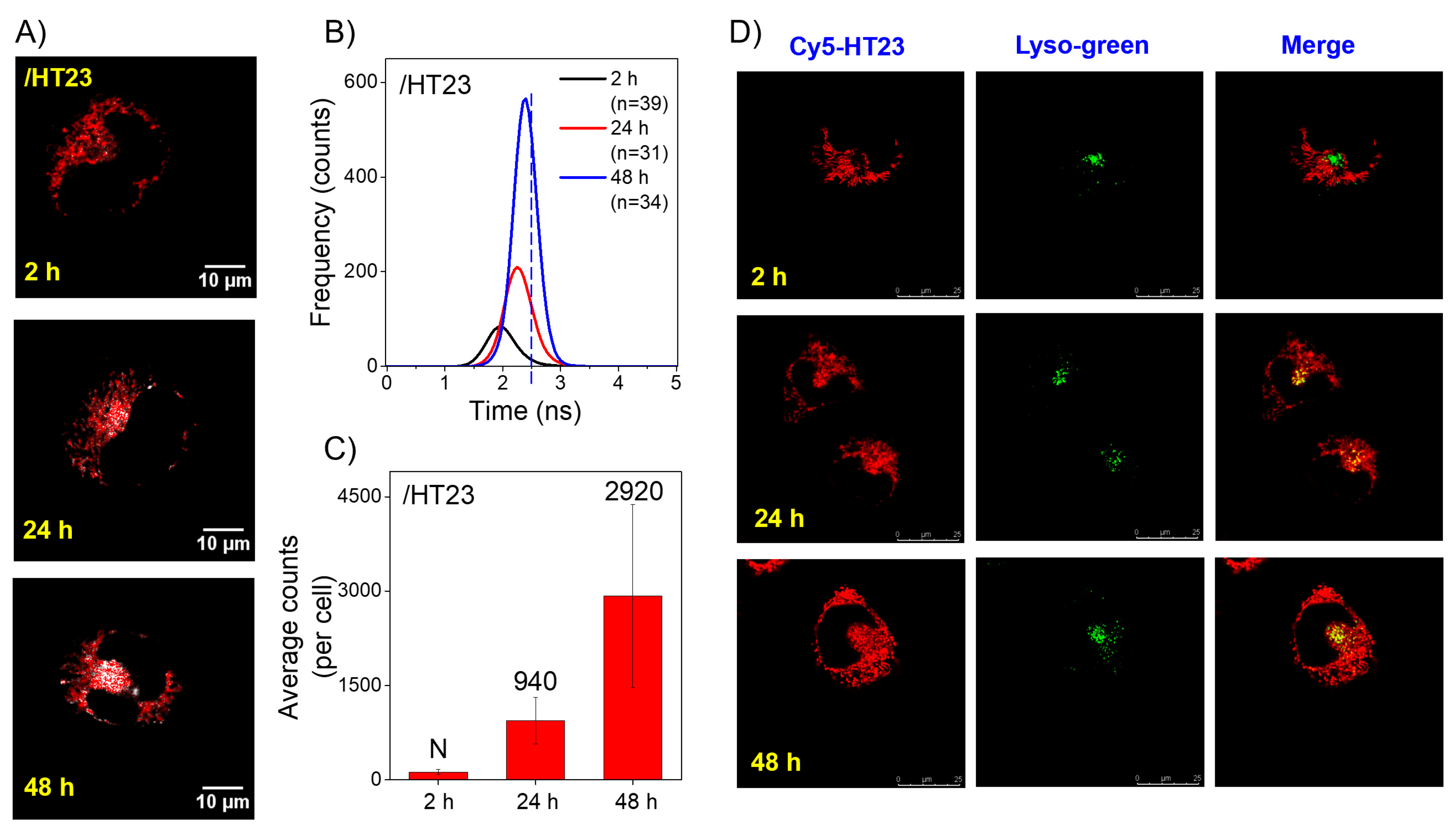
2.3. Imaging Study of the Exogenous HT23 G4s in CL1-0 Live Cells
2.4. Time-Gated FLIM Images of the Exogenous ss-GROs in CL1-0 Live Cells
3. Discussion
4. Materials and Methods
4.1. DNA Preparation
4.2. Circular Dichroism (CD)
4.3. Nuclear Magnetic Resonance (NMR) Spectroscopy
4.4. Polyacrylamide Gel Electrophoresis (PAGE)
4.5. Cell Cultures
4.6. Fluorescence Lifetime Imaging Microscopy (FLIM)
4.7. Confocal Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Neidle, S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017, 1, 0041. [Google Scholar] [CrossRef]
- Kosiol, N.; Juranek, S.; Brossart, P.; Heine, A.; Paeschke, K. G-quadruplexes: A promising target for cancer therapy. Mol. Cancer 2021, 20, 40. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Henderson, A.; Wu, Y.; Huang, Y.C.; Chavez, E.A.; Platt, J.; Johnson, F.B.; Brosh, R.M., Jr.; Sen, D.; Lansdorp, P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014, 42, 860–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tseng, T.Y.; Chien, C.H.; Chu, J.F.; Huang, W.C.; Lin, M.Y.; Chang, C.C.; Chang, T.C. Fluorescent probe for visualizing guanine-quadruplex DNA by fluorescence lifetime imaging microscopy. J. Biomed. Opt. 2013, 18, 101309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tseng, T.Y.; Chen, W.W.; Chu, I.T.; Wang, C.L.; Chang, C.C.; Lin, M.C.; Lou, P.J.; Chang, T.C. The G-quadruplex fluorescent probe 3,6-bis(1-methyl-2-vinyl-pyridinium) carbazole diiodide as a biosensor for human cancers. Sci. Rep. 2018, 8, 16082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tseng, T.Y.; Liu, S.Y.; Wang, C.L.; Chang, T.C. Antisense oligonucleotides used to identify telomeric G-quadruplexes in metaphase chromosomes and fixed cells by fluorescence lifetime imaging microscopy of o-BMVC foci. Molecules 2020, 25, 4083. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tannahill, D.; Miller, J.; Howat, W.J.; Balasubramanian, S. Elevated levels of G-quadruplex formation in human stomach and liver cancer tissues. PLoS ONE 2014, 9, e102711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.Y.; Liu, W.; Wang, K.N.; Zhu, B.C.; Xia, X.Y.; Ji, L.N.; Mao, Z.W. Quantitative detection of G-quadruplex DNA in live cells based on photon counts and complex structure discrimination. Angew. Chem. 2020, 59, 9719–9726. [Google Scholar] [CrossRef]
- Tseng, T.-Y.; Shih, S.-R.; Wang, C.-P.; Lin, S.-J.; Jan, I.-S.; Wang, C.-L.; Liu, S.-Y.; Chang, C.-C.; Lou, P.-J.; Chang, T.-C. Pilot imaging study of o-BMVC foci for discrimination of indeterminate cytology in diagnosing fine-needle aspiration of thyroid nodules. Sci. Rep. 2021, 11, 23475. [Google Scholar] [CrossRef]
