Cryo-Treatment Enhances the Embryogenicity of Mature Somatic Embryos via the lncRNA–miRNA–mRNA Network in White Spruce
Abstract
:1. Introduction
2. Results
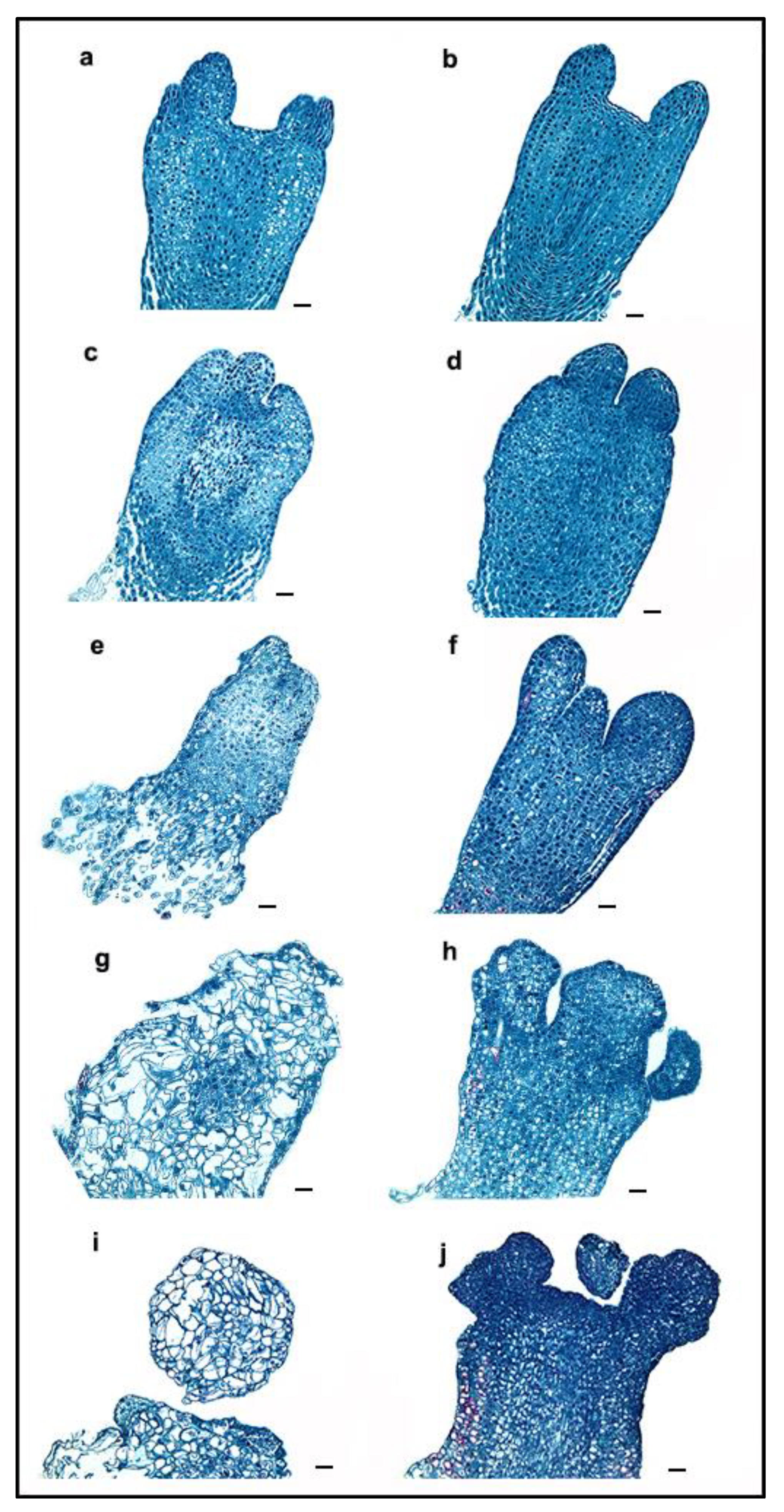
2.1. Morphological Changes during ET Induction
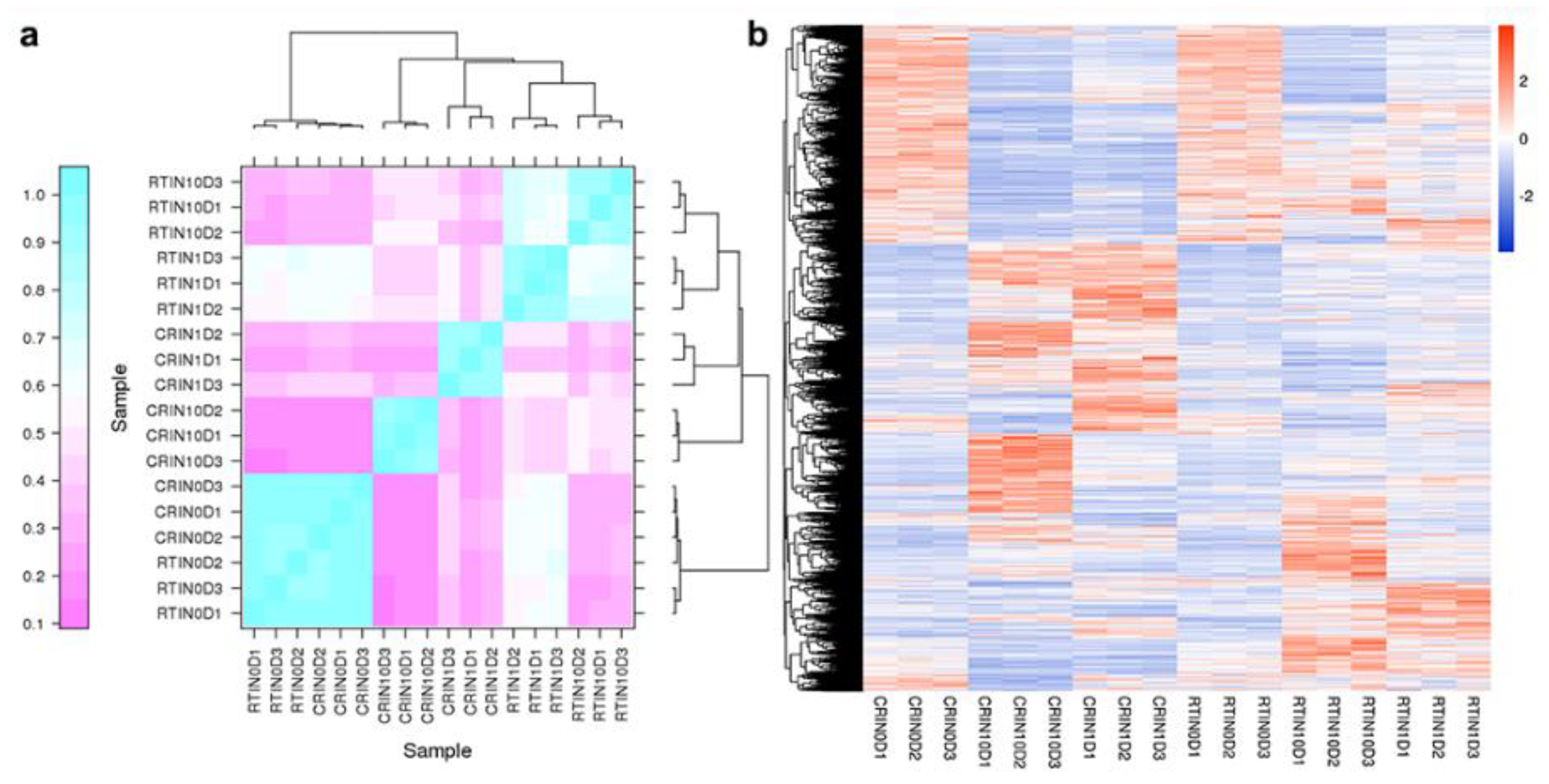
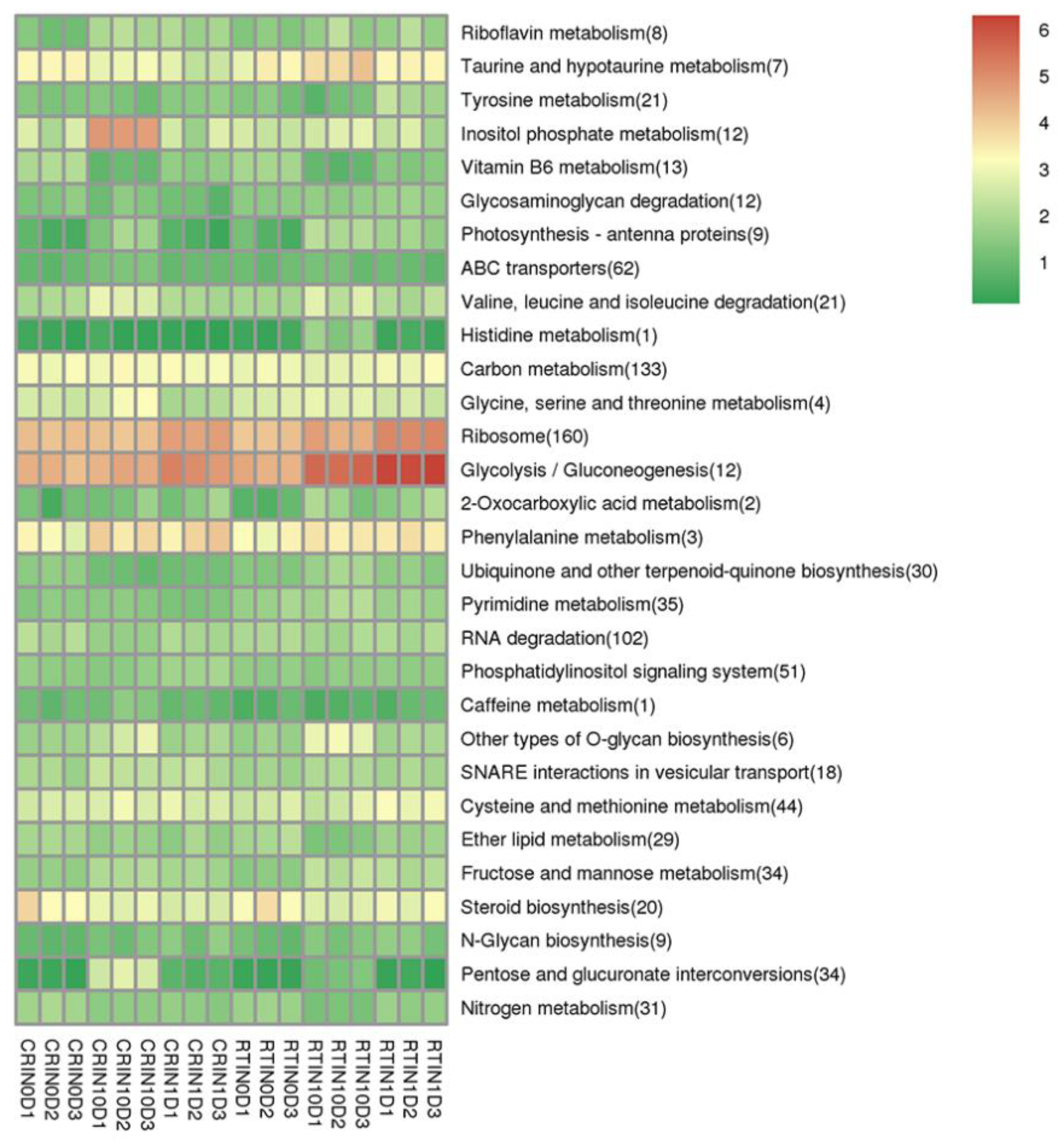
2.2. Differentially Expressed Genes (DEGs) and KEGG Analysis during the Induction Process
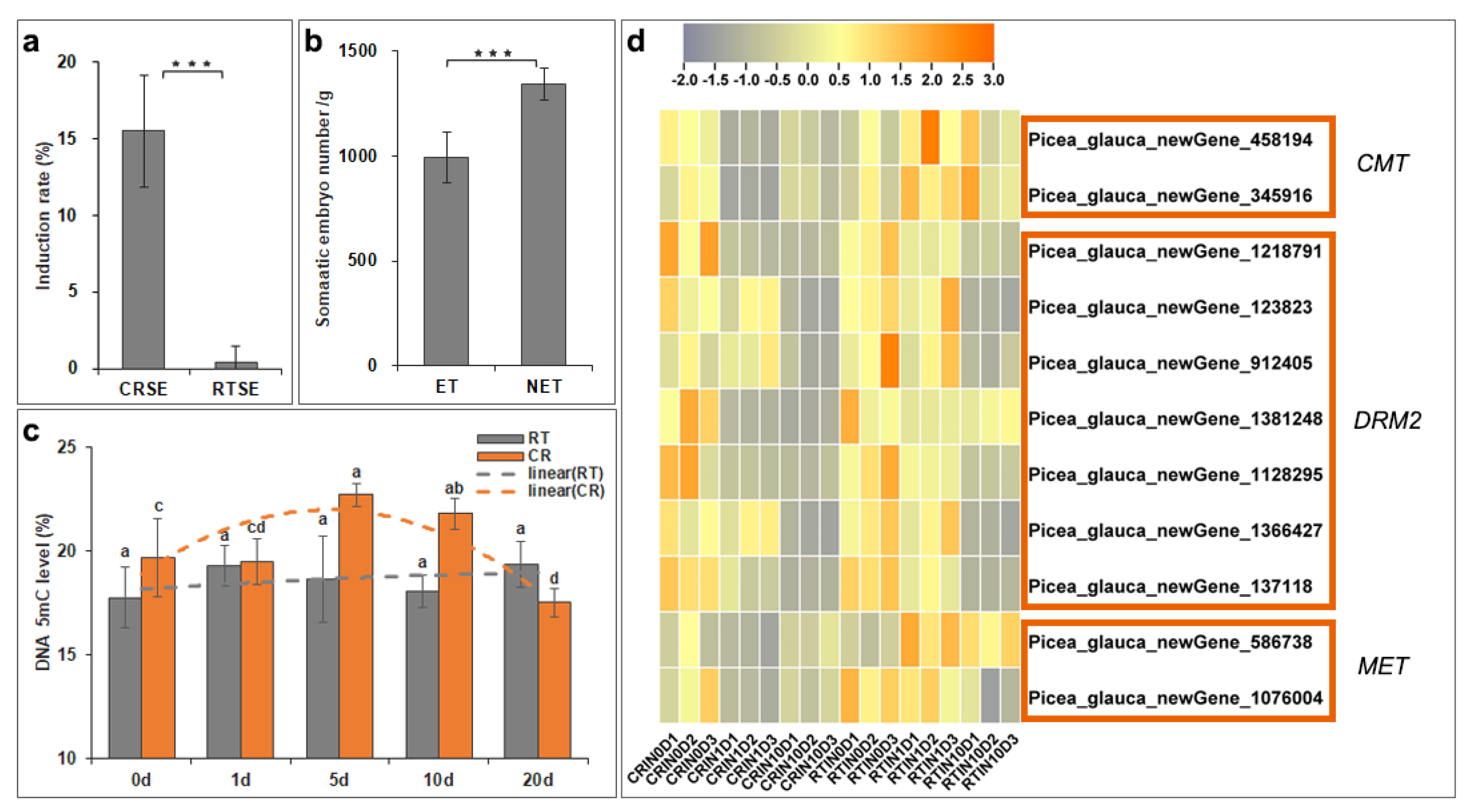
2.3. DNA Methylation Level Fluctuated during the Induction Process
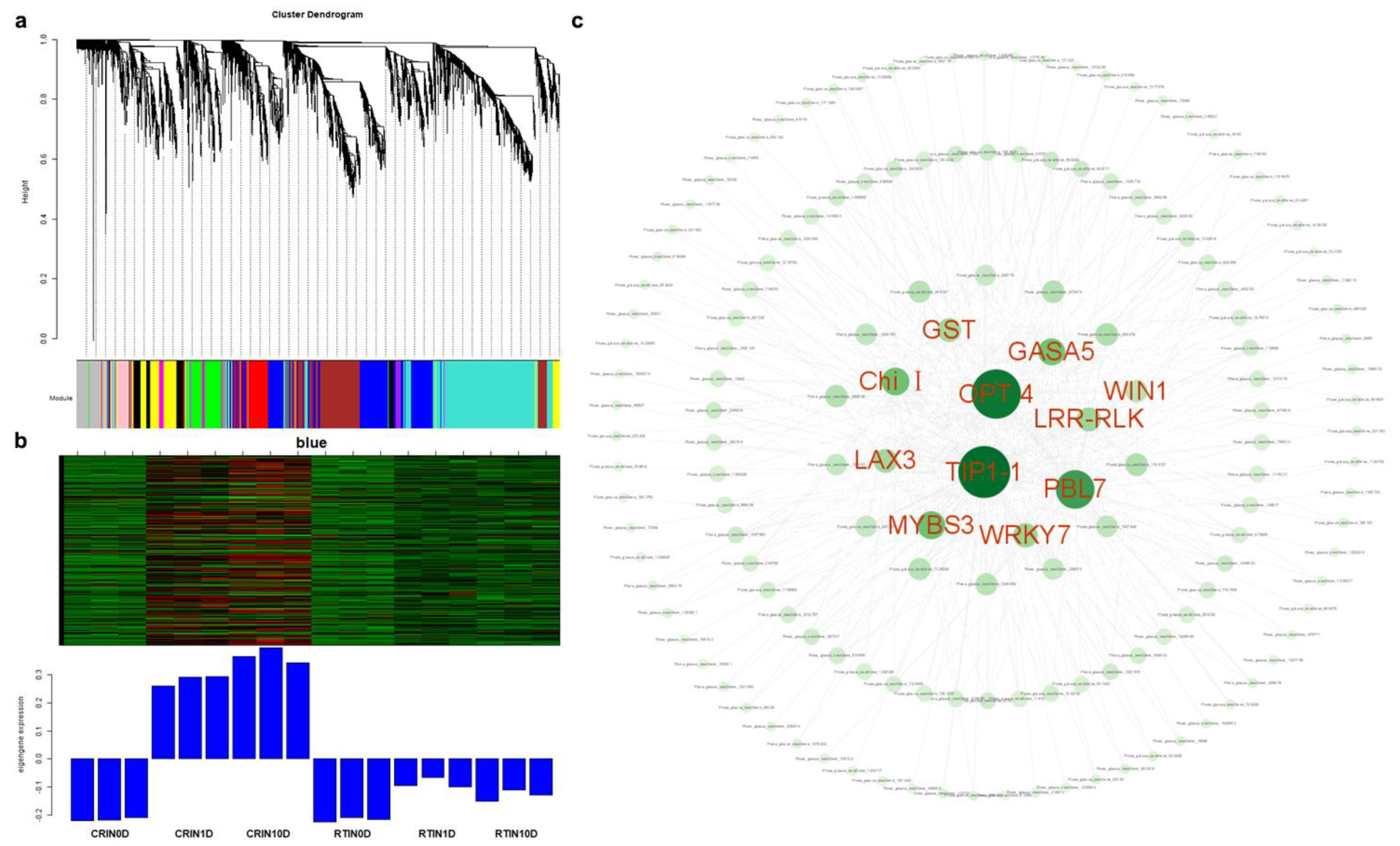
2.4. Analysis of Hub Genes via WGCNA and Network Construction
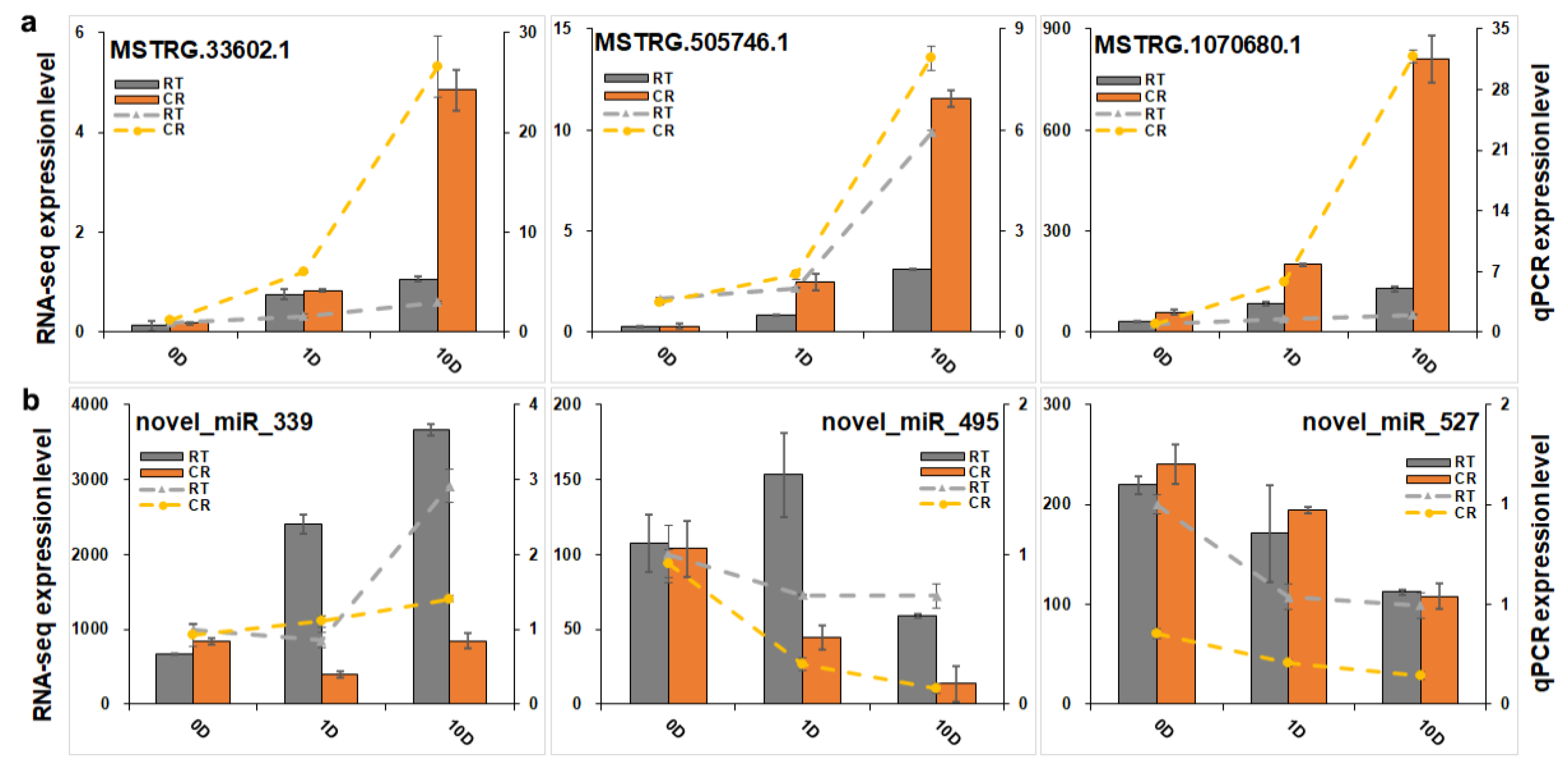
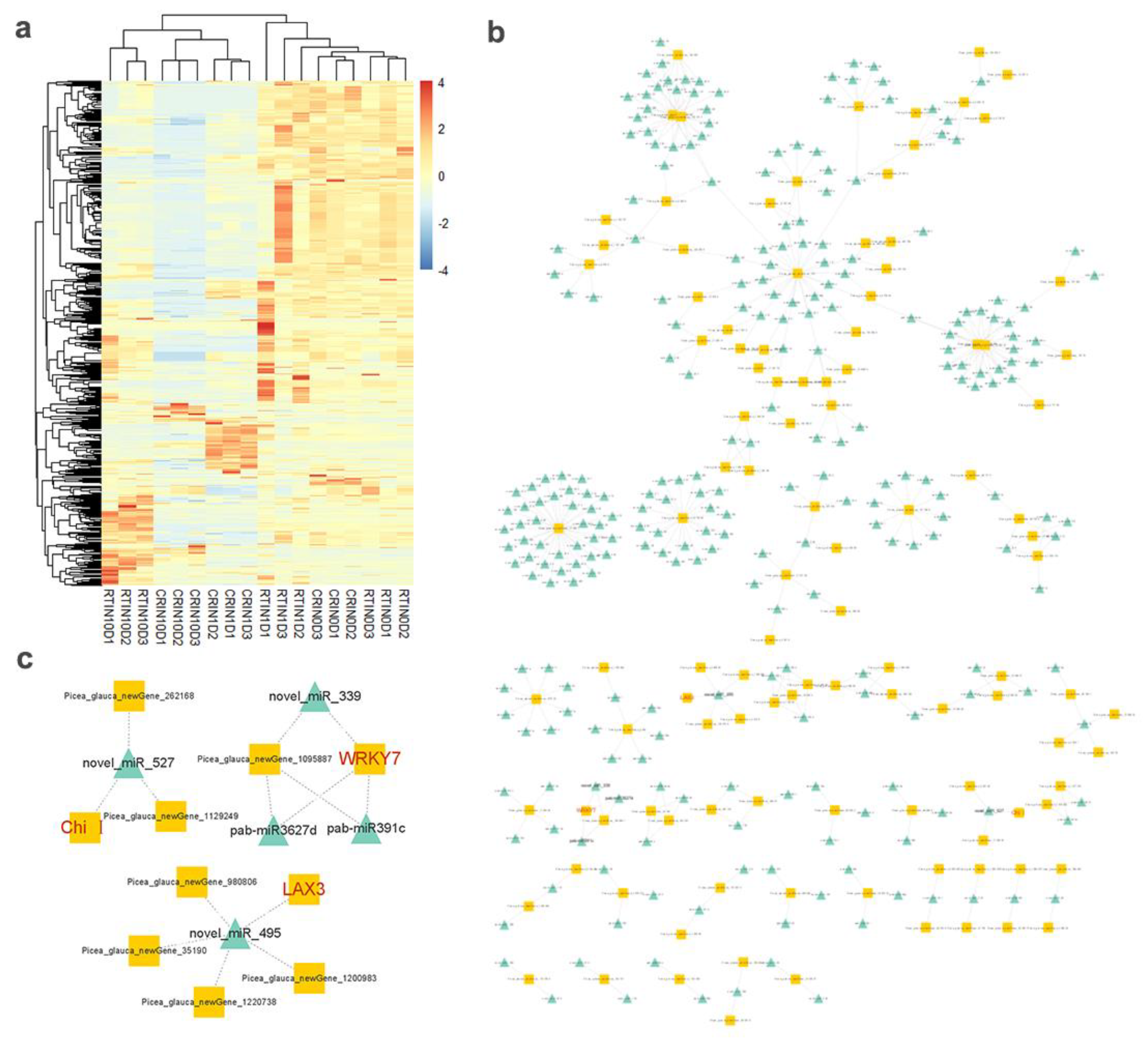
2.5. Expression Profiles of lncRNAs and Construction of the lncRNA–mRNA Co-Expression Network
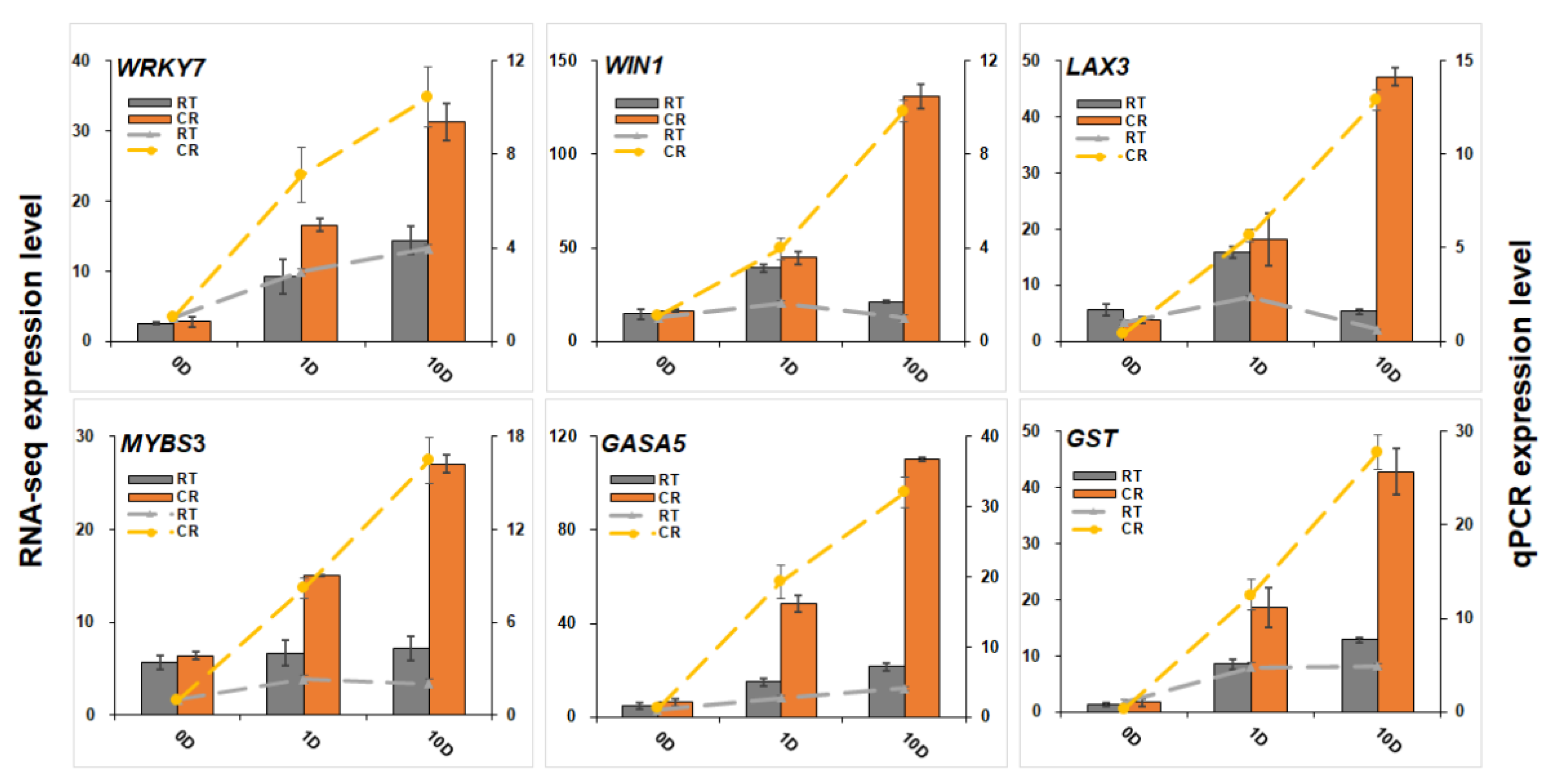
2.6. Expression Profiles of miRNAs and Analysis of Targeted mRNA in the Blue Module
3. Discussion
3.1. Application Value of Ultra-Low Temperature in Promoting ET Induction
3.2. Effects of Cryo-Treatment and Exogenous Hormones on Gene Expression during the Induction from CRSE
3.3. CeRNA Network of lncRNAs, miRNAs, mRNAs during the Induction from CRSE
4. Materials and Methods
4.1. Preparation of Plant Materials
4.2. Microscopic Observation
4.3. Detection of Induction Rate and Somatic Embryo Maturation Ability
4.4. Analysis of Global DNA Methylation
4.5. Whole-Transcriptome Sequencing
4.6. Transcriptomic Co-Expression Network Analysis
4.7. Quantitative PCR Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Attree, S.M.; Fowke, L.C. Embryogeny of gymnosperms—Advances in synthetic seed technology of conifers. Plant Cell Tissue Organ. Cult. 1993, 35, 31–35. [Google Scholar] [CrossRef]
- Park, Y.S.; Pond, S.E.; Bonga, J.M. Initiation of somatic embryogenesis in white spruce (Picea glauca): Genetic control, culture treatment effects, and implications for tree breeding. Theor. Appl. Genet. 1993, 86, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.S.; Pond, S.E.; Bonga, J.M. Somatic embryogenesis in white spruce (Picea glauca): Genetic control in somatic embryos exposed to storage, maturation treatments, germination, and cryopreservation. Theor. Appl. Genet. 1994, 89, 742–750. [Google Scholar] [CrossRef]
- Kong, L.; von Aderkas, P. A novel method of cryopreservation without a cryoprotectant for immature somatic embryos of conifer. Plant. Cell Tissue Organ. Cult. 2010, 106, 115–125. [Google Scholar] [CrossRef]
