The Role of Androgen Receptor and microRNA Interactions in Androgen-Dependent Diseases
Abstract
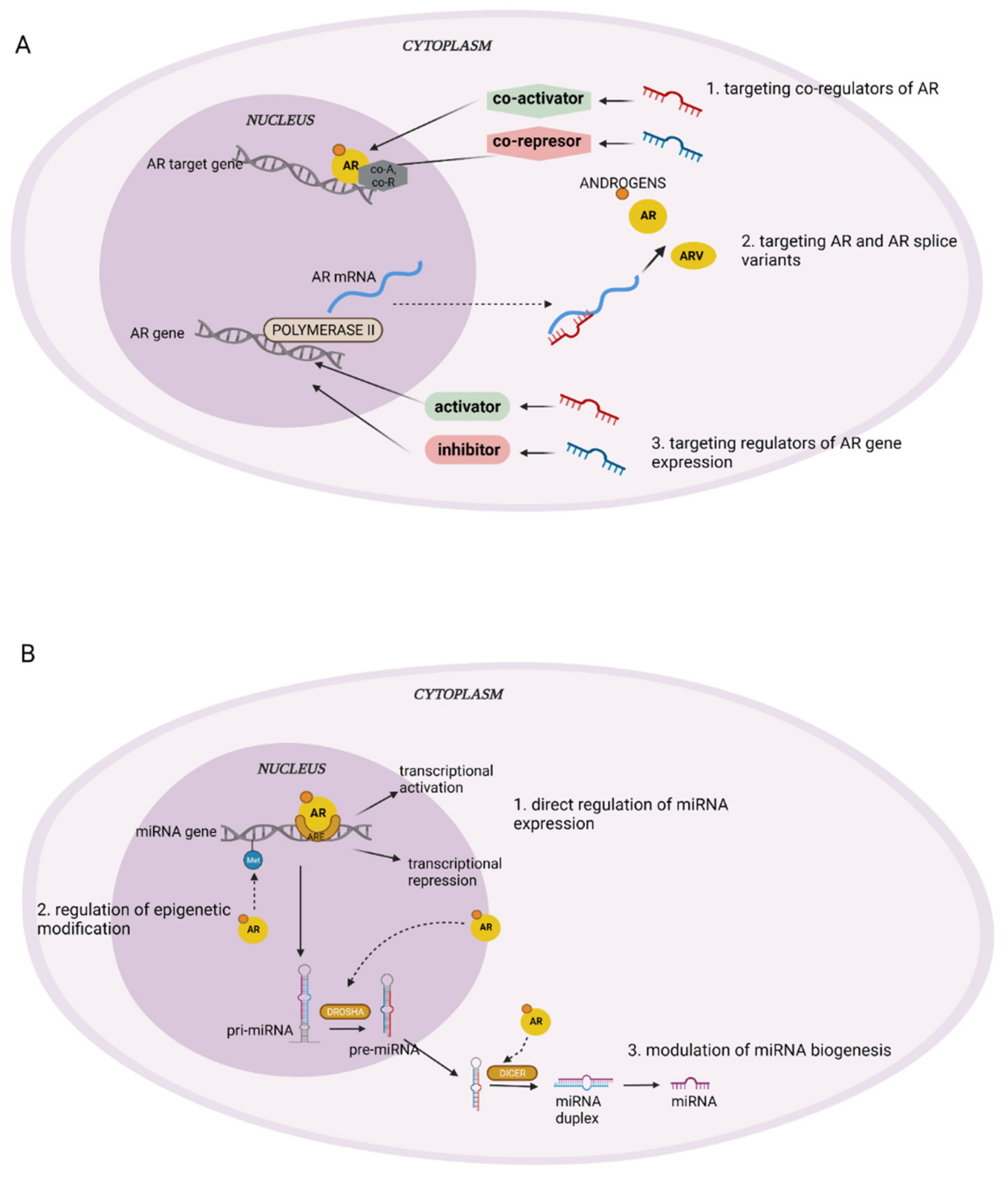
1. Introduction
2. miRNA
3. miRNA and AR
4. miRNA and AR in Various Diseases
4.1. AR and miRNA in Breast Cancer
4.2. AR and miRNA in Prostate Cancer
4.3. AR and miRNA in Other Genitourinary System Diseases
4.4. AR and miRNA in Liver Cancer
4.5. AR and miRNA in Thyroid and Head and Neck Cancer
4.6. AR and miRNA in Pancreatic Cancer
4.7. AR and miRNA in Lung Cancer
4.8. AR and miRNA in Cardiovascular Diseases
4.9. AR and miRNA in Ovarian Cancer and Polycystic Ovary Syndrome
4.10. AR and miRNA in Trophoblast and Placenta Development
4.11. AR and miRNA in Adipogenesis
5. Is There a Potential for miRNA-Based Therapy in AR-Dependent Malignancies?
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kuiper, G.G.; Faber, P.W.; van Rooij, H.C.; van der Korput, J.A.; Ris-Stalpers, C.; Klaassen, P.; Trapman, J.; Brinkmann, A.O. Structural Organization of the Human Androgen Receptor Gene. J. Mol. Endocrinol. 1989, 2, R1–R4. [Google Scholar] [CrossRef] [PubMed]
- Proverbs-Singh, T.; Feldman, J.L.; Morris, M.J.; Autio, K.A.; Traina, T.A. Targeting the Androgen Receptor in Prostate and Breast Cancer: Several New Agents in Development. Endocr. Relat. Cancer 2015, 22, R87–R106. [Google Scholar] [CrossRef] [PubMed]
- Gucalp, A.; Traina, T.A. The Androgen Receptor: Is It a Promising Target? Ann. Surg. Oncol. 2017, 24, 2876–2880. [Google Scholar] [CrossRef] [PubMed]
- Lubahn, D.B.; Joseph, D.R.; Sullivan, P.M.; Willard, H.F.; French, F.S.; Wilson, E.M. Cloning of Human Androgen Receptor Complementary DNA and Localization to the X Chromosome. Science 1988, 240, 327–330. [Google Scholar] [CrossRef]
- O’Hara, L.; Smith, L.B. Androgen Receptor Roles in Spermatogenesis and Infertility. Best Pract. Res. Clin. Endocrinol. Metab. 2015, 29, 595–605. [Google Scholar] [CrossRef]
- Anestis, A.; Zoi, I.; Papavassiliou, A.G.; Karamouzis, M.V. Androgen Receptor in Breast Cancer—Clinical and Preclinical Research Insights. Molecules 2020, 25, 358. [Google Scholar] [CrossRef]
- Davey, R.A.; Grossmann, M. Androgen Receptor Structure, Function and Biology: From Bench to Bedside. Clin. Biochem. Rev. 2016, 37, 3–15. [Google Scholar]
- Shukla, G.C.; Plaga, A.R.; Shankar, E.; Gupta, S. Androgen Receptor-Related Diseases: What Do We Know? Andrology 2016, 4, 366–381. [Google Scholar] [CrossRef]
- Huang, W. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. In Bioinformatics in MicroRNA Research; Methods in Molecular Biology; Huang, J., Borchert, G.M., Dou, D., Huan, J., Lan, W., Tan, M., Wu, B., Eds.; Springer: New York, NY, USA, 2017; Volume 1617, pp. 57–67. ISBN 978-1-4939-7044-5. [Google Scholar]
- Vishnoi, A.; Rani, S. MiRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol. Biol. Clifton N. J. 2017, 1509, 1–10. [Google Scholar] [CrossRef]
- Bhowmick, S.S.; Saha, I.; Bhattacharjee, D.; Genovese, L.M.; Geraci, F. Genome-Wide Analysis of NGS Data to Compile Cancer-Specific Panels of MiRNA Biomarkers. PLoS ONE 2018, 13, e0200353. [Google Scholar] [CrossRef]
- Felekkis, K.; Touvana, E.; Stefanou, C.; Deltas, C. MicroRNAs: A Newly Described Class of Encoded Molecules That Play a Role in Health and Disease. Hippokratia 2010, 14, 236–240. [Google Scholar]
- Ha, M.; Kim, V.N. Regulation of MicroRNA Biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from Repression to Activation: MicroRNAs Can up-Regulate Translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef]
- Doench, J.G.; Sharp, P.A. Specificity of MicroRNA Target Selection in Translational Repression. Genes Dev. 2004, 18, 504–511. [Google Scholar] [CrossRef]
- Rc, F.; Kk, F.; Cb, B.; Dp, B. Most Mammalian MRNAs Are Conserved Targets of MicroRNAs. Genome Res. 2008, 19, 92–105. [Google Scholar] [CrossRef]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of MiR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef]
- Tiwari, A.; Mukherjee, B.; Dixit, M. MicroRNA Key to Angiogenesis Regulation: MiRNA Biology and Therapy. Curr. Cancer Drug Targets 2018, 18, 266–277. [Google Scholar] [CrossRef]
- Vienberg, S.; Geiger, J.; Madsen, S.; Dalgaard, L.T. MicroRNAs in Metabolism. Acta Physiol. 2017, 219, 346–361. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent Deletions and Down-Regulation of Micro- RNA Genes MiR15 and MiR16 at 13q14 in Chronic Lymphocytic Leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating MicroRNAs as Potential Cancer Biomarkers: The Advantage and Disadvantage. Clin. Epigenetics 2018, 10, 59. [Google Scholar] [CrossRef]
- Macfarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef]
- Faruq, O.; Vecchione, A. MicroRNA: Diagnostic Perspective. Front. Med. 2015, 2. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA Biogenesis Pathways in Cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Banno, K.; Iida, M.; Yanokura, M.; Kisu, I.; Iwata, T.; Tominaga, E.; Tanaka, K.; Aoki, D. MicroRNA in Cervical Cancer: OncomiRs and Tumor Suppressor MiRs in Diagnosis and Treatment. Sci. World J. 2014, 2014, 178075. [Google Scholar] [CrossRef]
- Achkar, N.P.; Cambiagno, D.A.; Manavella, P.A. MiRNA Biogenesis: A Dynamic Pathway. Trends Plant Sci. 2016, 21, 1034–1044. [Google Scholar] [CrossRef]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and Regulation of the Let-7 MiRNAs and Their Functional Implications. Protein Cell 2016, 7, 100–113. [Google Scholar] [CrossRef]
- Culig, Z.; Santer, F.R. Androgen Receptor Signaling in Prostate Cancer. Cancer Metastasis Rev. 2014, 33, 413–427. [Google Scholar] [CrossRef]
- Waltering, K.K.; Porkka, K.P.; Jalava, S.E.; Urbanucci, A.; Kohonen, P.J.; Latonen, L.M.; Kallioniemi, O.P.; Jenster, G.; Visakorpi, T. Androgen Regulation of Micro-RNAs in Prostate Cancer. Prostate 2011, 71, 604–614. [Google Scholar] [CrossRef]
- Fletcher, C.E.; Dart, D.A.; Sita-Lumsden, A.; Cheng, H.; Rennie, P.S.; Bevan, C.L. Androgen-Regulated Processing of the Oncomir MiR-27a, Which Targets Prohibitin in Prostate Cancer. Hum. Mol. Genet. 2012, 21, 3112–3127. [Google Scholar] [CrossRef]
- Nilsson, E.M.; Laursen, K.B.; Whitchurch, J.; McWilliam, A.; Ødum, N.; Persson, J.L.; Heery, D.M.; Gudas, L.J.; Mongan, N.P. MiR137 Is an Androgen Regulated Repressor of an Extended Network of Transcriptional Coregulators. Oncotarget 2015, 6, 35710–35725. [Google Scholar] [CrossRef] [PubMed]