- Dai, J.; Carver, M.; Yang, D. Polymorphism of human telomeric quadruplex structures. Biochimie 2008, 90, 1172–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lane, A.N.; Chaires, J.B.; Gray, R.D.; Trent, J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008, 36, 5482–5515. [Google Scholar] [CrossRef] [Green Version]
- Phan, A.T. Human telomeric G-quadruplex: Structures of DNA and RNA sequences. FEBS 2010, 277, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Gray, R.D.; Trent, J.O.; Chaires, J.B. Folding and unfolding pathways of the human telomeric G-quadruplex. J. Mol. Biol. 2014, 428, 1629–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.F.; Li, M.H.; Hsu, S.T.; Chang, T.C. Structural basis of sodium-potassium exchange of a human telomeric DNA quadruplex without topological conversion. Nucleic Acids Res. 2014, 42, 4723–4733. [Google Scholar] [CrossRef]
- Bates, P.J.; Kahlon, J.B.; Thomas, S.D.; Trent, J.O.; Miller, D.M. Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J. Biol. Chem. 1999, 274, 26369–26377. [Google Scholar] [CrossRef] [Green Version]
- Jing, N.; Li, Y.; Xiong, W.; Sha, W.; Jing, L.; Tweardy, D.J. G-quartet oligonucleotides: A new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res. 2004, 64, 6603–6609. [Google Scholar] [CrossRef] [Green Version]
- Qi, H.; Lin, C.P.; Fu, X.; Wood, L.M.; Liu, A.A.; Tsai, Y.C.; Chen, Y.; Barbieri, C.M.; Pilch, D.S.; Liu, L.F. Gquadruplexes induces apoptosis in tumor cells. Cancer Res. 2006, 66, 11808–11816. [Google Scholar] [CrossRef] [Green Version]
- Sedoris, K.C.; Thomas, S.D.; Clarkson, C.R.; Muench, D.; Islam, A.; Singh, R.; Miller, D.M. Genomic c-Myc quadruplex DNA selectively kills leukemia. Mol. Cancer Ther. 2012, 11, 66–76. [Google Scholar] [CrossRef]
- Tseng, T.Y.; Wang, Z.F.; Chien, C.H.; Chang, T.C. In-cell optical imaging of exogenous G-quadruplex DNA by fluorogenic ligands. Nucleic Acids Res. 2013, 41, 10605–10618. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.Y.; Wang, C.L.; Huang, W.C.; Chang, T.C. Folding and unfolding of exogenous G-rich oligonucleotides in live cells by fluorescence lifetime imaging microscopy of o-BMVC fluorescent probe. Molecules 2022, 27, 140. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chu, I.T.; Yeh, T.A.; Chen, D.Y.; Wang, C.L.; Chang, T.C. Effects of length and loop composition on structural diversity and similarity of (G3TG3NmG3TG3) G-quadruplexes. Molecules 2020, 25, 1779. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, D.; Sugimoto, N. Molecular crowding effects on structure and stability of DNA. Biochimie 2008, 90, 1040–1051. [Google Scholar] [CrossRef] [PubMed]