- Tremblay, F.M. Somatic embryogenesis and plantlet regeneration from embryos isolated from stored seeds of Picea glauca. Can. J. Bot. 1990, 68, 236–242. [Google Scholar] [CrossRef]
- Tao, J.; Chen, S.G.; Qin, C.Y.; Li, Q.M.; Cai, J.F.; Sun, C.; Wang, W.; Weng, Y. Somatic embryogenesis in mature zygotic embryos of Picea pungens. Sci. Rep. 2021, 11, 19072. [Google Scholar] [CrossRef] [PubMed]
- Vondrakova, Z.; Cvikrova, M.; Eliasova, K.; Martincova, O.; Vagner, M. Cryotolerance in Norway spruce and its association with growth rates, anatomical features and polyamines of embryogenic cultures. Tree Physiol. 2010, 30, 1335–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Gao, Y.; Zhao, R.R.; Zhao, J.; Li, Y.X.; Qi, S.J.; Zhang, J.F.; Kong, L. Transcriptomic, metabolomic, and physiological analyses reveal that the culture temperatures modulate the cryotolerance and embryogenicity of developing somatic embryos in Picea glauca. Front. Plant Sci. 2021, 12, 694229. [Google Scholar] [CrossRef]
- Klimaszewska, K.; Hargreaves, C.; Lelu-Walter, M.A.; Trontin, J.F. Advances in Conifer Somatic Embryogenesis Since Year 2000; Maria, A.G., Maurizio, L., Eds.; Springer Press: New York, NY, USA, 2016; Volume 1359, pp. 131–166. [Google Scholar]
- Tretyakova, I.N.; Mineev, V.V. Reproductive potential of conifers, somatic embryogenesis and apomixis. Russ. J. Dev. Biol. 2021, 52, 75–86. [Google Scholar] [CrossRef]
- Ikeda-Iwai, M.; Umehara, M.; Satoh, S.; Kamada, H. Stress-induced somatic embryogenesis in vegetative tissues of Arabidopsis thaliana. Plant J. 2003, 34, 107–114. [Google Scholar] [CrossRef]
- Pais, M.S. Somatic embryogenesis induction in woody species: The future after OMICs data assessment. Front. Plant Sci. 2019, 10, 240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariel, F.; Romero-Barrios, N.; Jegu, T.; Benhamed, M.; Crespi, M. Battles and hijacks, noncoding transcription in plants. Trends Plant Sci. 2015, 20, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Wang, Z.K.; Zhang, J.J.; Zhao, X.Y.; Xu, P.; Liu, X.; Li, M.; Lv, C.; Song, X. Crosstalk of mRNA, miRNA, lncRNA, and circRNA and their regulatory pattern in Pulmonary fibrosis. Mol. Ther. Nucleic Acids 2019, 18, 204–218. [Google Scholar] [CrossRef] [Green Version]