- Nadiminty, N.; Tummala, R.; Lou, W.; Zhu, Y.; Zhang, J.; Chen, X.; de Vere White, R.W.; Kung, H.-J.; Evans, C.P.; Gao, A.C. MicroRNA Let-7c Suppresses Androgen Receptor Expression and Activity via Regulation of Myc Expression in Prostate Cancer Cells*. J. Biol. Chem. 2012, 287, 1527–1537. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.-B.; Ma, A.-H.; Xue, L.; Li, M.; Nguyen, H.G.; Yang, J.C.; Tepper, C.G.; Gandour-Edwards, R.; Evans, C.P.; Kung, H.-J.; et al. MiR-124 and Androgen Receptor Signaling Inhibitors Repress Prostate Cancer Growth by Downregulating Androgen Receptor Splice Variants, EZH2, and Src. Cancer Res. 2015, 75, 5309–5317. [Google Scholar] [CrossRef] [PubMed]
- Sikand, K.; Slaibi, J.E.; Singh, R.; Slane, S.D.; Shukla, G.C. MiR 488* Inhibits Androgen Receptor Expression in Prostate Carcinoma Cells. Int. J. Cancer 2011, 129, 810–819. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Z.; Cheng, Q.; Wang, G.; Bai, Z. Androgen Receptor Suppresses Prostate Cancer Cell Invasion via Altering the MiR-4496/β-Catenin Signals. Biochem. Biophys. Res. Commun. 2018, 504, 82–88. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, J.; Zhang, Y.; Wan, X.; Zhang, C.; Huang, X.; Huang, W.; Pu, H.; Pei, C.; Wu, H.; et al. KDM1A Triggers Androgen-Induced MiRNA Transcription via H3K4me2 Demethylation and DNA Oxidation. The Prostate 2015, 75, 936–946. [Google Scholar] [CrossRef]
- Pasqualini, L.; Bu, H.; Puhr, M.; Narisu, N.; Rainer, J.; Schlick, B.; Schäfer, G.; Angelova, M.; Trajanoski, Z.; Börno, S.T.; et al. MiR-22 and MiR-29a Are Members of the Androgen Receptor Cistrome Modulating LAMC1 and Mcl-1 in Prostate Cancer. Mol. Endocrinol. 2015, 29, 1037–1054. [Google Scholar] [CrossRef]
- Mo, W.; Zhang, J.; Li, X.; Meng, D.; Gao, Y.; Yang, S.; Wan, X.; Zhou, C.; Guo, F.; Huang, Y.; et al. Identification of Novel AR-Targeted MicroRNAs Mediating Androgen Signalling through Critical Pathways to Regulate Cell Viability in Prostate Cancer. PLoS ONE 2013, 8, e56592. [Google Scholar] [CrossRef]
- Kalinina, T.S.; Kononchuk, V.V.; Yakovleva, A.K.; Alekseenok, E.Y.; Sidorov, S.V.; Gulyaeva, L.F. Association between Lymph Node Status and Expression Levels of Androgen Receptor, MiR-185, MiR-205, and MiR-21 in Breast Cancer Subtypes. Int. J. Breast Cancer 2020, 2020, 3259393. [Google Scholar] [CrossRef]
- Casaburi, I.; Cesario, G.M.; Donà, A.; Rizza, P.; Aquila, S.; Avena, P.; Lanzino, M.; Pellegrino, M.; Vivacqua, A.; Tucci, P.; et al. Androgens Downregulate MiR-21 Expression in Breast Cancer Cells Underlining the Protective Role of Androgen Receptor. Oncotarget 2016, 7, 12651–12661. [Google Scholar] [CrossRef]
- Ahram, M.; Mustafa, E.; Zaza, R.; Abu Hammad, S.; Alhudhud, M.; Bawadi, R.; Zihlif, M. Differential Expression and Androgen Regulation of MicroRNAs and Metalloprotease 13 in Breast Cancer Cells. Cell Biol. Int. 2017, 41, 1345–1355. [Google Scholar] [CrossRef]
- Al-Othman, N.; Hammad, H.; Ahram, M. Dihydrotestosterone Regulates Expression of CD44 via MiR-328-3p in Triple-Negative Breast Cancer Cells. Gene 2018, 675, 128–135. [Google Scholar] [CrossRef]
- Al-Momany, B.; Hammad, H.; Ahram, M. Dihydrotestosterone Induces Chemo-Resistance of Triple-Negative Breast MDA-MB-231 Cancer Cells towards Doxorubicin Independent of ABCG2 and MiR-328-3p. Curr. Mol. Pharmacol. 2021. [Google Scholar] [CrossRef]
- Bandini, E.; Fanini, F.; Vannini, I.; Rossi, T.; Plousiou, M.; Tumedei, M.M.; Limarzi, F.; Maltoni, R.; Fabbri, F.; Hrelia, S.; et al. MiR-9-5p as a Regulator of the Androgen Receptor Pathway in Breast Cancer Cell Lines. Front. Cell Dev. Biol. 2020, 8, 579160. [Google Scholar] [CrossRef]
- Shi, Y.; Yang, F.; Sun, Z.; Zhang, W.; Gu, J.; Guan, X. Differential MicroRNA Expression Is Associated with Androgen Receptor Expression in Breast Cancer. Mol. Med. Rep. 2017, 15, 29–36. [Google Scholar] [CrossRef][Green Version]
- Wang, S.; Yang, Y.; Cao, Y.-D.; Tang, X.-X.; Du, P. Androgen Downregulation of MiR-760 Promotes Prostate Cancer Cell Growth by Regulating IL6. Asian J. Androl. 2021, 23, 85–90. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, W.; Li, H.; Tang, X.; Xu, S.; Wu, M.; Wan, L.; Su, F.; Zhang, Y. Five Circular RNAs in Metabolism Pathways Related to Prostate Cancer. Front. Genet. 2021, 12, 636419. [Google Scholar] [CrossRef]
- Lin, Y.; Miao, Z.; Zhang, X.; Wei, X.; Hou, J.; Huang, Y.; Shen, B. Identification of Key MicroRNAs and Mechanisms in Prostate Cancer Evolution Based on Biomarker Prioritization Model and Carcinogenic Survey. Front. Genet. 2021, 11, 596826. [Google Scholar] [CrossRef]
- Martínez-González, L.J.; Sánchez-Conde, V.; González-Cabezuelo, J.M.; Antunez-Rodríguez, A.; Andrés-León, E.; Robles-Fernandez, I.; Lorente, J.A.; Vázquez-Alonso, F.; Alvarez-Cubero, M.J. Identification of MicroRNAs as Viable Aggressiveness Biomarkers for Prostate Cancer. Biomedicines 2021, 9, 646. [Google Scholar] [CrossRef]
- Rönnau, C.G.H.; Fussek, S.; Smit, F.P.; Aalders, T.W.; van Hooij, O.; Pinto, P.M.C.; Burchardt, M.; Schalken, J.A.; Verhaegh, G.W. Upregulation of MiR-3195, MiR-3687 and MiR-4417 Is Associated with Castration-Resistant Prostate Cancer. World J. Urol. 2021, 39, 3789–3797. [Google Scholar] [CrossRef]
- Naidoo, M.; Levine, F.; Gillot, T.; Orunmuyi, A.T.; Olapade-Olaopa, E.O.; Ali, T.; Krampis, K.; Pan, C.; Dorsaint, P.; Sboner, A.; et al. MicroRNA-1205 Regulation of FRYL in Prostate Cancer. Front. Cell Dev. Biol. 2021, 9, 647485. [Google Scholar] [CrossRef]
- Huang, Q.; Sun, Y.; Ma, X.; Gao, Y.; Li, X.; Niu, Y.; Zhang, X.; Chang, C. Androgen Receptor Increases Hematogenous Metastasis yet Decreases Lymphatic Metastasis of Renal Cell Carcinoma. Nat. Commun. 2017, 8, 918. [Google Scholar] [CrossRef]
- Huang, Q.; Sun, Y.; Zhai, W.; Ma, X.; Shen, D.; Du, S.; You, B.; Niu, Y.; Huang, C.-P.; Zhang, X.; et al. Androgen Receptor Modulates Metastatic Routes of VHL Wild-Type Clear Cell Renal Cell Carcinoma in an Oxygen-Dependent Manner. Oncogene 2020, 39, 6677–6691. [Google Scholar] [CrossRef]
