Divergent Contribution of the Golgi Apparatus to Microtubule Organization in Related Cell Lines
Abstract
:1. Introduction
2. Results
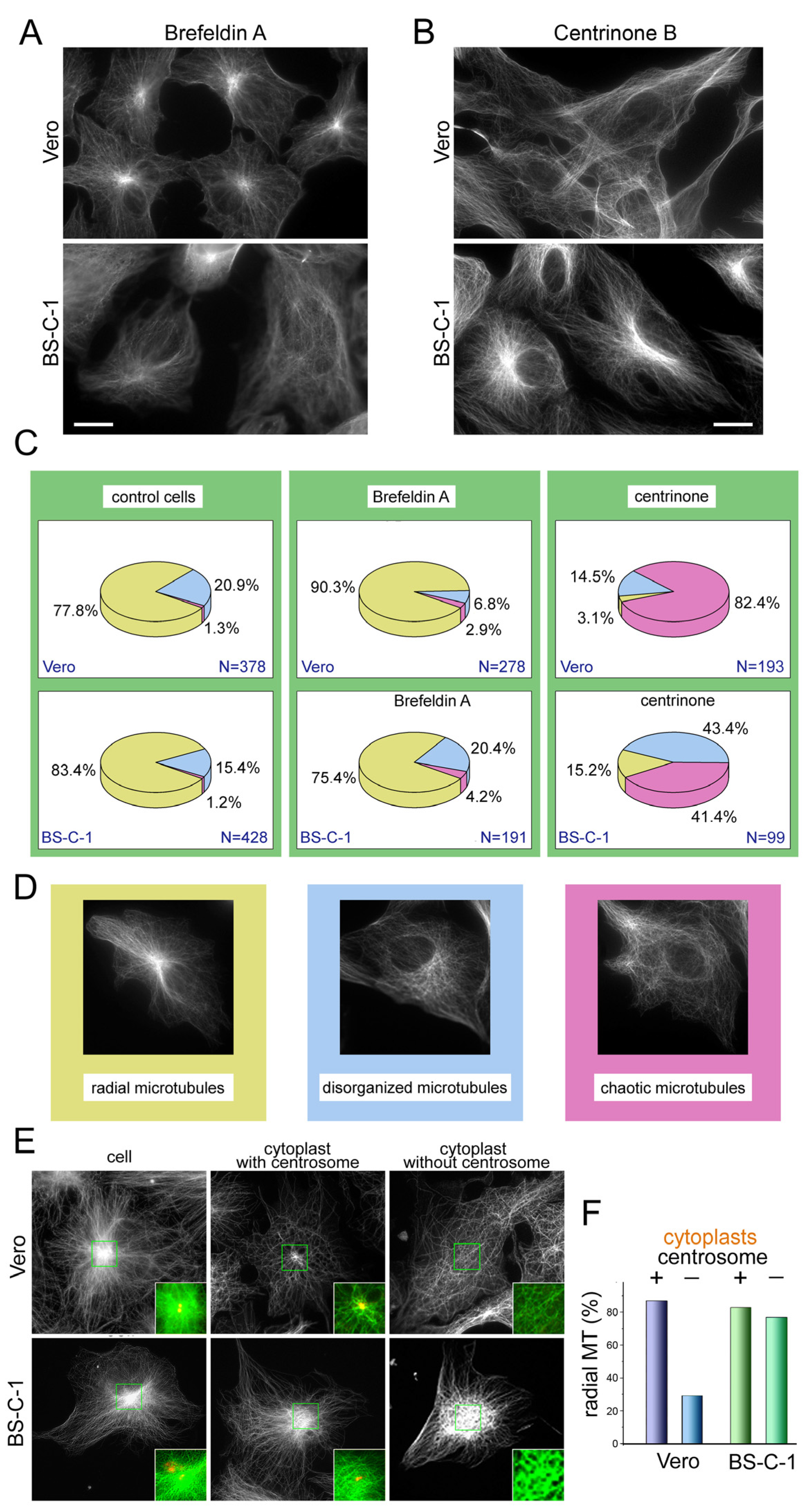
2.1. Morphological Analysis of the Microtubule System Revealed a Difference in Microtubule Architecture between the Two Lines of Green Monkey Kidney Cells
2.2. The Centrosome and the Golgi Have Different Impacts on Microtubule Organization in the Vero and BS-C-1 Cells
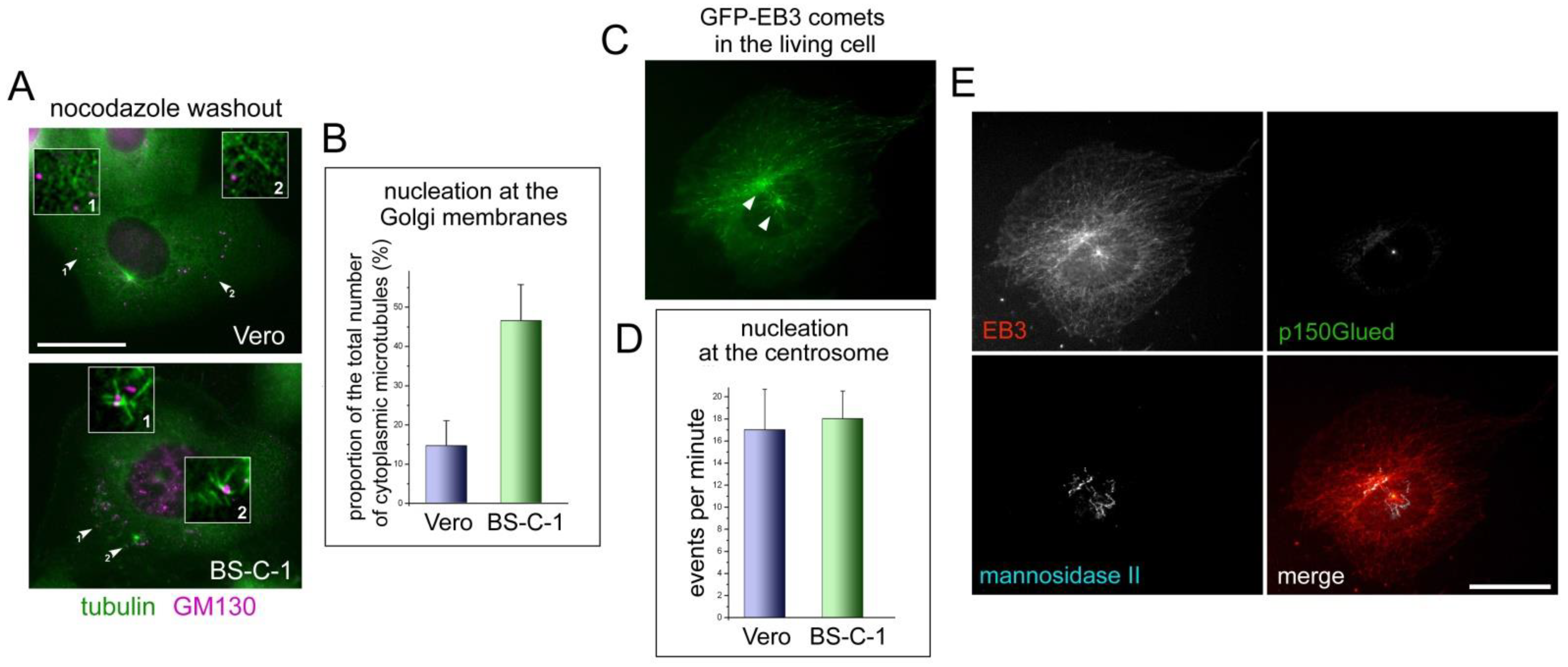
2.3. Different Nucleation Activity at the Golgi Membranes in the Vero and BS-C-1 Cells Coexists with the Same Activity of Their Centrosomes
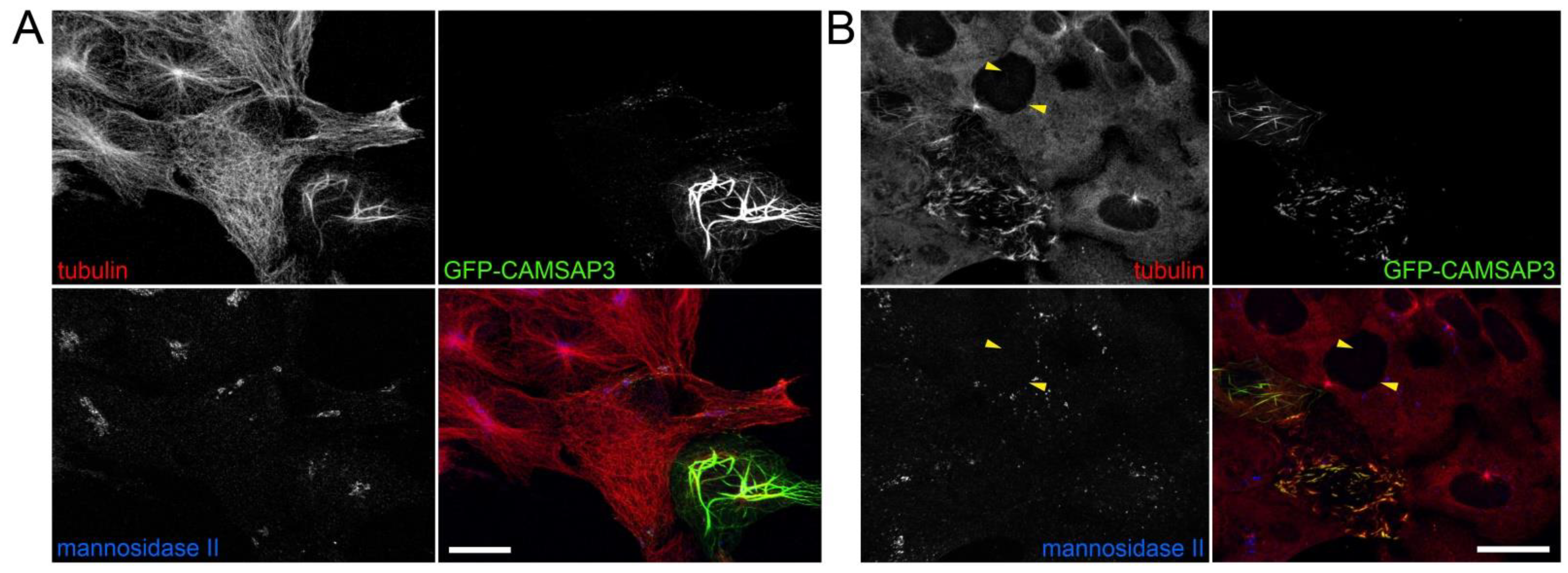
2.4. CAMSAP3 Overexpression Alters Microtubule Organization in the Golgi-Independent Manner
2.5. The GCC185-CLASP2 Axis Is Not Determining for the Golgi Activity as an MTOC
2.6. Transcriptomic Analysis Reveals Several Hitherto Unknown Potential Factors That May Influence the Golgi Functioning as a Microtubule Organizer
3. Discussion
4. Materials and Methods
4.1. Cultured Cells, Transfection, Inhibitors, Microtubule Regrowth, Cytoplast Preparation, Fixation
4.2. Antibodies
4.3. Immunofluorescent Staining and Microscopy, Live Imaging and Data Analysis
4.4. DNA Constructs
4.5. SDS-PAGE and Immunoblotting
4.6. RNA Sequencing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fokin, A.I.; Zhapparova, O.N.; Burakov, A.V.; Nadezhdina, E.S. Centrosome-derived microtubule radial array, PCM-1 protein, and primary cilia formation. Protoplasma 2019, 256, 1361–1373. [Google Scholar] [CrossRef] [PubMed]
- Conduit, P.T.; Wainman, A.; Raff, J.W. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 2015, 16, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Chabin-Brion, K.; Marceiller, J.; Perez, F.; Settegrana, C.; Drechou, A.; Durand, G.; Poüs, C. The Golgi complex is a microtubule-organizing organelle. Mol. Biol. Cell 2001, 12, 2047–2060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efimov, A.; Kharitonov, A.; Efimova, N.; Loncarek, J.; Miller, P.M.; Andreyeva, N.; Gleeson, P.; Galjart, N.; Maia, A.R.R.; McLeod, I.X.; et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell 2007, 12, 917–930. [Google Scholar] [CrossRef] [Green Version]