- D’Ario, M.; Griffiths-Jones, S.; Kim, M. Small RNAs: Big impact on plant development. Trends Plant Sci. 2017, 22, 1056–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, P.C.; Liu, S.; Nie, X.J.; Song, W.S.; Wu, L. Conservation analysis of long non-coding RNAs in plants. Sci. China Life Sci. 2018, 61, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J. Circular RNA expression: Its potential regulation and function. Trends Genet. 2016, 32, 309–316. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.X.; Cui, J.; Meng, J.; Luan, Y.S. Interactions and links among the noncoding RNAs in plants under stresses. Theor. Appl. Genet. 2020, 133, 3235–3248. [Google Scholar] [CrossRef]
- Chen, D.; He, B.; Zheng, P.; Wang, S.; Zhao, X.; Liu, J.; Yang, X.; Cheng, W. Identification of mRNA-, circRNA- and lncRNA- associated ceRNA networks and potential biomarkers for preeclampsia from umbilical vein endothelial cells. Front. Mol. Biosci. 2021, 8, 652250. [Google Scholar] [CrossRef]
- Chen, Y.; Li, X.; Su, L.; Chen, X.; Zhang, S.; Xu, X.; Zhang, Z.; Chen, Y.; XuHan, X.; Lin, Y.; et al. Genome-wide identification and characterization of long non-coding RNAs involved in the early somatic embryogenesis in Dimocarpus longan Lour. BMC Genom. 2018, 19, 805. [Google Scholar] [CrossRef] [Green Version]
- Song, J.H.; Cao, J.S. The progress of non-coding RNA in plants. Biotechnol. Bull. 2009, 5, 5–8. [Google Scholar]
- Cao, P.; Fan, W.; Li, P.; Hu, Y. Genome-wide profiling of long noncoding RNAs involved in wheat spike development. BMC Genom. 2021, 22, 493. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.J.; Tao, J.J.; Cheng, T.; Bian, X.H.; Wei, W.; Zhang, W.K.; Ma, B.; Chen, S.Y.; Zhang, J.S. Soybean miR172a improves salt tolerance and can function as a long-distance signal. Mol. Plant. 2016, 9, 1337–1340. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Sun, W.; Guo, Z.Y.; Zhang, J.C.; Yu, H.G.; Liu, B. Mechanisms of lncRNA/microRNA interactions in angiogenesis. Life Sci. 2020, 254, 116900. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Li, J.; Luo, M.; Li, H.; Chen, Q.; Wang, L.; Song, S.; Zhao, L.; Xu, W.; Zhang, C.; et al. Characterization and cloning of grape circular RNAs identified the cold resistance-related Vv-circATS1. Plant Physiol. 2019, 180, 966–985. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.M.; Liu, M.Y.; Ge, X.X.; Xu, Q.A.; Guo, W.W. Stage and tissue-specific modulation of ten conserved miRNAs and their targets during somatic embryogenesis of Valencia sweet orange. Planta 2011, 233, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.H.; Liu, Y.B.; Zhou, C.; Li, X.M.; Zhang, X.S. The microRNA167 controls somatic embryogenesis in Arabidopsis through regulating its target genes ARF6 and ARF8. Plant Cell Tissue Organ. Cult. 2016, 124, 405–417. [Google Scholar] [CrossRef]
