Transcriptional Regulation of Yin-Yang 1 Expression through the Hypoxia Inducible Factor-1 in Pediatric Acute Lymphoblastic Leukemia
Abstract
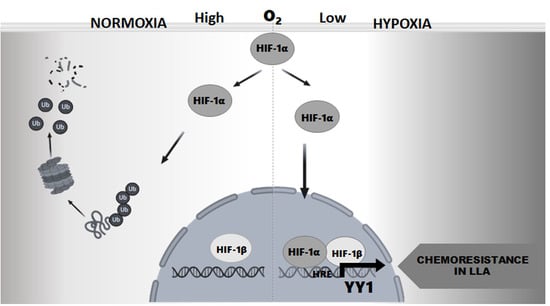
:1. Introduction
2. Results
2.1. Transcriptional Regulation of the YY1 Protein by HIF-1α in Leukemia Cell Lines
2.2. Induction or Inhibition of HIF-1α Expression Affects in the YY1 Expression
2.3. Correlation of HIF-1α and YY1 Expression in Pediatric Patients with ALL
2.4. Network Analysis of HIF-1α/YY1 and Correlation between HIF-1α and YY1 Expression in ALL
3. Discussion
4. Materials and Methods
4.1. Ethical and Biosecurity Aspects
4.2. Patients
4.3. Cell Culture
4.4. Determination of the Putative Binding Sites of HIF1α in the YY1 Promoter
4.5. Cloning of the Promoter Region of YY1
4.6. Site-Direct Mutagenesis in Putative Binding Sites for HIF-1α of the Promoter YY1
4.7. Transfection of Cell Lines with the Construct Generated
4.8. Chromatin Immunoprecipitation
4.9. Treatments and Exposure of ALL Cells to Hypoxic Conditions
4.10. Immunocytochemistry
4.11. Real-Time RT-PCR Assays
4.12. Cell Viability Assays
4.13. Network Analysis of HIF-1α/YY1 and the Construction of Gene Networks Related to Function
4.14. Bioinformatics Analysis of and Correlation between HIF-1α and YY1 Gene Expression in ALL
4.15. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fajardo-Gutierrez, A.; Juarez-Ocana, S.; Gonzalez-Miranda, G.; Palma-Padilla, V.; Carreon-Cruz, R.; Ortega-Alvarez, M.C.; Mejia-Arangure, J.M. Incidence of cancer in children residing in ten jurisdictions of the Mexican Republic: Importance of the Cancer registry (a population-based study). BMC Cancer 2007, 7, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fajardo-Gutierrez, A.; Rendon-Macias, M.E.; Mejia-Arangure, J.M. Cancer epidemiology in Mexican children. Overall results. Rev. Med. Del Inst. Mex. Seguro Soc. 2011, 49 (Suppl. 1), S43–S70. [Google Scholar]
- Mejia-Arangure, J.M.; Fajardo-Gutierrez, A.; Bernaldez-Rios, R.; Paredes-Aguilera, R.; Flores-Aguilar, H.; Martinez-Garcia, M.C. Incidence of acute leukemia in children in Mexico City, from 1982 to 1991. Salud Publica Mex 2000, 42, 431–437. [Google Scholar] [PubMed]
- Perez-Saldivar, M.L.; Ortega-Alvarez, M.C.; Fajardo-Gutierrez, A.; Bernaldez-Rios, R.; Del Campo-Martinez Mde, L.; Medina-Sanson, A.; Palomo-Colli, M.A.; Paredes-Aguilera, R.; Martinez-Avalos, A.; Borja-Aburto, V.H.; et al. Father’s occupational exposure to carcinogenic agents and childhood acute leukemia: A new method to assess exposure (a case-control study). BMC Cancer 2008, 8, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pui, C.H. Update on Pediatric ALL. Clin. Adv. Hematol. Oncol. 2006, 4, 884–886. [Google Scholar] [PubMed]
- Pui, C.H. Central nervous system disease in acute lymphoblastic leukemia: Prophylaxis and treatment. Hematol. Am. Soc. Hematol. Educ. Progr. 2006, 142–146. [Google Scholar] [CrossRef] [Green Version]
- Kourti, M.; Vavatsi, N.; Gombakis, N.; Sidi, V.; Tzimagiorgis, G.; Papageorgiou, T.; Koliouskas, D.; Athanassiadou, F. Expression of multidrug resistance 1 (MDR1), multidrug resistance-related protein 1 (MRP1), lung resistance protein (LRP), and breast cancer resistance protein (BCRP) genes and clinical outcome in childhood acute lymphoblastic leukemia. Int. J. Hematol. 2007, 86, 166–173. [Google Scholar] [CrossRef]