- Tassin, A.M.; Maro, B.; Bornens, M. Fate of microtubule-organizing centers during myogenesis in vitro. J. Cell Biol. 1985, 100, 35–46. [Google Scholar] [CrossRef] [Green Version]
- Bugnard, E.; Zaal, K.J.; Ralston, E. Reorganization of microtubule nucleation during muscle differentiation. Cell Motil. Cytoskelet. 2004, 60, 1–13. [Google Scholar] [CrossRef]
- Sanchez, A.D.; Feldman, J.L. Microtubule-organizing centers: From the centrosome to non-centrosomal sites. Curr. Opin. Cell Biol. 2017, 44, 93–101. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Akhmanova, A. Microtubule-Organizing Centers. Annu. Rev. Cell Dev. Biol. 2017, 33, 51–75. [Google Scholar] [CrossRef]
- Vinogradova, T.; Paul, R.; Grimaldi, A.D.; Loncarek, J.; Miller, P.M.; Yampolsky, D.; Magidson, V.; Khodjakov, A.; Mogilner, A.; Kaverina, I. Concerted effort of centrosomal and Golgi-derived microtubules is required for proper Golgi complex assembly but not for maintenance. Mol. Biol. Cell 2012, 23, 820–833. [Google Scholar] [CrossRef]
- Petry, S.; Vale, R.D. Microtubule nucleation at the centrosome and beyond. Nat. Cell Biol. 2015, 17, 1089–1093. [Google Scholar] [CrossRef]
- Sanders, A.A.; Kaverina, I. Nucleation and Dynamics of Golgi-derived Microtubules. Front. Neurosci. 2015, 9, 431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogalski, A.A.; Singer, S.J. Associations of elements of the Golgi apparatus with microtubules. J. Cell Biol. 1984, 99, 1092–1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thyberg, J.; Moskalewski, S. Microtubules and the organization of the Golgi complex. Exp. Cell Res. 1985, 159, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, A.; Meservey, L.; Nguyen, H.; Fu, M.-M. Golgi Outposts Nucleate Microtubules in Cells with Specialized Shapes. Trends Cell Biol. 2020, 30, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Sulimenko, V.; Hájková, Z.; Klebanovych, A.; Dráber, P. Regulation of microtubule nucleation mediated by γ-tubulin complexes. Protoplasma 2017, 254, 1187–1199. [Google Scholar] [CrossRef] [PubMed]
- Ríos, R.M.; Sanchís, A.; Tassin, A.M.; Fedriani, C.; Bornens, M. GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell 2004, 118, 323–335. [Google Scholar] [CrossRef] [Green Version]