- Lujan-Soto, E.; Juarez-Gonzalez, V.T.; Reyes, J.L.; Dinkova, T.D. MicroRNA Zma-miR528 versatile regulation on target mRNAs during maize somatic embryogenesis. Int. J. Mol. Sci. 2021, 22, 5310. [Google Scholar] [CrossRef]
- Zhang, J.H.; Wu, T.; Li, L.; Han, S.Y.; Li, X.M.; Zhang, S.; Qi, L. Dynamic expression of small RNA populations in larch (Larix leptolepis). Planta 2013, 237, 89–101. [Google Scholar] [CrossRef]
- Yakovlev, I.A.; Fossdal, C.G. In silico analysis of small RNAs suggest roles for novel and conserved miRNAs in the formation of epigenetic memory in somatic embryos of Norway spruce. Front. Physiol. 2017, 8, 674. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.F.; Deng, C.; Zhu, T.Q.; Ling, J.J.; Zhang, H.G.; Kong, L.; Zhang, S.; Wang, J.; Chen, X. Dynamics of physiological and miRNA changes after long-term proliferation in somatic embryogenesis of Picea balfouriana. Trees 2019, 33, 469–480. [Google Scholar] [CrossRef]
- Hu, X.G.; Zhou, S.S.; Yang, Y.; Liu, H.; Anil, S.; Wang, Q.; Zhao, W.; Gao, Q.; El-Kassaby, Y.A.; Wang, T.; et al. Transcriptome-wide identification and profiling of miRNAs in a stress-tolerant conifer Sabina chinensis. J. Biosci. 2020, 45, 41. [Google Scholar] [CrossRef]
- Perdiguero, P.; Rodrigues, A.S.; Chaves, I.; Costa, B.; Alves, A.; de Maria, N.; Vélez, M.D.; Díaz-Sala, C.; Cervera, M.T.; Miguel, C.M. Comprehensive analysis of the isomiRome in the vegetative organs of the conifer Pinus pinaster under contrasting water availability. Plant Cell Environ. 2021, 44, 706–728. [Google Scholar] [CrossRef] [PubMed]
- Varhanikova, M.; Uvackova, L.; Skultety, L.; Pretova, A.; Obert, B.; Hajduch, M. Comparative quantitative proteomic analysis of embryogenic and non-embryogenic calli in maize suggests the role of oxylipins in plant totipotency. J. Proteom. 2014, 104, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Li, W.; Parris, S.; West, M.; Lawson, J.; Smathers, M.; Li, Z.; Jones, D.; Jin, S.; Saski, C.A. Transcriptomic profiles of non-embryogenic and embryogenic callus cells in a highly regenerative upland cotton line (Gossypium hirsutum L.). BMC Dev. Biol. 2020, 20, 25. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Park, O.S.; Lee, H.G.; Seo, P.J. The ASHR3 SET-domain protein is a pivotal upstream coordinator for wound-induced callus formation in Arabidopsis. J. Plant Biol. 2020, 63, 361–368. [Google Scholar] [CrossRef]