- Wang, K.; Sun, Y.; Guo, C.; Liu, T.; Fei, X.; Chang, C. Androgen Receptor Regulates ASS1P3/MiR-34a-5p/ASS1 Signaling to Promote Renal Cell Carcinoma Cell Growth. Cell Death Dis. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Chen, Y.; Sun, Y.; Rao, Q.; Xu, H.; Li, L.; Chang, C. Androgen Receptor (AR) Suppresses MiRNA-145 to Promote Renal Cell Carcinoma (RCC) Progression Independent of VHL Status. Oncotarget 2015, 6, 31203–31215. [Google Scholar] [CrossRef]
- Wang, K.; Sun, Y.; Tao, W.; Fei, X.; Chang, C. Androgen Receptor (AR) Promotes Clear Cell Renal Cell Carcinoma (CcRCC) Migration and Invasion via Altering the CircHIAT1/MiR-195-5p/29a-3p/29c-3p/CDC42 Signals. Cancer Lett. 2017, 394, 1–12. [Google Scholar] [CrossRef]
- Zhai, W.; Sun, Y.; Guo, C.; Hu, G.; Wang, M.; Zheng, J.; Lin, W.; Huang, Q.; Li, G.; Zheng, J.; et al. LncRNA-SARCC Suppresses Renal Cell Carcinoma (RCC) Progression via Altering the Androgen Receptor(AR)/MiRNA-143-3p Signals. Cell Death Differ. 2017, 24, 1502–1517. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, J.; Xie, H.; Liu, T.; Chen, Y.; Ma, Z.; Pei, X.; Yang, W.; Li, L. Androgen Receptor Suppresses Prostate Cancer Metastasis but Promotes Bladder Cancer Metastasis via Differentially Altering MiRNA525-5p/SLPI-Mediated Vasculogenic Mimicry Formation. Cancer Lett. 2020, 473, 118–129. [Google Scholar] [CrossRef]
- Xiong, Y.; Wang, L.; Li, Y.; Chen, M.; He, W.; Qi, L. The Long Non-Coding RNA XIST Interacted with MiR-124 to Modulate Bladder Cancer Growth, Invasion and Migration by Targeting Androgen Receptor (AR). Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 43, 405–418. [Google Scholar] [CrossRef]
- Chen, C.-C.; Hsieh, T.-F.; Huang, C.-P.; Yu, A.-L.; Chang, W.-L.; Shyr, C.-R. Androgen Receptor Expands the Population of Cancer Stem Cells in Upper Urinary Tract Urothelial Cell Carcinoma Cells. Am. J. Cancer Res. 2016, 6, 238–248. [Google Scholar] [CrossRef]
- Chen, P.-J.; Yeh, S.-H.; Liu, W.-H.; Lin, C.-C.; Huang, H.-C.; Chen, C.-L.; Chen, D.-S.; Chen, P.-J. Androgen Pathway Stimulates MicroRNA-216a Transcription to Suppress the Tumor Suppressor in Lung Cancer-1 Gene in Early Hepatocarcinogenesis. Hepatol. Baltim. Md 2012, 56, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, X.; Yao, L.; Liu, G.; Liu, S.; Gong, L.; Xiao, Y. Loss of Androgen Receptor Promotes HCC Invasion and Metastasis via Activating Circ-LNPEP/MiR-532–3p/RAB9A Signal under Hypoxia. Biochem. Biophys. Res. Commun. 2021, 557, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.-L.; Huang, X.-Y.; Huang, J.; Huang, X.-Y.; Xu, Y.-H.; Zhou, J.; Tang, Z.-Y. Multiple Omics Integration Reveals Key Circular RNAs in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 1141. [Google Scholar] [CrossRef] [PubMed]
- Stanley, J.A.; Aruldhas, M.M.; Chandrasekaran, M.; Neelamohan, R.; Suthagar, E.; Annapoorna, K.; Sharmila, S.; Jayakumar, J.; Jayaraman, G.; Srinivasan, N.; et al. Androgen Receptor Expression in Human Thyroid Cancer Tissues: A Potential Mechanism Underlying the Gender Bias in the Incidence of Thyroid Cancers. J. Steroid Biochem. Mol. Biol. 2012, 130, 105–124. [Google Scholar] [CrossRef] [PubMed]
- Lubov, J.; Maschietto, M.; Ibrahim, I.; Mlynarek, A.; Hier, M.; Kowalski, L.P.; Alaoui-Jamali, M.A.; Silva, S.D. da Meta-Analysis of MicroRNAs Expression in Head and Neck Cancer: Uncovering Association with Outcome and Mechanisms. Oncotarget 2017, 8, 55511–55524. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Kim, M.; Hwang, D.; Park, M.; Kim, W.K.; Kim, S.K.; Shin, J.; Park, E.S.; Kang, C.M.; Paik, Y.-K.; et al. Characterization of Gene Expression and Activated Signaling Pathways in Solid-Pseudopapillary Neoplasm of Pancreas. Mod. Pathol. 2014, 27, 580–593. [Google Scholar] [CrossRef]
- Liang, L.; Wei, D.-M.; Li, J.-J.; Luo, D.-Z.; Chen, G.; Dang, Y.-W.; Cai, X.-Y. Prognostic MicroRNAs and Their Potential Molecular Mechanism in Pancreatic Cancer: A Study Based on The Cancer Genome Atlas and Bioinformatics Investigation. Mol. Med. Rep. 2018, 17, 939–951. [Google Scholar] [CrossRef]
- Jin, X.; Guan, Y.; Sheng, H.; Liu, Y. Crosstalk in Competing Endogenous RNA Network Reveals the Complex Molecular Mechanism Underlying Lung Cancer. Oncotarget 2017, 8, 91270–91280. [Google Scholar] [CrossRef]
- Fabro, A.T.; Machado-Rugolo, J.; Baldavira, C.M.; Prieto, T.G.; Farhat, C.; Rotea ManGone, F.R.; Batah, S.S.; Cruvinel, H.R.; Aldá, M.A.; Monteiro, J.S.; et al. Circulating Plasma MiRNA and Clinical/Hemodynamic Characteristics Provide Additional Predictive Information About Acute Pulmonary Thromboembolism, Chronic Thromboembolic Pulmonary Hypertension and Idiopathic Pulmonary Hypertension. Front. Pharmacol. 2021, 12, 648769. [Google Scholar] [CrossRef]
- Miao, R.; Wang, Y.; Wan, J.; Leng, D.; Gong, J.; Li, J.; Zhang, Y.; Pang, W.; Zhai, Z.; Yang, Y. Microarray Analysis and Detection of MicroRNAs Associated with Chronic Thromboembolic Pulmonary Hypertension. BioMed Res. Int. 2017, 2017, 8529796. [Google Scholar] [CrossRef]
- Pandey, R.; Woo, H.-H.; Varghese, F.; Zhou, M.; Chambers, S.K. Circulating MiRNA Profiling of Women at High Risk for Ovarian Cancer. Transl. Oncol. 2019, 12, 714–725. [Google Scholar] [CrossRef]
- Murri, M.; Insenser, M.; Fernández-Durán, E.; San-Millán, J.L.; Luque-Ramírez, M.; Escobar-Morreale, H.F. Non-Targeted Profiling of Circulating MicroRNAs in Women with Polycystic Ovary Syndrome (PCOS): Effects of Obesity and Sex Hormones. Metabolism 2018, 86, 49–60. [Google Scholar] [CrossRef]
- Butler, A.E.; Ramachandran, V.; Cunningham, T.K.; David, R.; Gooderham, N.J.; Benurwar, M.; Dargham, S.R.; Hayat, S.; Sathyapalan, T.; Najafi-Shoushtari, S.H.; et al. Increased MicroRNA Levels in Women With Polycystic Ovarian Syndrome but Without Insulin Resistance: A Pilot Prospective Study. Front. Endocrinol. 2020, 11, 571357. [Google Scholar] [CrossRef]
- Shao, X.; Liu, Y.; Liu, M.; Wang, Y.; Yan, L.; Wang, H.; Ma, L.; Li, Y.-X.; Zhao, Y.; Wang, Y.-L. Testosterone Represses Estrogen Signaling by Upregulating MiR-22: A Mechanism for Imbalanced Steroid Hormone Production in Preeclampsia. Hypertens. Dallas Tex 1979 2017, 69, 721–730. [Google Scholar] [CrossRef]
- McWhorter, E.S.; West, R.C.; Russ, J.E.; Ali, A.; Winger, Q.A.; Bouma, G.J. LIN28B Regulates Androgen Receptor in Human Trophoblast Cells through Let-7c. Mol. Reprod. Dev. 2019, 86, 1086–1093. [Google Scholar] [CrossRef]