- Mascanzoni, F.; Iannitti, R.; Colanzi, A. Functional Coordination among the Golgi Complex, the Centrosome and the Microtubule Cytoskeleton during the Cell Cycle. Cells 2022, 11, 354. [Google Scholar] [CrossRef]
- Al-Bassam, J.; Kim, H.; Brouhard, G.; van Oijen, A.; Harrison, S.C.; Chang, F. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev. Cell 2010, 19, 245–258. [Google Scholar] [CrossRef] [Green Version]
- Matsuoka, R.; Miki, M.; Mizuno, S.; Ito, Y.; Yamada, C.; Suzuki, A. MTCL2 promotes asymmetric microtubule organization by crosslinking microtubules on the Golgi membrane. J. Cell Sci. 2022, 135, jcs259374. [Google Scholar] [CrossRef]
- Wu, J.; de Heus, C.; Liu, Q.; Bouchet, B.P.; Noordstra, I.; Jiang, K.; Hua, S.; Martin, M.; Yang, C.; Grigoriev, I.; et al. Molecular Pathway of Microtubule Organization at the Golgi Apparatus. Dev. Cell 2016, 39, 44–60. [Google Scholar] [CrossRef]
- Ayala, I.; Crispino, R.; Colanzi, A. GRASP65 controls Golgi position and structure during G2/M transition by regulating the stability of microtubules. Traffic 2019, 20, 785–802. [Google Scholar] [CrossRef] [PubMed]
- Sanders, A.A.W.M.; Chang, K.; Zhu, X.; Thoppil, R.J.; Holmes, W.R.; Kaverina, I. Nonrandom γ-TuNA-dependent spatial pattern of microtubule nucleation at the Golgi. Am. Soc. Cell Biol. 2017, 28, 3181–3192. [Google Scholar] [CrossRef]
- Hao, H.; Niu, J.; Xue, B.; Su, Q.P.; Liu, M.; Yang, J.; Qin, J.; Zhao, S.; Wu, C.; Sun, Y. Golgi-associated microtubules are fast cargo tracks and required for persistent cell migration. EMBO Rep. 2020, 21, e48385. [Google Scholar] [CrossRef] [PubMed]
- Ronchi, P.; Tischer, C.; Acehan, D.; Pepperkok, R. Positive feedback between Golgi membranes, microtubules and ER exit sites directs de novo biogenesis of the Golgi. J. Cell Sci. 2014, 127, 4620–4633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, Y.L.; Anzola, J.V.; Davis, R.L.; Yoon, M.; Motamedi, A.; Kroll, A.; Seo, C.P.; Hsia, J.E.; Kim, S.K.; Mitchell, J.W.; et al. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science 2015, 348, 1155–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodionov, V.; Nadezhdina, E.; Borisy, G. Centrosomal control of microtubule dynamics. Proc. Natl. Acad. Sci. USA 1999, 96, 115–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malikov, V.; Cytrynbaum, E.N.; Kashina, A.; Mogilner, A.; Rodionov, V. Centering of a radial microtubule array by translocation along microtubules spontaneously nucleated in the cytoplasm. Nat. Cell Biol. 2005, 7, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, H.; Jiang, Y.; Takahashi, M.; Takeichi, M.; Meng, W. CAMSAP3-dependent microtubule dynamics regulates Golgi assembly in epithelial cells. J. Genet. Genom. 2017, 44, 39–49. [Google Scholar] [CrossRef]
- Coquand, L.; Victoria, G.S.; Tata, A.; Carpentieri, J.A.; Brault, J.-B.; Guimiot, F.; Fraisier, V.; Baffet, A.D. CAMSAPs organize an acentrosomal microtubule network from basal varicosities in radial glial cells. J. Cell Biol. 2021, 220, e202003151. [Google Scholar] [CrossRef]
- Munro, S.; Nichols, B.J. The GRIP domain—a novel Golgi-targeting domain found in several coiled-coil proteins. Curr. Biol. 1999, 9, 377–380. [Google Scholar] [CrossRef]
- Brown, D.L.; Heimann, K.; Lock, J.; Kjer-Nielsen, L.; van Vliet, C.; Stow, J.L.; Gleeson, P.A. The GRIP domain is a specific targeting sequence for a population of trans-Golgi network derived tubulo-vesicular carriers. Traffic 2001, 2, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Dráberová, E.; Sulimenko, V.; Vinopal, S.; Sulimenko, T.; Sládková, V.; D’Agostino, L.; Sobol, M.; Hozák, P.; Křen, L.; Katsetos, C.D.; et al. Differential expression of human γ-tubulin isotypes during neuronal development and oxidative stress points to a γ-tubulin-2 prosurvival function. FASEB J. 2017, 31, 1828–1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelletier, L.; Jokitalo, E.; Warren, G. The effect of Golgi depletion on exocytic transport. Nat. Cell Biol. 2000, 2, 840–846. [Google Scholar] [CrossRef]
- Gavilan, M.P.; Gandolfo, P.; Balestra, F.R.; Arias, F.; Bornens, M.; Rios, R.M. The dual role of the centrosome in organizing the microtubule network in interphase. EMBO Rep. 2018, 19, e45942. [Google Scholar] [CrossRef]