- Hely, H.; Anne, J.; Jana, K.; Anneli, K.; Karoliina, N.; Aronen, T. Somatic embryogenesis of Scots pine: Cold treatment and characteristics of explants affecting induction. J. Exp. Bot. 1999, 50, 1769–1778. [Google Scholar]
- Lan, Z.; Xu, Y.H.; Wang, J.B. DNA-methylation changes induced by salt stress in wheat Triticum aestivum. Afr. J. Biotechnol. 2009, 8, 6201–6207. [Google Scholar] [CrossRef]
- Joel, G.A.J. Epigenetic responses to drought stress in rice (Oryza sativa L.). Physiol. Mol. Biol. Plants 2013, 19, 379–387. [Google Scholar]
- Falahi, A.; Zarei, L.; Cheghamirza, K. Most drought-induced DNA methylation changes switched to pre-stress state after re-irrigation in barley (Hordeum vulgare L.) cultivars. Cereal Res. Commun. 2021, 10. [Google Scholar] [CrossRef]
- Lease, K.A.; Lau, N.Y.; Schuster, R.A.; Torii, K.U.; Walker, J.C. Receptor serine/threonine protein kinases in signalling: Analysis of the erecta receptor-like kinase of Arabidopsis thaliana. New Phytol. 2001, 151, 133–143. [Google Scholar] [CrossRef]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant Microb. Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef] [Green Version]
- Khodakovskaya, M.; Sword, C.; Wu, Q.; Perera, I.Y.; Boss, W.F.; Brown, C.S.; Winter Sederoff, H. Increasing inositol (1,4,5)-trisphosphate metabolism affects drought tolerance, carbohydrate metabolism and phosphate-sensitive biomass increases in tomato. Plant Biotechnol. J. 2010, 8, 170–183. [Google Scholar] [CrossRef] [PubMed]
- Su, C.F.; Wang, Y.C.; Hsieh, T.H.; Lu, C.A.; Tseng, T.H.; Yu, S.M. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 2010, 153, 145–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Zhao, X.Q.; Wang, W.S.; Pan, Y.J.; Huang, L.Y.; Liu, X.; Zong, Y.; Zhu, L.; Yang, D.; Fu, B. Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS ONE 2012, 7, e43274. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Li, J.J.; Chen, Y.; Yang, J.; Chen, X.Y. Genome-wide analysis of the WRKY gene family and its response to abiotic stress in buckwheat (Fagopyrum Tataricum). Open Life Sci. 2019, 14, 80–96. [Google Scholar] [CrossRef]
- Baillo, E.H.; Hanif, M.S.; Guo, Y.; Zhang, Z.; Xu, P.; Algam, S.A. Genome-wide Identification of WRKY transcription factor family members in sorghum (Sorghum bicolor (L.) moench). PLoS ONE 2020, 15, e0236651. [Google Scholar]
- Stanford, A.; Bevan, M.; Northcote, D. Differential expression within a family of novel wound-induced genes in potato. Mol. Gen. Genet. 1989, 215, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.N.; Pu, L.M.; Qu, Y.; Yang, Y.; Bai, Z.W.; Guan, R.P.; Zhang, Q.; Cui, X.M.; Liu, D.Q. Cloning and expression analysis of a chitinase gene PnCHI1 from Panax notoginseng. China J. Chin. Mater. Med. 2016, 41, 20. [Google Scholar]