- Heddi, B.; Phan, A.T. Structure of Human Telomeric DNA in Crowded Solution. J. Am. Chem. Soc. 2011, 133, 9824–9833. [Google Scholar] [CrossRef]
- Xu, H.; Ren, D. Lysosomal Physiology. Annu. Rev. Physiol. 2015, 77, 57–80. [Google Scholar] [CrossRef] [Green Version]
- Jeger, J.L. Endosomes, lysosomes, and the role of endosomal and lysosomal biogenesis in cancer development. Mol. Biol. Rep. 2020, 47, 9801–9810. [Google Scholar] [CrossRef]
- Pu, J.; Guardia, C.M.; Keren-Kaplan, T.; Bonifacino, J.S. Mechanisms and functions of lysosome positioning. J. Cell Sci. 2016, 129, 4329–4339. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, R.E.; Zoncu, R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 2019, 21, 133–142. [Google Scholar] [CrossRef]
- Ballabio, A.; Bonifacino, J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 101–118. [Google Scholar] [CrossRef]
- Audano, M.; Schneider, A.; Mitro, N. Mitochondria, lysosomes, and dysfunction: Their meaning in neurodegeneration. J. Neurochem. 2018, 147, 291–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Fang, H.; Shao, X.; Tian, Z.; Geng, S.; Zhang, Y.; Fan, H.; Xiang, P.; Zhang, J.; Tian, X.; et al. A dual-labeling probe to track functional mitochondria-lysosome interactions in live cells. Nat. Common. 2020, 11, 6290. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.C.; Luh, K.T.; Wu, R.; Wu, C.W. Characterization of the mucin differentiation in human lung adenocarcinoma cell lines. Am. J. Respir. Cell Mol. Biol. 1992, 7, 161–171. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tseng, T.-Y.; Wang, C.-L.; Chang, T.-C. Structural Change from Nonparallel to Parallel G-Quadruplex Structures in Live Cancer Cells Detected in the Lysosomes Using Fluorescence Lifetime Imaging Microscopy. Int. J. Mol. Sci. 2022, 23, 15799. https://doi.org/10.3390/ijms232415799
Tseng T-Y, Wang C-L, Chang T-C. Structural Change from Nonparallel to Parallel G-Quadruplex Structures in Live Cancer Cells Detected in the Lysosomes Using Fluorescence Lifetime Imaging Microscopy. International Journal of Molecular Sciences. 2022; 23(24):15799. https://doi.org/10.3390/ijms232415799
Chicago/Turabian StyleTseng, Ting-Yuan, Chiung-Lin Wang, and Ta-Chau Chang. 2022. "Structural Change from Nonparallel to Parallel G-Quadruplex Structures in Live Cancer Cells Detected in the Lysosomes Using Fluorescence Lifetime Imaging Microscopy" International Journal of Molecular Sciences 23, no. 24: 15799. https://doi.org/10.3390/ijms232415799