- Greither, T.; Wenzel, C.; Jansen, J.; Kraus, M.; Wabitsch, M.; Behre, H.M. MiR-130a in the Adipogenesis of Human SGBS Preadipocytes and Its Susceptibility to Androgen Regulation. Adipocyte 2020, 9, 197–205. [Google Scholar] [CrossRef]
- Kraus, M.; Greither, T.; Wenzel, C.; Bräuer-Hartmann, D.; Wabitsch, M.; Behre, H.M. Inhibition of Adipogenic Differentiation of Human SGBS Preadipocytes by Androgen-Regulated MicroRNA MiR-375. Mol. Cell. Endocrinol. 2015, 414, 177–185. [Google Scholar] [CrossRef]
- Pujar, M.K.; Vastrad, B.; Vastrad, C. Integrative Analyses of Genes Associated with Subcutaneous Insulin Resistance. Biomolecules 2019, 9, 37. [Google Scholar] [CrossRef]
- Xie, H.; Xiao, R.; He, Y.; He, L.; Xie, C.; Chen, J.; Hong, Y. MicroRNA-100 Inhibits Breast Cancer Cell Proliferation, Invasion and Migration by Targeting FOXA1. Oncol. Lett. 2021, 22, 1–10. [Google Scholar] [CrossRef]
- Yang, Y.; Jia, B.; Zhao, X.; Wang, Y.; Ye, W. MiR-93-5p May Be an Important Oncogene in Prostate Cancer by Bioinformatics Analysis. J. Cell. Biochem. 2019, 120, 10463–10483. [Google Scholar] [CrossRef]
- Ye, D.; Shen, Z.; Zhou, S. Function of MicroRNA-145 and Mechanisms Underlying Its Role in Malignant Tumor Diagnosis and Treatment. Cancer Manag. Res. 2019, 11, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, Q.; Xu, B.; Wang, P.; Fan, W.; Cai, Y.; Gu, X.; Meng, F. MiR-124 Exerts Tumor Suppressive Functions on the Cell Proliferation, Motility and Angiogenesis of Bladder Cancer by Fine-Tuning UHRF1. FEBS J. 2015, 282, 4376–4388. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Z.; Wang, L.; Sun, L.; Liu, Z.; Li, Q.; Yao, B.; Chen, T.; Wang, C.; Yang, W.; et al. MiR-532-3p Promotes Hepatocellular Carcinoma Progression by Targeting PTPRT. Biomed. Pharmacother. Biomedecine Pharmacother. 2019, 109, 991–999. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, L.; Zhang, S. MicroRNA-124-3p Inhibits Tumourigenesis by Targeting Mitogen-Activated Protein Kinase 4 in Papillary Thyroid Carcinoma. Cell Biochem. Funct. 2020, 38, 1017–1024. [Google Scholar] [CrossRef]
- Chen, Z.; Jin, Y.; Yu, D.; Wang, A.; Mahjabeen, I.; Wang, C.; Liu, X.; Zhou, X. Down-Regulation of the MicroRNA-99 Family Members in Head and Neck Squamous Cell Carcinoma. Oral Oncol. 2012, 48, 686–691. [Google Scholar] [CrossRef]
- Diaz-Riascos, Z.V.; Ginesta, M.M.; Fabregat, J.; Serrano, T.; Busquets, J.; Buscail, L.; Cordelier, P.; Capellá, G. Expression and Role of MicroRNAs from the MiR-200 Family in the Tumor Formation and Metastatic Propensity of Pancreatic Cancer. Mol. Ther. Nucleic Acids 2019, 17, 491–503. [Google Scholar] [CrossRef]
- Sun, B.; Hua, J.; Cui, H.; Liu, H.; Zhang, K.; Zhou, H. MicroRNA-1197 Downregulation Inhibits Proliferation and Migration in Human Non- Small Cell Lung Cancer Cells by Upregulating HOXC11. Biomed. Pharmacother. 2019, 117, 109041. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, S.; Xiu, Y.-L.; Sun, K.-X.; Zong, Z.-H.; Zhao, Y. RhoC Is a Major Target of MicroRNA-93-5P in Epithelial Ovarian Carcinoma Tumorigenesis and Progression. Mol. Cancer 2015, 14, 31. [Google Scholar] [CrossRef][Green Version]
- Burger, H.G. Androgen Production in Women. Fertil. Steril. 2002, 77, 3–5. [Google Scholar] [CrossRef]
- Bienenfeld, A.; Azarchi, S.; Lo Sicco, K.; Marchbein, S.; Shapiro, J.; Nagler, A.R. Androgens in Women: Androgen-Mediated Skin Disease and Patient Evaluation. J. Am. Acad. Dermatol. 2019, 80, 1497–1506. [Google Scholar] [CrossRef]
- Niemeier, L.A.; Dabbs, D.J.; Beriwal, S.; Striebel, J.M.; Bhargava, R. Androgen Receptor in Breast Cancer: Expression in Estrogen Receptor-Positive Tumors and in Estrogen Receptor-Negative Tumors with Apocrine Differentiation. Mod. Pathol. 2010, 23, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Koo, J.; Park, H.S.; Kim, J.-H.; Choi, S.-Y.; Lee, J.H.; Park, B.-W.; Lee, K.S. Expression of Androgen Receptors in Primary Breast Cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2010, 21, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Bockhorn, J.; Yee, K.; Chang, Y.-F.; Prat, A.; Huo, D.; Nwachukwu, C.; Dalton, R.; Huang, S.; Swanson, K.E.; Perou, C.M.; et al. MicroRNA-30c Targets Cytoskeleton Genes Involved in Breast Cancer Cell Invasion. Breast Cancer Res. Treat. 2013, 137, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Giovannelli, P.; Di Donato, M.; Galasso, G.; Di Zazzo, E.; Bilancio, A.; Migliaccio, A. The Androgen Receptor in Breast Cancer. Front. Endocrinol. 2018, 9, 492. [Google Scholar] [CrossRef]
- Al-Othman, N.; Ahram, M.; Alqaraleh, M. Role of Androgen and MicroRNA in Triple-Negative Breast Cancer. Breast Dis. 2020, 39, 15–27. [Google Scholar] [CrossRef]
- Hagman, Z.; Haflidadóttir, B.S.; Ceder, J.A.; Larne, O.; Bjartell, A.; Lilja, H.; Edsjö, A.; Ceder, Y. MiR-205 Negatively Regulates the Androgen Receptor and Is Associated with Adverse Outcome of Prostate Cancer Patients. Br. J. Cancer 2013, 108, 1668–1676. [Google Scholar] [CrossRef]
- Liu, C.; Chen, Z.; Hu, X.; Wang, L.; Li, C.; Xue, J.; Zhang, P.; Chen, W.; Jiang, A. MicroRNA-185 Downregulates Androgen Receptor Expression in the LNCaP Prostate Carcinoma Cell Line. Mol. Med. Rep. 2015, 11, 4625–4632. [Google Scholar] [CrossRef]
- Ribas, J.; Ni, X.; Haffner, M.; Wentzel, E.A.; Salmasi, A.H.; Chowdhury, W.H.; Kudrolli, T.A.; Yegnasubramanian, S.; Luo, J.; Rodriguez, R.; et al. MiR-21: An Androgen Receptor–Regulated MicroRNA That Promotes Hormone-Dependent and Hormone-Independent Prostate Cancer Growth. Cancer Res. 2009, 69, 7165–7169. [Google Scholar] [CrossRef]
- Yan, L.X.; Wu, Q.N.; Zhang, Y.; Li, Y.Y.; Liao, D.Z.; Hou, J.H.; Fu, J.; Zeng, M.S.; Yun, J.P.; Wu, Q.L.; et al. Knockdown of MiR-21 in Human Breast Cancer Cell Lines Inhibits Proliferation, in Vitro Migration and in Vivo Tumor Growth. Breast Cancer Res. BCR 2011, 13, R2. [Google Scholar] [CrossRef]
- Guan, C.; Zhang, L.; Wang, S.; Long, L.; Zhou, H.; Qian, S.; Ma, M.; Bai, F.; Meng, Q.H.; Lyu, J. Upregulation of MicroRNA-21 Promotes Tumorigenesis of Prostate Cancer Cells by Targeting KLF5. Cancer Biol. Ther. 2019, 20, 1149–1161. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs - MicroRNAs with a Role in Cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Nohata, N.; Hanazawa, T.; Enokida, H.; Seki, N. MicroRNA-1/133a and MicroRNA-206/133b Clusters: Dysregulation and Functional Roles in Human Cancers. Oncotarget 2012, 3, 9–21. [Google Scholar] [CrossRef]