- Rajninec, M.; Jopcik, M.; Danchenko, M.; Libantova, J. Biochemical and antifungal characteristics of recombinant class I chitinase from Drosera rotundifolia. Int. J. Biol. Macromol. 2020, 161, 854–863. [Google Scholar] [CrossRef] [PubMed]
- Kragh, K.M.; Hendriks, T.; de Jong, A.J.; Lo Schiavo, F.; Bucherna, N.; Højrup, P.; Mikkelsen, J.D.; de Vries, S.C. Characterization of chitinases able to rescue somatic embryos of the temperature-sensitive carrot variant ts 11. Plant Mol. Biol. 1996, 31, 631–645. [Google Scholar] [CrossRef] [PubMed]
- Mohsenzadeh, S.; Esmaeili, M.; Moosavi, F.; Shahrtash, M.; Saffari, B.; Mohabatkar, H. Plant glutathione S-transferase classification, structure and evolution. Afr. J. Biotechnol. 2011, 10, 8160–8165. [Google Scholar]
- Wang, J.R.; Zhang, Z.; Wu, J.H.; Han, X.Y.; Wang-Pruski, G.F.; Zhang, Z. Genome-wide identification, characterization, and expression analysis related to autotoxicity of the GST gene family in Cucumis melo L. Plant Physiol. Biochem. 2020, 155, 59–69. [Google Scholar] [CrossRef]
- Medina, S.; Vicente, R.; Nieto-Taladriz, M.T.; Aparicio, N.; Chairi, F.; Vergara-Diaz, O.; Araus, J.L. The plant-transpiration response to vapor pressure deficit (VPD) in durum wheat is associated with differential yield performance and specific expression of genes involved in primary metabolism and water transport. Front. Plant Sci. 2018, 9, 1994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomolplitinant, K.M.; Saier, M.H.J. Evolution of the oligopeptide transporter family. J. Membrane Biol. 2011, 240, 89–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, Q.J.; Shen, K.Y.; Yu, X.M.; Zhao, K.; Gu, Y.F.; Zhang, X.; Chen, X.; Chen, Q. Analysis of the oligopeptide transporter gene family in Ganoderma lucidum: Structure, phylogeny, and expression patterns. Genome 2017, 60, 293–302. [Google Scholar] [CrossRef]
- Swarup, K.; Benkova, E.; Swarup, R.; Casimiro, I.; Péret, B.; Yang, Y.; Parry, G.; Nielsen, E.; De Smet, I.; Vanneste, S.; et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 2008, 10, 946–954. [Google Scholar] [CrossRef]
- Shen, C.J.; Bai, Y.H.; Wang, S.K.; Zhang, S.N.; Wu, Y.R.; Chen, M.; Jiang, D.; Qi, Y. Expression profile of PIN, AUX/LAX and PGP auxin transporter gene families in Sorghum bicolor under phytohormone and abiotic stress. FEBS J. 2010, 277, 2954–2969. [Google Scholar] [CrossRef]