- Kourti, M.; Vavatsi, N.; Gombakis, N.; Tzimagiorgis, G.; Sidi, V.; Koliouskas, D.; Athanassiadou, F. Increased expression of multidrug resistance gene (MDR1) at relapse in a child with acute lymphoblastic leukemia. Pediatr. Hematol. Oncol. 2006, 23, 489–494. [Google Scholar] [CrossRef]
- Li, Y.P.; Tian, F.G.; Shi, P.C.; Guo, L.Y.; Wu, H.M.; Chen, R.Q.; Xue, J.M. 4-Hydroxynonenal promotes growth and angiogenesis of breast cancer cells through HIF-1alpha stabilization. Asian Pac. J. Cancer Prev. 2014, 15, 10151–10156. [Google Scholar] [CrossRef] [Green Version]
- Lu, G.W.; Shao, G. Hypoxic preconditioning: Effect, mechanism and clinical implication (Part 1). Zhongguo Ying Yong Sheng Li Xue Za Zhi 2014, 30, 489–501. [Google Scholar]
- Niu, F.; Li, Y.; Lai, F.F.; Chen, X.G. Research progress of hypoxia-inducible factor 1 inhibitors against tumors. Yao Xue Xue Bao 2014, 49, 832–836. [Google Scholar]
- Ye, Y.; Wang, M.; Li, J.; Shi, Y.; Zhang, X.; Zhou, Y.; Zhao, C.; Wen, J. Hypoxia-inducible factor-1alpha G polymorphism and the risk of cancer: A meta-analysis. Tumori 2014, 100, e257-65. [Google Scholar] [CrossRef]
- Zhu, J.; Cheng, X.; Xie, R.; Chen, Z.; Li, Y.; Lin, G.; Liu, J.; Yang, Y. Genetic association between the HIF-1alpha P582S polymorphism and cervical cancer risk: A meta analysis. Int. J. Clin. Exp. Pathol. 2014, 7, 6085–6090. [Google Scholar] [PubMed]
- Moroz, E.; Carlin, S.; Dyomina, K.; Burke, S.; Thaler, H.T.; Blasberg, R.; Serganova, I. Real-time imaging of HIF-1alpha stabilization and degradation. PLoS ONE 2009, 4, e5077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brader, S.; Eccles, S.A. Phosphoinositide 3-kinase signalling pathways in tumor progression, invasion and angiogenesis. Tumori 2004, 90, 2–8. [Google Scholar] [CrossRef]
- Maxwell, P.H.; Pugh, C.W.; Ratcliffe, P.J. Activation of the HIF pathway in cancer. Curr. Opin. Genet. Dev. 2001, 11, 293–299. [Google Scholar] [CrossRef]
- Wellmann, S.; Guschmann, M.; Griethe, W.; Eckert, C.; von Stackelberg, A.; Lottaz, C.; Moderegger, E.; Einsiedel, H.G.; Eckardt, K.U.; Henze, G.; et al. Activation of the HIF pathway in childhood ALL, prognostic implications of VEGF. Leukemia 2004, 18, 926–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonneveld, P. Multidrug resistance in haematological malignancies. J. Intern. Med. 2000, 247, 521–534. [Google Scholar] [CrossRef]
- van den Heuvel-Eibrink, M.M.; Sonneveld, P.; Pieters, R. The prognostic significance of membrane transport-associated multidrug resistance (MDR) proteins in leukemia. Int. J. Clin. Pharmacol. Ther. 2000, 38, 94–110. [Google Scholar] [CrossRef]
- Fardel, O.; Lecureur, V.; Guillouzo, A. The P-glycoprotein multidrug transporter. Gen. Pharmacol. 1996, 27, 1283–1291. [Google Scholar] [CrossRef]
- Sugawara, I. Expression and functions of P-glycoprotein (mdr1 gene product) in normal and malignant tissues. Acta Pathol. Jpn. 1990, 40, 545–553. [Google Scholar]
- Comerford, K.M.; Wallace, T.J.; Karhausen, J.; Louis, N.A.; Montalto, M.C.; Colgan, S.P. Hypoxia-inducible factor-1-dependent regulation of the multidrug resistance (MDR1) gene. Cancer Res. 2002, 62, 3387–3394. [Google Scholar]
- Gordon, S.; Akopyan, G.; Garban, H.; Bonavida, B. Transcription factor YY1: Structure, function, and therapeutic implications in cancer biology. Oncogene 2006, 25, 1125–1142. [Google Scholar] [CrossRef] [Green Version]