- Li, X.; Zeng, Z.; Wang, J.; Wu, Y.; Chen, W.; Zheng, L.; Xi, T.; Wang, A.; Lu, Y. MicroRNA-9 and Breast Cancer. Biomed. Pharmacother. 2020, 122, 109687. [Google Scholar] [CrossRef]
- Castellano, L.; Giamas, G.; Jacob, J.; Coombes, R.C.; Lucchesi, W.; Thiruchelvam, P.; Barton, G.; Jiao, L.R.; Wait, R.; Waxman, J.; et al. The Estrogen Receptor-α-Induced MicroRNA Signature Regulates Itself and Its Transcriptional Response. Proc. Natl. Acad. Sci. USA 2009. [Google Scholar] [CrossRef]
- Leung, C.-M.; Chen, T.-W.; Li, S.-C.; Ho, M.-R.; Hu, L.-Y.; Liu, W.-S.; Wu, T.T.; Hsu, P.-C.; Chang, H.-T.; Tsai, K.-W. MicroRNA Expression Profiles in Human Breast Cancer Cells after Multifraction and Single-Dose Radiation Treatment. Oncol. Rep. 2014, 31, 2147–2156. [Google Scholar] [CrossRef]
- Ottman, R.; Levy, J.; Grizzle, W.E.; Chakrabarti, R. The Other Face of MiR-17-92a Cluster, Exhibiting Tumor Suppressor Effects in Prostate Cancer. Oncotarget 2016, 7, 73739–73753. [Google Scholar] [CrossRef]
- Walter, B.A.; Valera, V.A.; Pinto, P.A.; Merino, M.J. Comprehensive MicroRNA Profiling of Prostate Cancer. J. Cancer 2013, 4, 350–357. [Google Scholar] [CrossRef]
- Epis, M.R.; Giles, K.M.; Barker, A.; Kendrick, T.S.; Leedman, P.J. MiR-331-3p Regulates ERBB-2 Expression and Androgen Receptor Signaling in Prostate Cancer*. J. Biol. Chem. 2009, 284, 24696–24704. [Google Scholar] [CrossRef]
- Stope, M.B.; Bradl, J.; Peters, S.; Streitbörger, A.; Weiss, M.; Zimmermann, U.; Walther, R.; Lillig, C.H.; Burchardt, M. Shortened Isoforms of the Androgen Receptor Are Regulated by the Cytoprotective Heat-Shock Protein HSPB1 and the Tumor-Suppressive MicroRNA MiR-1 in Prostate Cancer Cells. Anticancer Res. 2013, 33, 4921–4926. [Google Scholar]
- Stope, M.B.; Stender, C.; Schubert, T.; Peters, S.; Weiss, M.; Ziegler, P.; Zimmermann, U.; Walther, R.; Burchardt, M. Heat-Shock Protein HSPB1 Attenuates MicroRNA MiR-1 Expression Thereby Restoring Oncogenic Pathways in Prostate Cancer Cells. Anticancer Res. 2014, 34, 3475–3480. [Google Scholar]
- Stope, M.B.; Peters, S.; Großebrummel, H.; Zimmermann, U.; Walther, R.; Burchardt, M. Androgen Receptor (AR) Inhibitor ErbB3-Binding Protein-1 (Ebp1) Is Not Targeted by the Newly Identified AR Controlling Signaling Axis Heat-Shock Protein HSP27 and MicroRNA MiR-1 in Prostate Cancer Cells. World J. Urol. 2015, 33, 323–327. [Google Scholar] [CrossRef]
- Matin, F.; Jeet, V.; Srinivasan, S.; Cristino, A.S.; Panchadsaram, J.; Clements, J.A.; Batra, J. On behalf of Australian Prostate Cancer BioResource MicroRNA-3162-5p-Mediated Crosstalk between Kallikrein Family Members Including Prostate-Specific Antigen in Prostate Cancer. Clin. Chem. 2019, 65, 771–780. [Google Scholar] [CrossRef]
- Larne, O.; Hagman, Z.; Lilja, H.; Bjartell, A.; Edsjö, A.; Ceder, Y. MiR-145 Suppress the Androgen Receptor in Prostate Cancer Cells and Correlates to Prostate Cancer Prognosis. Carcinogenesis 2015, 36, 858–866. [Google Scholar] [CrossRef]
- Xie, Z.-C.; Huang, J.-C.; Zhang, L.-J.; Gan, B.-L.; Wen, D.-Y.; Chen, G.; Li, S.-H.; Yan, H.-B. Exploration of the Diagnostic Value and Molecular Mechanism of MiR-1 in Prostate Cancer: A Study Based on Meta-Analyses and Bioinformatics. Mol. Med. Rep. 2018, 18, 5630–5646. [Google Scholar] [CrossRef]
- Hudson, R.S.; Yi, M.; Esposito, D.; Watkins, S.K.; Hurwitz, A.A.; Yfantis, H.G.; Lee, D.H.; Borin, J.F.; Naslund, M.J.; Alexander, R.B.; et al. MicroRNA-1 Is a Candidate Tumor Suppressor and Prognostic Marker in Human Prostate Cancer. Nucleic Acids Res. 2012, 40, 3689–3703. [Google Scholar] [CrossRef]
- Xu, Y.; Qin, S.; An, T.; Tang, Y.; Huang, Y.; Zheng, L. MiR-145 Detection in Urinary Extracellular Vesicles Increase Diagnostic Efficiency of Prostate Cancer Based on Hydrostatic Filtration Dialysis Method. Prostate 2017, 77, 1167–1175. [Google Scholar] [CrossRef]
- Chandrasekar, T.; Yang, J.C.; Gao, A.C.; Evans, C.P. Mechanisms of Resistance in Castration-Resistant Prostate Cancer (CRPC). Transl. Androl. Urol. 2015, 4, 365–380. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. CircRNA Biogenesis Competes with Pre-MRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The Multilayered Complexity of CeRNA Crosstalk and Competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient MicroRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hu, B.; Wang, Y.; Li, Z.; Wu, J.; Yang, Y.; Wei, Y.; Peng, X.; Chen, H.; Chen, R.; et al. MiR-216a-5p Inhibits Malignant Progression in Small Cell Lung Cancer: Involvement of the Bcl-2 Family Proteins. Cancer Manag. Res. 2018, 10, 4735–4745. [Google Scholar] [CrossRef] [PubMed]
- Mekhail, S.M.; Yousef, P.G.; Jackinsky, S.W.; Pasic, M.; Yousef, G.M. MiRNA in Prostate Cancer: New Prospects for Old Challenges. EJIFCC 2014, 25, 79–98. [Google Scholar] [PubMed]
- Catto, J.W.F.; Alcaraz, A.; Bjartell, A.S.; De Vere White, R.; Evans, C.P.; Fussel, S.; Hamdy, F.C.; Kallioniemi, O.; Mengual, L.; Schlomm, T.; et al. MicroRNA in Prostate, Bladder, and Kidney Cancer: A Systematic Review. Eur. Urol. 2011, 59, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Fendler, A.; Stephan, C.; Yousef, G.M.; Jung, K. MicroRNAs as Regulators of Signal Transduction in Urological Tumors. Clin. Chem. 2011, 57, 954–968. [Google Scholar] [CrossRef]
- Chow, W.-H.; Dong, L.M.; Devesa, S.S. Epidemiology and Risk Factors for Kidney Cancer. Nat. Rev. Urol. 2010, 7, 245–257. [Google Scholar] [CrossRef]
- Jian, Y.; Yang, K.; Sun, X.; Zhao, J.; Huang, K.; Aldanakh, A.; Xu, Z.; Wu, H.; Xu, Q.; Zhang, L.; et al. Current Advance of Immune Evasion Mechanisms and Emerging Immunotherapies in Renal Cell Carcinoma. Front. Immunol. 2021, 12, 639636. [Google Scholar] [CrossRef]
- Yu, G.; Li, H.; Wang, J.; Gumireddy, K.; Li, A.; Yao, W.; Tang, K.; Xiao, W.; Hu, J.; Xiao, H.; et al. MiRNA-34a Suppresses Cell Proliferation and Metastasis by Targeting CD44 in Human Renal Carcinoma Cells. J. Urol. 2014, 192, 1229–1237. [Google Scholar] [CrossRef]