- Pongrakhananon, V.; Saito, H.; Hiver, S.; Abe, T.; Shioi, G.; Meng, W.; Takeichi, M. CAMSAP3 maintains neuronal polarity through regulation of microtubule stability. Proc. Natl. Acad. Sci. USA 2018, 115, 9750–9755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhmanova, A.; Hoogenraad, C.C.; Drabek, K.; Stepanova, T.; Dortland, B.; Verkerk, T.; Vermeulen, W.; Burgering, B.M.; De Zeeuw, C.I.; Grosveld, F.; et al. CLASPs Are CLIP-115 and -170 Associating Proteins Involved in the Regional Regulation of Microtubule Dynamics in Motile Fibroblasts. Cell 2001, 104, 923–935. [Google Scholar] [CrossRef]
- Cruz-Garcia, D.; Vazquez-Martinez, R.; Peinado, J.; Anouar, Y.; Tonon, M.; Vaudry, H.; Castaño, J.P.; Malagon, M.M. Identification and characterization of two novel (neuro)endocrine long coiled-coil proteins. FEBS Lett. 2007, 581, 3149–3156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steindler, C.; Li, Z.; Algarté, M.; Alcover, A.; Libri, V.; Ragimbeau, J.; Pellegrini, S. Jamip1 (Marlin-1) Defines a Family of Proteins Interacting with Janus Kinases and Microtubules. J. Biol. Chem. 2004, 279, 43168–43177. [Google Scholar] [CrossRef] [Green Version]
- Cruz-García, D.; Diaz-Ruiz, A.; Rabanal-Ruiz, Y.; Peinado, J.R.; Gracia-Navarro, F.; Castaño, J.P.; Montero-Hadjadje, M.; Tonon, M.; Vaudry, H.; Anouar, Y.; et al. The Golgi-associated long coiled-coil protein NECC1 participates in the control of the regulated secretory pathway in PC12 cells. Biochem. J. 2012, 443, 387–396. [Google Scholar] [CrossRef] [Green Version]
- Vidal, R.L.; I Valenzuela, J.; Luján, R.; Couve, A. Cellular and subcellular localization of Marlin-1 in the brain. BMC Neurosci. 2009, 10, 37. [Google Scholar] [CrossRef]
- Díaz-Ruiz, A.; Rabanal-Ruiz, Y.; Trávez, A.; Gracia-Navarro, F.; Cruz-García, D.; Montero-Hadjadje, M.; Anouar, Y.; Gasman, S.; Vitale, N.; Vázquez-Martínez, R.; et al. The long coiled-coil protein NECC2 is associated to caveolae and modulates NGF/TrkA signaling in PC12 cells. PLoS ONE 2013, 8, e73668. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Chiang, T.C.; Liu, Y.T.; Tsai, Y.T.; Jang, L.T.; Lee, F.J.S. ARL4A acts with GCC185 to modulate Golgi complex organization. J. Cell Sci. 2011, 124, 4014–4026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brodsky, I.B.; Burakov, A.V.; Nadezhdina, E.S. Microtubules’ interaction with cell cortex is required for their radial organization, but not for centrosome positioning. Cell Motil. Cytoskelet. 