- Nicholson, S.; Whitehouse, H.; Naidoo, K.; Byers, R.J. Yin Yang 1 in human cancer. Crit. Rev. Oncog. 2011, 16, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Stovall, D.B.; Inoue, K.; Sui, G. The oncogenic role of Yin Yang 1. Crit. Rev. Oncog. 2011, 16, 163–197. [Google Scholar] [CrossRef]
- Deng, Z.; Cao, P.; Wan, M.M.; Sui, G. Yin Yang 1: A multifaceted protein beyond a transcription factor. Transcription 2010, 1, 81–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castellano, G.; Torrisi, E.; Ligresti, G.; Nicoletti, F.; Malaponte, G.; Traval, S.; McCubrey, J.A.; Canevari, S.; Libra, M. Yin Yang 1 overexpression in diffuse large B-cell lymphoma is associated with B-cell transformation and tumor progression. Cell Cycle 2010, 9, 557–563. [Google Scholar] [CrossRef] [Green Version]
- Antonio-Andrés, G.; Rangel-Santiago, J.; Tirado-Rodríguez, B.; Martinez-Ruiz, G.U.; Klunder-Klunder, M.; Vega, M.I.; Lopez-Martinez, B.; Jiménez-Hernández, E.; Torres Nava, J.; Medina-Sanson, A.; et al. Role of Yin Yang-1 (YY1) in the transcription regulation of the multi-drug resistance (MDR1) gene. Leuk. Lymphoma 2018, 59, 2628–2638. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Torres, E.; López-Pérez, T.V.; Morales-Martínez, M.; Huerta-Yepez, S. Yin-yang-1 decreases fas-induced apoptosis in acute lymphoblastic leukemia under hypoxic conditions: Its implications in immune evasion. Bol. Med. Hosp. Infant. Mex. 2020, 77, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Bonavida, B. Therapeutic YY1 Inhibitors in Cancer: ALL in ONE. Crit. Rev. Oncog. 2017, 22, 37–47. [Google Scholar] [CrossRef]
- Bonavida, B.; Huerta-Yepez, S.; Baritaki, S.; Vega, M.; Liu, H.; Chen, H.; Berenson, J. Overexpression of Yin Yang 1 in the pathogenesis of human hematopoietic malignancies. Crit. Rev. Oncog. 2011, 16, 261–267. [Google Scholar] [CrossRef]
- STRING. Available online: https://string-db.org (accessed on 12 December 2021).
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Andersson, A.; Ritz, C.; Lindgren, D.; Eden, P.; Lassen, C.; Heldrup, J.; Olofsson, T.; Rade, J.; Fontes, M.; Porwit-Macdonald, A.; et al. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: Prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia 2007, 21, 1198–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Semenza, G.L. Oxygen homeostasis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 336–361. [Google Scholar] [CrossRef]
- Schito, L.; Semenza, G.L. Hypoxia-Inducible Factors: Master Regulators of Cancer Progression. Trends Cancer 2016, 2, 758–770. [Google Scholar] [CrossRef] [Green Version]
- Wigerup, C.; Pahlman, S.; Bexell, D. Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer. Pharmacol. Ther. 2016, 164, 152–169. [Google Scholar] [CrossRef] [Green Version]
- Austen, M.; Cerni, C.; Luscher-Firzlaff, J.M.; Luscher, B. YY1 can inhibit c-Myc function through a mechanism requiring DNA binding of YY1 but neither its transactivation domain nor direct interaction with c-Myc. Oncogene 1998, 17, 511–520. [Google Scholar] [CrossRef] [Green Version]
- Eltzschig, H.K.; Bratton, D.L.; Colgan, S.P. Targeting hypoxia signalling for the treatment of ischaemic and inflammatory diseases. Nat. Rev. Drug Discov. 2014, 13, 852–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chua, Y.S.; Chua, Y.L.; Hagen, T. Structure activity analysis of 2-methoxyestradiol analogues reveals targeting of microtubules as the major mechanism of antiproliferative and proapoptotic activity. Mol. Cancer Ther. 2010, 9, 224–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fotsis, T.; Zhang, Y.; Pepper, M.S.; Adlercreutz, H.; Montesano, R.; Nawroth, P.P.; Schweigerer, L. The endogenous oestrogen metabolite 2-methoxyoestradiol inhibits angiogenesis and suppresses tumour growth. Nature 1994, 368, 237–239. [Google Scholar] [CrossRef]