- Yamamura, S.; Saini, S.; Majid, S.; Hirata, H.; Ueno, K.; Chang, I.; Tanaka, Y.; Gupta, A.; Dahiya, R. MicroRNA-34a Suppresses Malignant Transformation by Targeting c-Myc Transcriptional Complexes in Human Renal Cell Carcinoma. Carcinogenesis 2012, 33, 294–300. [Google Scholar] [CrossRef]
- Evan, A.P.; Worcester, E.M.; Coe, F.L.; Williams, J.; Lingeman, J.E. Mechanisms of Human Kidney Stone Formation. Urolithiasis 2015, 43, 19–32. [Google Scholar] [CrossRef]
- Zhu, W.; Zhao, Z.; Chou, F.; Zuo, L.; Liu, T.; Yeh, S.; Bushinsky, D.; Zeng, G.; Chang, C. Loss of the Androgen Receptor Suppresses Intrarenal Calcium Oxalate Crystals Deposition via Altering Macrophage Recruitment/M2 Polarization with Change of the MiR-185-5p/CSF-1 Signals. Cell Death Dis. 2019, 10, 275. [Google Scholar] [CrossRef]
- Zhang, H.; Li, X.-X.; Yang, Y.; Zhang, Y.; Wang, H.-Y.; Zheng, X.F.S. Significance and Mechanism of Androgen Receptor Overexpression and Androgen Receptor/Mechanistic Target of Rapamycin Cross-Talk in Hepatocellular Carcinoma. Hepatology 2018, 67, 2271–2286. [Google Scholar] [CrossRef]
- Tian, Y.; Xie, X.; Lin, Y.; Tan, G.; Zhong, W. Androgen Receptor in Hepatocarcinogenesis: Recent Developments and Perspectives (Review). Oncol. Lett. 2015, 9, 1983–1988. [Google Scholar] [CrossRef][Green Version]
- Sun, J.; Lu, H.; Wang, X.; Jin, H. MicroRNAs in Hepatocellular Carcinoma: Regulation, Function, and Clinical Implications. Sci. World J. 2013, 2013, e924206. [Google Scholar] [CrossRef]
- Du, X.; Zhang, J.; Wang, J.; Lin, X.; Ding, F. Role of MiRNA in Lung Cancer-Potential Biomarkers and Therapies. Curr. Pharm. Des. 2018, 23, 5997–6010. [Google Scholar] [CrossRef]
- Bouhaddioui, W.; Provost, P.R.; Tremblay, Y. Expression Profile of Androgen-Modulated MicroRNAs in the Fetal Murine Lung. Biol. Sex Differ. 2016, 7, 20. [Google Scholar] [CrossRef]
- Beattie, C.W.; Hansen, N.W.; Thomas, P.A. Steroid Receptors in Human Lung Cancer. Cancer Res. 1985, 45, 4206–4214. [Google Scholar]
- Kaiser, U.; Hofmann, J.; Schilli, M.; Wegmann, B.; Klotz, U.; Wedel, S.; Virmani, A.K.; Wollmer, E.; Branscheid, D.; Gazdar, A.F.; et al. Steroid-Hormone Receptors in Cell Lines and Tumor Biopsies of Human Lung Cancer. Int. J. Cancer 1996, 67, 357–364. [Google Scholar] [CrossRef]
- Wu, M.; Wang, G.; Tian, W.; Deng, Y.; Xu, Y. MiRNA-Based Therapeutics for Lung Cancer. Curr. Pharm. Des. 2018, 23, 5989–5996. [Google Scholar] [CrossRef]
- Kalayinia, S.; Arjmand, F.; Maleki, M.; Malakootian, M.; Singh, C.P. MicroRNAs: Roles in Cardiovascular Development and Disease. Cardiovasc. Pathol. 2021, 50, 107296. [Google Scholar] [CrossRef]
- Huang, C.-K.; Lee, S.O.; Chang, E.; Pang, H.; Chang, C. Androgen Receptor (AR) in Cardiovascular Diseases. J. Endocrinol. 2016, 229, R1–R16. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ma, W.; Lu, S.; Yan, L.; Hu, F.; Wang, Z.; Cheng, B. Androgen Receptor Regulates Cardiac Fibrosis in Mice with Experimental Autoimmune Myocarditis by Increasing MicroRNA-125b Expression. Biochem. Biophys. Res. Commun. 2018, 506, 130–136. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, C.; Montani, D.; Savale, L.; Sitbon, O.; Parent, F.; Seferian, A.; Bulifon, S.; Fadel, E.; Mercier, O.; Mussot, S.; et al. Chronic Thromboembolic Pulmonary Hypertension. Presse Médicale 2015, 44, e409–e416. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Zhao, X.-D.; Shi, K.-H.; Ding, X.-S.; Tao, H. MiR-21–3p Triggers Cardiac Fibroblasts Pyroptosis in Diabetic Cardiac Fibrosis via Inhibiting Androgen Receptor. Exp. Cell Res. 2021, 399, 112464. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.-M.; Chen, L.; Chang, W.-C.; Su, S.-Y.; Hung, Y.-C.; Ma, W.-L. Androgen/Androgen Receptor Signaling in Ovarian Cancer: Molecular Regulation and Therapeutic Potentials. Int. J. Mol. Sci. 2021, 22, 7748. [Google Scholar] [CrossRef]
- Gibson, D.A.; Simitsidellis, I.; Collins, F.; Saunders, P.T.K. Evidence of Androgen Action in Endometrial and Ovarian Cancers. Endocr. Relat. Cancer 2014, 21, T203–T218. [Google Scholar] [CrossRef]
- Zheng, H.; Kavanagh, J.J.; Hu, W.; Liao, Q.; Fu, S. Hormonal Therapy in Ovarian Cancer. Int. J. Gynecol. Cancer Off. J. Int. Gynecol. Cancer Soc. 2007, 17, 325–338. [Google Scholar] [CrossRef]
- Tchernof, A.; Brochu, D.; Maltais-Payette, I.; Mansour, M.F.; Marchand, G.B.; Carreau, A.-M.; Kapeluto, J. Androgens and the Regulation of Adiposity and Body Fat Distribution in Humans. Compr. Physiol. 2018, 8, 1253–1290. [Google Scholar] [CrossRef]
- Jansen, J.; Greither, T.; Behre, H.M. Androgen-Regulated MicroRNAs (AndroMiRs) as Novel Players in Adipogenesis. Int. J. Mol. Sci. 2019, 20, 5767. [Google Scholar] [CrossRef]
- He, S.; Shi, J.; Mao, J.; Luo, X.; Liu, W.; Liu, R.; Yang, F. The Expression of MiR-375 in Prostate Cancer: A Study Based on GEO, TCGA Data and Bioinformatics Analysis. Pathol. Res. Pract. 2019, 215, 152375. [Google Scholar] [CrossRef]
- Jia-yuan, X.; Wei, S.; Fang-fang, L.; Zhi-jian, D.; Long-he, C.; Sen, L. MiR-375 Inhibits the Proliferation and Invasion of Nasopharyngeal Carcinoma Cells by Suppressing PDK1. BioMed Res. Int. 2020, 2020, 9704245. [Google Scholar] [CrossRef]
- Li, X. MiR-375, a MicroRNA Related to Diabetes. Gene 2014, 533, 1–4. [Google Scholar] [CrossRef]
- Smith, U.; Kahn, B.B. Adipose Tissue Regulates Insulin Sensitivity: Role of Adipogenesis, de Novo Lipogenesis and Novel Lipids. J. Intern. Med. 2016, 280, 465–475. [Google Scholar] [CrossRef]
- Baumann, V.; Winkler, J. MiRNA-Based Therapies: Strategies and Delivery Platforms for Oligonucleotide and Non-Oligonucleotide Agents. Future Med. Chem. 2014, 6, 1967–1984. [Google Scholar] [CrossRef]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef]
- Stenvang, J.; Kauppinen, S. MicroRNAs as Targets for Antisense-Based Therapeutics. Expert Opin. Biol. Ther. 2008, 8, 59–81. [Google Scholar] [CrossRef]
- Devulapally, R.; Sekar, N.M.; Sekar, T.V.; Foygel, K.; Massoud, T.F.; Willmann, J.K.; Paulmurugan, R. Polymer Nanoparticles Mediated Codelivery of AntimiR-10b and AntimiR-21 for Achieving Triple Negative Breast Cancer Therapy. ACS Nano 2015, 9, 2290–2302. [Google Scholar] [CrossRef]
- Alhasan, A.H.; Patel, P.C.; Choi, C.H.J.; Mirkin, C.A. Exosome Encased Spherical Nucleic Acid Gold Nanoparticle Conjugates As Potent MicroRNA Regulation Agents. Small Weinh. Bergstr. Ger. 2014, 10, 186–192. [Google Scholar] [CrossRef]