2007, 64, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Burakov, A.; Kovalenko, O.; Semenova, I.; Zhapparova, O.; Nadezhdina, E.; Rodionov, V. Cytoplasmic Dynein is Involved in the Retention of Microtubules at the Centrosome in Interphase Cells. Traffic 2008, 9, 472–480. [Google Scholar] [CrossRef]


| Name | Alternative Name | BS-C-1 | Vero | BS-C-1/Vero |
|---|---|---|---|---|
| Reads | Reads | |||
| AKAP9 | AKAP450 | 1754 | 1871 | 0.94 |
| CAMSAP1 | 1215 | 1737 | 0.7 | |
| CAMSAP2 | 2338 | 1909 | 1.22 | |
| CAMSAP3 | 200 | 54 | 3.75 | |
| CDK5RAP1 | 574 | 866 | 0.66 | |
| CDK5RAP2 | 747 | 880 | 0.85 | |
| CDK5RAP3 | 1194 | 1669 | 0.72 | |
| CEP350 | 1584 | 1552 | 1.02 | |
| CLASP1 | 1224 | 1038 | 1.18 | |
| CLASP2 | 1276 | 1338 | 0.95 | |
| CLASRP | 300 | 530 | 0.57 | |
| GCC2 | GCC185 | 1529 | 1056 | 1.45 |
| GOLGA2 | GM130 | 1181 | 989 | 1.19 |
| GORASP1 | GRASP65 | 1678 | 940 | 1.79 |
| MMG * | LOC103247162 | 0 | 0 | 0 |
| LOC103224073 | 5 | 2 | 2.5 | |
| LOC103224075 | 0.5 | 0.5 | 1 | |
| MTCL1 | CCDC165 | 1413 | 871 | 1.62 |
| NIN | ninein | 1610 | 1539 | 1.05 |
| TRIP11 | GMAP210 | 1062 | 1366 | 0.78 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brodsky, I.B.; Fokin, A.I.; Efremov, A.A.; Nadezhdina, E.S.; Burakov, A.V. Divergent Contribution of the Golgi Apparatus to Microtubule Organization in Related Cell Lines. Int. J. Mol. Sci. 2022, 23, 16178. https://doi.org/10.3390/ijms232416178
Brodsky IB, Fokin AI, Efremov AA, Nadezhdina ES, Burakov AV. Divergent Contribution of the Golgi Apparatus to Microtubule Organization in Related Cell Lines. International Journal of Molecular Sciences. 2022; 23(24):16178. https://doi.org/10.3390/ijms232416178
Chicago/Turabian StyleBrodsky, Ilya B., Artem I. Fokin, Aleksei A. Efremov, Elena S. Nadezhdina, and Anton V. Burakov. 2022. "Divergent Contribution of the Golgi Apparatus to Microtubule Organization in Related Cell Lines" International Journal of Molecular Sciences 23, no. 24: 16178. https://doi.org/10.3390/ijms232416178