- Zhou, N.N.; Zhu, X.F.; Zhou, J.M.; Li, M.Z.; Zhang, X.S.; Huang, P.; Jiang, W.Q. 2-Methoxyestradiol induces cell cycle arrest and apoptosis of nasopharyngeal carcinoma cells. Acta Pharmacol. Sin. 2004, 25, 1515–1520. [Google Scholar]
- Hijiya, N.; Thomson, B.; Isakoff, M.S.; Silverman, L.B.; Steinherz, P.G.; Borowitz, M.J.; Kadota, R.; Cooper, T.; Shen, V.; Dahl, G.; et al. Phase 2 trial of clofarabine in combination with etoposide and cyclophosphamide in pediatric patients with refractory or relapsed acute lymphoblastic leukemia. Blood 2011, 118, 6043–6049. [Google Scholar] [CrossRef] [Green Version]
- Zhe, N.; Chen, S.; Zhou, Z.; Liu, P.; Lin, X.; Yu, M.; Cheng, B.; Zhang, Y.; Wang, J. HIF-1alpha inhibition by 2-methoxyestradiol induces cell death via activation of the mitochondrial apoptotic pathway in acute myeloid leukemia. Cancer Biol. Ther. 2016, 17, 625–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vukovic, M.; Guitart, A.V.; Sepulveda, C.; Villacreces, A.; O’Duibhir, E.; Panagopoulou, T.I.; Ivens, A.; Menendez-Gonzalez, J.; Iglesias, J.M.; Allen, L.; et al. Hif-1α and Hif-2α synergize to suppress AML development but are dispensable for disease maintenance. J. Exp. Med. 2015, 212, 2223–2234. [Google Scholar] [CrossRef]
- Sui, G.; El Bachir, A.; Shi, Y.; Brignone, C.; Wall, N.R.; Yin, P.; Donohoe, M.; Luke, M.P.; Calvo, D.; Grossman, S.R.; et al. Yin Yang 1 Is a Negative Regulator of p53. Cell 2004, 117, 859–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.; Kasim, V.; Kano, M.R.; Tanaka, S.; Ohba, S.; Miura, Y.; Miyata, K.; Liu, X.; Matsuhashi, A.; Chung, U.-I.; et al. Transcription Factor YY1 Contributes to Tumor Growth by Stabilizing Hypoxia Factor HIF-1 in a p53-Independent Manner. Cancer Res. 2013, 73, 1787–1799. [Google Scholar] [CrossRef] [Green Version]
- Forristal, C.E.; Brown, A.L.; Helwani, F.M.; Winkler, I.G.; Nowlan, B.; Barbier, V.; Powell, R.J.; Engler, G.A.; Diakiw, S.M.; Zannettino, A.C.; et al. Hypoxia inducible factor (HIF)-2alpha accelerates disease progression in mouse models of leukemia and lymphoma but is not a poor prognosis factor in human AML. Leukemia 2015, 29, 2075–2085. [Google Scholar] [CrossRef] [PubMed]
- Frolova, O.; Samudio, I.; Benito, J.M.; Jacamo, R.; Kornblau, S.M.; Markovic, A.; Schober, W.; Lu, H.; Qiu, Y.H.; Buglio, D.; et al. Regulation of HIF-1alpha signaling and chemoresistance in acute lymphocytic leukemia under hypoxic conditions of the bone marrow microenvironment. Cancer Biol. Ther. 2012, 13, 858–870. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.; Huang, L.E. Suppression of hypoxia-inducible factor 1alpha (HIF-1alpha) transcriptional activity by the HIF prolyl hydroxylase EGLN1. J. Biol. Chem. 2005, 280, 38102–38107. [Google Scholar] [CrossRef] [Green Version]
- Meier, E.L.K. Nonparametric Estimation from Incomplete Observations. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar]
- Ranstam, J.; Cook, J.A. Kaplan-Meier curve. Br. J. Surg. 2017, 104, 442. [Google Scholar] [CrossRef] [PubMed]
- Thermo Fisher Scientific Inc. Oncomine Research Edition.