- Cubillos-Ruiz, J.R.; Baird, J.R.; Tesone, A.J.; Rutkowski, M.R.; Scarlett, U.K.; Camposeco-Jacobs, A.L.; Anadon-Arnillas, J.; Harwood, N.M.; Korc, M.; Fiering, S.N.; et al. Reprogramming Tumor-Associated Dendritic Cells In Vivo Using Microrna Mimetics Triggers Protective Immunity Against Ovarian Cancer. Cancer Res. 2012, 72, 1683–1693. [Google Scholar] [CrossRef]
- Seviour, E.G.; Sehgal, V.; Lu, Y.; Luo, Z.; Moss, T.; Zhang, F.; Hill, S.M.; Liu, W.; Maiti, S.N.; Cooper, L.; et al. Functional Proteomics Identifies MiRNAs to Target a P27/Myc/Phospho-Rb Signature in Breast and Ovarian Cancer. Oncogene 2016, 35, 691–701. [Google Scholar] [CrossRef]
- Montgomery, R.L.; Yu, G.; Latimer, P.A.; Stack, C.; Robinson, K.; Dalby, C.M.; Kaminski, N.; van Rooij, E. MicroRNA Mimicry Blocks Pulmonary Fibrosis. EMBO Mol. Med. 2014, 6, 1347–1356. [Google Scholar] [CrossRef]
- Backes, C.; Meese, E.; Keller, A. Specific MiRNA Disease Biomarkers in Blood, Serum and Plasma: Challenges and Prospects. Mol. Diagn. Ther. 2016, 20, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Shoorei, H.; Taheri, M. MiRNA Profile in Ovarian Cancer. Exp. Mol. Pathol. 2020, 113, 104381. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Dutta, A. MicroRNAs in Cancer. Annu. Rev. Pathol. 2009, 4, 199–227. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Ghaffari, S.H. Why Have MicroRNA Biomarkers Not Been Translated from Bench to Clinic? Future Oncol. 2019, 15, 801–803. [Google Scholar] [CrossRef]
- Kaudewitz, D.; Zampetaki, A.; Mayr, M. MicroRNA Biomarkers for Coronary Artery Disease? Curr. Atheroscler. Rep. 2015, 17, 70. [Google Scholar] [CrossRef]
- Snow, O.; Lallous, N.; Singh, K.; Lack, N.; Rennie, P.; Cherkasov, A. Androgen Receptor Plasticity and Its Implications for Prostate Cancer Therapy. Cancer Treat. Rev. 2019, 81, 101871. [Google Scholar] [CrossRef]
- Munkley, J.; Vodak, D.; Livermore, K.E.; James, K.; Wilson, B.T.; Knight, B.; Mccullagh, P.; Mcgrath, J.; Crundwell, M.; Harries, L.W.; et al. Glycosylation Is an Androgen-Regulated Process Essential for Prostate Cancer Cell Viability. EBioMedicine 2016, 8, 103–116. [Google Scholar] [CrossRef]
- Kunnumakkara, A.B.; Bordoloi, D.; Padmavathi, G.; Monisha, J.; Roy, N.K.; Prasad, S.; Aggarwal, B.B. Curcumin, the Golden Nutraceutical: Multitargeting for Multiple Chronic Diseases. Br. J. Pharmacol. 2017, 174, 1325–1348. [Google Scholar] [CrossRef]
- Lodi, A.; Saha, A.; Lu, X.; Wang, B.; Sentandreu, E.; Collins, M.; Kolonin, M.G.; DiGiovanni, J.; Tiziani, S. Combinatorial Treatment with Natural Compounds in Prostate Cancer Inhibits Prostate Tumor Growth and Leads to Key Modulations of Cancer Cell Metabolism. NPJ Precis. Oncol. 2017, 1, 18. [Google Scholar] [CrossRef]
- Javed, Z.; Khan, K.; Rasheed, A.; Sadia, H.; Shahwani, M.N.; Irshad, A.; Raza, S.; Salehi, B.; Sharifi-Rad, J.; Suleria, H.A.R.; et al. Targeting Androgen Receptor Signaling with MicroRNAs and Curcumin: A Promising Therapeutic Approach for Prostate Cancer Prevention and Intervention. Cancer Cell Int. 2021, 21, 77. [Google Scholar] [CrossRef]
- Fletcher, C.E.; Sulpice, E.; Combe, S.; Shibakawa, A.; Leach, D.A.; Hamilton, M.P.; Chrysostomou, S.L.; Sharp, A.; Welti, J.; Yuan, W.; et al. Androgen Receptor-Modulatory MicroRNAs Provide Insight into Therapy Resistance and Therapeutic Targets in Advanced Prostate Cancer. Oncogene 2019, 38, 5700–5724. [Google Scholar] [CrossRef]
- Lin, P.-C.; Chiu, Y.-L.; Banerjee, S.; Park, K.; Mosquera, J.M.; Giannopoulou, E.; Alves, P.; Tewari, A.K.; Gerstein, M.B.; Beltran, H.; et al. Epigenetic Repression of MiR-31 Disrupts Androgen Receptor Homeostasis and Contributes to Prostate Cancer Progression. Cancer Res. 2013, 73, 1232–1244. [Google Scholar] [CrossRef]
- Zheng, L.; Kang, Y.; Zhang, L.; Zou, W. MiR-133a-5p Inhibits Androgen Receptor (AR)-Induced Proliferation in Prostate Cancer Cells via Targeting FUsed in Sarcoma (FUS) and AR. Cancer Biol. Ther. 2020, 21, 34–42. [Google Scholar] [CrossRef]
- Lyu, S.; Yu, Q.; Ying, G.; Wang, S.; Wang, Y.; Zhang, J.; Niu, Y. Androgen Receptor Decreases CMYC and KRAS Expression by Upregulating Let-7a Expression in ER-, PR-, AR+ Breast Cancer. Int. J. Oncol. 2014, 44, 229–237. [Google Scholar] [CrossRef]
- Bao, S.; Jin, S.; Wang, C.; Tu, P.; Hu, K.; Lu, J. Androgen Receptor Suppresses Vasculogenic Mimicry in Hepatocellular Carcinoma via CircRNA7/MiRNA7-5p/VE-Cadherin/Notch4 Signalling. J. Cell. Mol. Med. 2020, 24, 14110–14120. [Google Scholar] [CrossRef]

| Condition | miRNA | Significant Findings and Implications Concerning AR–miRNA Interactions | Year of Publication | Reference |
|---|---|---|---|---|
| Breast cancer | miR-185, miR-205, miR-21 | Disturbance of AR, miR-205, miR-185, and miR-21 expression may be the marker for the presence of metastases depends on the tumour subtype. | 2020 | [40] |
| miR-21 | AR downregulates miR-21 expression | 2016 | [41] | |
| miR-100, miR-125 | AR regulates the extracellular release of MMP13 via the regulation of miR-100 and miR-125 | 2017 | [42] | |
| miR-328-3p | DHT regulates miR-328-3p expression via AR | 2018 | [43] | |
| DHT controls chemo-response independently of ABCG2 and miR-328-3p | 2021 | [44] | ||
| miR-9-5p | miR-9-5p acts as a tumour suppressor and downregulates AR | 2020 | [45] | |
| 153 DE miRNAs in AR- positive BC including miR-933, miR-5793, miR-4792 | miRNAs promote AR-mediated signalling BC progression | 2017 | [46] | |
| Prostate cancer | miR-760 | Downregulation of miR-760 promotes cancer cell growth by regulating IL-6 | 2021 | [47] |
| miR-216a-5p, miR-183-5p, miR-206, miR-3160-5p, miRNA-204-5p | Target prediction analysis for 5 circRNAs related to the AR signalling pathway showed circRNA—miRNA regulatory network with more than 200 interactions | 2021 | [48] | |
| miR-1-3p, miR-125b-5p, miR-145-5p, miR-182-5p, miR-198, miR-24-3p, miR-34a-5p, miR-22-3p, miR-499a-5p | miR-145-5p/NDRG2/AR and miR-145-5p/KLF5/AR axis were found to be potential mechanisms in PCa development | 2021 | [49] | |
| miR-210-3p, miR-23c, miR-592, miR-93-5 | miR-210-3p, miR-23c, miR-592, and miR-93-5 as a potential diagnostic and aggressiveness biomarkers for PCa | 2021 | [50] | |