- Vandenbussche, F.; Petrasek, J.; Zadnikova, P.; Hoyerová, K.; Pesek, B.; Raz, V.; Swarup, R.; Bennett, M.; Zažímalová, E.; Benková, E.; et al. The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 2010, 137, 597–606. [Google Scholar] [CrossRef] [Green Version]
- da Costa, C.T.; Offringa, R.; Fett-Neto, A.G. The role of auxin transporters and receptors in adventitious rooting of Arabidopsis thaliana pre-etiolated flooded seedlings. Plant Sci. 2020, 290, 110294. [Google Scholar] [CrossRef]
- Qu, J.; Kang, S.G.; Hah, C.; Jang, J.C. Molecular and cellular characterization of GA-stimulated transcripts GASA4 and GASA6 in Arabidopsis thaliana. Plant Sci. 2016, 246, 1–10. [Google Scholar] [CrossRef]
- Zhong, C.; Patra, B.; Tang, Y.; Li, X.; Yuan, L.; Wang, X. A transcriptional hub integrating gibberellin-brassinosteroid signals to promote seed germination in Arabidopsis. J. Exp. Bot. 2021, 72, 4708–4720. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.; Xu, Q.; Guo, F.; Lv, Y.; Song, C.; Feng, M.; Yu, J.; Zhang, D.; Cang, J. Identification and characterization of long non-coding RNAs as competing endogenous RNAs in the cold stress response of Triticum aestivum. Plant Biol. 2020, 22, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Lv, X.B.; Pan, G.K.; Lu, Y.J.; Chen, W.X.; He, W.; Lei, X.; Zhang, H.; Liu, M. Long noncoding RNA MPRL promotes mitochondrial fission and cisplatin chemosensitivity via disruption of pre-miRNA processing. Clin. Cancer Res. 2019, 25, 3673–3688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoyerová, K.; Perry, L.; Hand, P.; Lanková, M.; Kocábek, T.; May, S.; Kottová, J.; Paces, J.; Napier, R.; Zazimalova, E. Functional characterization of PaLAX1, a putative auxin permease, in heterologous plant systems. Plant Physiol. 2008, 146, 1128–1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litvay, J.D.; Verma, D.C.; Johnson, M.A. Influence of a loblolly pine (Pinus taeda L.) Culture medium and its components on growth and somatic embryogenesis of the wild carrot (Daucus carota L.). Plant Cell Rep. 1985, 4, 325–328. [Google Scholar] [CrossRef]
- Kong, L.; von Aderkas, P. Genotype effects on ABA consumption and somatic embryo maturation in interior spruce (Picea glauca × engelmanni). J. Exp. Bot. 2007, 58, 1525–1531. [Google Scholar] [CrossRef]
- Chen, H.M.; Rui, J.S.; Tang, J.H. Safranin O and fast green FCF. J. Biol. 1989, 2, 33–34. [Google Scholar]