- The Gene Ontology Resource. Available online: http://geneontology.org (accessed on 12 December 2021).







| Total Number | 108 |
|---|---|
| Gender | |
| Female | 45 |
| Male | 63 |
| Age (years) | 7.8 (0.1–16) |
| Phenotype | |
| B | 32 |
| Pre-B | 40 |
| Pro-B | 13 |
| T | 11 |
| Unknown | 12 |
| Survivor | 79 |
| Deaths | 29 |
| Name | Sequence |
|---|---|
| YY1pro_Sen (BglI underlined) | CGAGATCTGCTTTTTTGAACAGAGAGCC |
| YY1Pro_Ant (BamH1 underlined) | GAGGATCCGGGTGCAAACCG |
| YY1mutAant (Mutation in bold) | GCCCGCGGCGAAGAACGTCAGCGCGCCGCCGCC |
| YY1mutAsen (Mutation in bold) | GGCGGCGGCGCGCTGACGTTCTTCGCCGCGGGC |
| YY1mutBant (Mutation in bold) | GGCGGGGCGGCTCGAGAACGCCCTGGCTGGCC |
| YY1mutBsen (Mutation in bold) | GGCCAGCCAGGGCGTTCTCGAGCCGCCCCGCC |
| YY1mutCant (Mutation in bold) | GTGGTGGTGGCCGGAAGACCCGTGGCCGCCCC |
| YY1mutCsen (Mutation in bold) | GGGGCGGCCACGGGTCTTCCGGCCACCACCAC |
| Ch(PromYY)_HIFABsen | TTTTGTGGCTGTTGCACCG |
| Ch(PromYY)_HIFABant | AATCGATCTGTCCGCTGGC |
| Ch(PromYY)_HIFCsen | AGACCATCGAGACCACAGTGGTGG |
| Ch(PromYY)_HIFant | CTGCAGAGCGATCATGGGCG |
| Ch(PromTel)_HIFposSen | AGCGCTGCGTCCTGCT |
| Ch(PromTel)_HIFposAnt | AGCACCTCGCGGTAGTGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antonio-Andres, G.; Martinez-Ruiz, G.U.; Morales-Martinez, M.; Jiménez-Hernandez, E.; Martinez-Torres, E.; Lopez-Perez, T.V.; Estrada-Abreo, L.A.; Patino-Lopez, G.; Juarez-Mendez, S.; Davila-Borja, V.M.; et al. Transcriptional Regulation of Yin-Yang 1 Expression through the Hypoxia Inducible Factor-1 in Pediatric Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2022, 23, 1728. https://doi.org/10.3390/ijms23031728
Antonio-Andres G, Martinez-Ruiz GU, Morales-Martinez M, Jiménez-Hernandez E, Martinez-Torres E, Lopez-Perez TV, Estrada-Abreo LA, Patino-Lopez G, Juarez-Mendez S, Davila-Borja VM, et al. Transcriptional Regulation of Yin-Yang 1 Expression through the Hypoxia Inducible Factor-1 in Pediatric Acute Lymphoblastic Leukemia. International Journal of Molecular Sciences. 2022; 23(3):1728. https://doi.org/10.3390/ijms23031728
Chicago/Turabian StyleAntonio-Andres, Gabriela, Gustavo U. Martinez-Ruiz, Mario Morales-Martinez, Elva Jiménez-Hernandez, Estefany Martinez-Torres, Tania V. Lopez-Perez, Laura A. Estrada-Abreo, Genaro Patino-Lopez, Sergio Juarez-Mendez, Víctor M. Davila-Borja, and et al. 2022. "Transcriptional Regulation of Yin-Yang 1 Expression through the Hypoxia Inducible Factor-1 in Pediatric Acute Lymphoblastic Leukemia" International Journal of Molecular Sciences 23, no. 3: 1728. https://doi.org/10.3390/ijms23031728