| miR-3195, miR-3687, miR-4417 | Upregulation of miR-3195, miR-3687, and miR-4417 in PCa | 2021 | [51] | |
| miR-1205 | miR-1205 act as a tumour suppressor through the regulation of FRYL | 2021 | [52] | |
| Renal cell carcinoma | miR-185-5p | AR elevates the expression of miR-185-5p, which suppresses VEGF-C and increases HIF2α/VEGF-A expression | 2017 | [53] |
| AR affects RCC metastasis via regulation of miR-185-5p | 2020 | [54] | ||
| miR-34a-5p | AR increases proliferation of RCC cells through regulation of ASS1P3/miR-34a-5p/ASS1 signalling | 2019 | [55] | |
| miR-145 | AR negatively regulates miR-145, which enhances RCC cell invasion and proliferation | 2015 | [56] | |
| miR-195-5p, 29a-3p, 29c-3p | AR promotes RCC cell migration and invasion by regulating circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals | 2017 | [57] | |
| miR-143-3p | lncRNA-SARCC suppresses RCC progression via altering AR/miRNA-143-3p signalling | 2017 | [58] | |
| Bladder cancer | miR-525-5p | AR binds to different AREs on the miR-525-5p promoter region and increases metastasis in bladder cancer | 2020 | [59] |
| miR-124 | XIST inhibits miR-124 expression; miR-124 regulates AR expression | 2017 | [60] | |
| Urothelial carcinoma | miR-27a, miR-125b, miR-145, miR-200b, miR200c | AR promote expansion of CSC | 2016 | [61] |
| Liver cancer | miR-216a, miR-224 | miR-216a and miR-224 are upregulated in HCC tissues | 2012 | [62] |
| miR-532-3p | AR/circ-LNPEP/miR-532-3p/RAB9A signalling axis may be committed to hypoxia-induced cell invasion of HCC cells | 2021 | [63] | |
| miR-6511a-5p, miR-4667-5p | Competing endogenous RNA network analysis indicates that some miRNAs and circRNAs are connected to AR in HCC | 2021 | [64] | |
| Thyroid cancer | miR-124a | AR is a target for miR-124a; miR-124a determines the expression of AR gene in human thyroid cancer tissues | 2012 | [65] |
| Head and neck cancer | 53 DE miRNAs | A total of 16 miRNAs might be involved in the regulation of AR in head and neck cancer | 2017 | [66] |
| Pancreatic cancer | 232 DE miRNAs including the miR-200 family and miR-192/215 | AR is targeted by miR-376b | 2014 | [67] |
| 494 miRNAs | PPI network analysis of target genes for miR-376b and miR-376c showed AR as a hub gene | 2018 | [68] | |
| Lung cancer | 59 DE miRNAs | Transcriptional factor regulatory network showed miR-657 as regulator of AR expression | 2017 | [69] |
| IPAH, CTEPH, APTE | 21 DE miRNAs including let-7i-5p, miR-320a miR-320b-1, miR-320b-2, miR-1291 | AR is a target gene for let-7i-5p and miR-320a | 2021 | [70] |
| CTEPH | 46 DE miRNAs including miR-3148 | AR is a target for miR-3148 | 2017 | [71] |
| Ovarian cancer | 137 DE miRNAs including miR-93-5p, miR-19a-3p, miR-22-3p, miR-362-5p, miR-210-3p | Most of tested miRNA target genes were connected to hypoxia and androgen pathways | 2019 | [72] |
| PCO | 38 DE miRNAs included miR-30c-5p, miR-34c-5p, miR-142-3p, miR-199a-3p, miR-224-3p, miR-548d-3p, miR-597-5p, miR-598-3p, miR-1468-5p, miR-107, miR-151a-3p, miR-199a-5p, miR-1539 | AR is a target of miR-30c-5p, miR-199-5p, and miR-597; other miRNAs possibly involved in AR signalling | 2018 | [73] |
| miR-1260a, miR-18b-5p, miR-424-5p, and miR-let-7b-3p | miR-1260a corelate with androgen levels | 2020 | [74] | |
| Early-onset preeclampsia placentas | miR-22 | Production of androgen and estrogen is modulated by miR-22 | 2017 | [75] |
| Placenta development | let-7c | LIN28 regulates AR expression via let-7c | 2019 | [76] |
| Adipogenesis | miR-130a, miR-301 | miR-130a is upregulated under androgen stimulation in the adipogenesis; AR is a target gene for miR-130a | 2020 | [77] |
| miR-375 | miR-375 is upregulated during adipogenic differentiation and is downregulated after androgen treatment | 2015 | [78] | |
| Insulin resistance | miRNA profile | PPI network indicated that AR was regulated by 96 different miRNAs in subcutaneous insulin resistance | 2019 | [79] |
| Cancer Type | miRNA | miRNA Type | Reference |
|---|---|---|---|
| Breast cancer | miR-185 | suppresor | [40] |
| miR-21 | oncomiR | [41] | |
| miR-100 | suppresor | [80] | |
| miR-328-3p | suppresor | [44] | |
| miR-9-5p | suppresor | [45] | |
| Prostate cancer | miR-760 | suppresor | [47] |
| miR-204-5p | suppresor | [48] | |
| miR-34a-5p, miR-145-5p | suppresor | [49] | |
| miR-93-5 | oncomiR | [50,81] | |
| miR-1205 | suppresor | [52] | |
| Renal cell carcinoma | miR-185-5p | oncomiR | [53] |
| miR-34a-5p | suppresor/oncomiR | [55] | |
| miR-145 | suppresor | [56,82] | |
| miR-195-5p, 29a-3p, 29c-3p | suppresor | [57] | |
| miR-143-3p | suppresor | [58] | |
| Bladder cancer | miR-525-5p | suppresor | [59] |
| miR-124 | suppresor | [60,83] | |
| Urothelial carcinoma | miR-27a, miR-125b | oncomiR | [61] |
| miR-145, miR-200b, miR-200c | suppresor | [61] | |
| Liver cancer | miR-216a | oncomiR | [62] |
| miR-532-3p | oncomiR | [63,84] | |
| Thyroid cancer | miR-124a | suppresor | [65,85] |
| Head and neck cancer | miR-100 | suppresor | [66,86] |
| Pancreatic cancer | miR-200 family | suppresor | [67,87] |
| Lung cancer | miR-1197 | oncomiR | [88] |
| Ovarian cancer | miR-93-5p | suppresor | [72,89] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bielska, A.; Skwarska, A.; Kretowski, A.; Niemira, M. The Role of Androgen Receptor and microRNA Interactions in Androgen-Dependent Diseases. Int. J. Mol. Sci. 2022, 23, 1553. https://doi.org/10.3390/ijms23031553
Bielska A, Skwarska A, Kretowski A, Niemira M. The Role of Androgen Receptor and microRNA Interactions in Androgen-Dependent Diseases. International Journal of Molecular Sciences. 2022; 23(3):1553. https://doi.org/10.3390/ijms23031553
Chicago/Turabian StyleBielska, Agnieszka, Anna Skwarska, Adam Kretowski, and Magdalena Niemira. 2022. "The Role of Androgen Receptor and microRNA Interactions in Androgen-Dependent Diseases" International Journal of Molecular Sciences 23, no. 3: 1553. https://doi.org/10.3390/ijms23031553
APA StyleBielska, A., Skwarska, A., Kretowski, A., & Niemira, M. (2022). The Role of Androgen Receptor and microRNA Interactions in Androgen-Dependent Diseases. International Journal of Molecular Sciences, 23(3), 1553. https://doi.org/10.3390/ijms23031553