- Gao, Y.; Hao, J.L.; Wang, Z.; Song, K.J.; Ye, J.H.; Zheng, X.Q.; Liang, Y.R.; Lu, J.L. DNA methylation levels in different tissues in tea plant via an optimized HPLC method. Horticult. Environ. Biotech. 2019, 60, 967–974. [Google Scholar] [CrossRef]
- Li, J.; Ma, W.; Zeng, P.; Wang, J.; Geng, B.; Yang, J.; Cui, Q. LncTar: A tool for predicting the RNA targets of long noncoding RNAs. Brief. Bioinform. 2014, 16, 806–812. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuosku, J.; Sarjala, T.; Jokela, A.; Sutela, S.; Saaskilahti, M.; Suorsa, M.; Läärä, E.; Häggman, H. One tissue, two fates: Different roles of megagametophyte cells during Scots pine embryogenesis. J. Exp. Bot. 2009, 60, 1375–1386. [Google Scholar] [CrossRef] [Green Version]
- Vestman, D.; Larsson, E.; Uddenberg, D.; Cairney, J.; Clapham, D.; Sundberg, E.; von Arnold, S. Important processes during differentiation and early development of somatic embryos of Norway spruce as revealed by changes in global gene expression. Tree Genet. Genomes 2011, 7, 347–362. [Google Scholar] [CrossRef]
- Alakarppa, E.; Salo, H.M.; Valledor, L.; Canal, M.J.; Haggman, H.; Vuosku, J. Natural variation of DNA methylation and gene expression may determine local adaptations of Scots pine populations. J. Exp. Bot. 2018, 69, 5293–5305. [Google Scholar] [CrossRef]
- Shi, R.; Yang, C.; Sederoff, R.; Chiang, V. Validation of artificial microRNA expression by poly(A) tailing-based RT-PCR. Nat. Protoc. 2012. [Google Scholar] [CrossRef]
- Yakovlev, I.A.; Fossdal, C.G.; Johnsen, Ø. MicroRNAs, the epigenetic memory and climatic adaptation in Norway spruce. New Phytol. 2010, 187, 1154–1169. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; Cui, Y.; Zhao, R.; Chen, X.; Zhang, J.; Zhao, J.; Kong, L. Cryo-Treatment Enhances the Embryogenicity of Mature Somatic Embryos via the lncRNA–miRNA–mRNA Network in White Spruce. Int. J. Mol. Sci. 2022, 23, 1111. https://doi.org/10.3390/ijms23031111
Gao Y, Cui Y, Zhao R, Chen X, Zhang J, Zhao J, Kong L. Cryo-Treatment Enhances the Embryogenicity of Mature Somatic Embryos via the lncRNA–miRNA–mRNA Network in White Spruce. International Journal of Molecular Sciences. 2022; 23(3):1111. https://doi.org/10.3390/ijms23031111
Chicago/Turabian StyleGao, Ying, Ying Cui, Ruirui Zhao, Xiaoyi Chen, Jinfeng Zhang, Jian Zhao, and Lisheng Kong. 2022. "Cryo-Treatment Enhances the Embryogenicity of Mature Somatic Embryos via the lncRNA–miRNA–mRNA Network in White Spruce" International Journal of Molecular Sciences 23, no. 3: 1111. https://doi.org/10.3390/ijms23031111




