Uncovering a Hub Signaling Pathway of Antimicrobial-Antifungal-Anticancer Peptides’ Axis on Short Cationic Peptides via Network Pharmacology Study
Abstract
:1. Introduction
2. Results
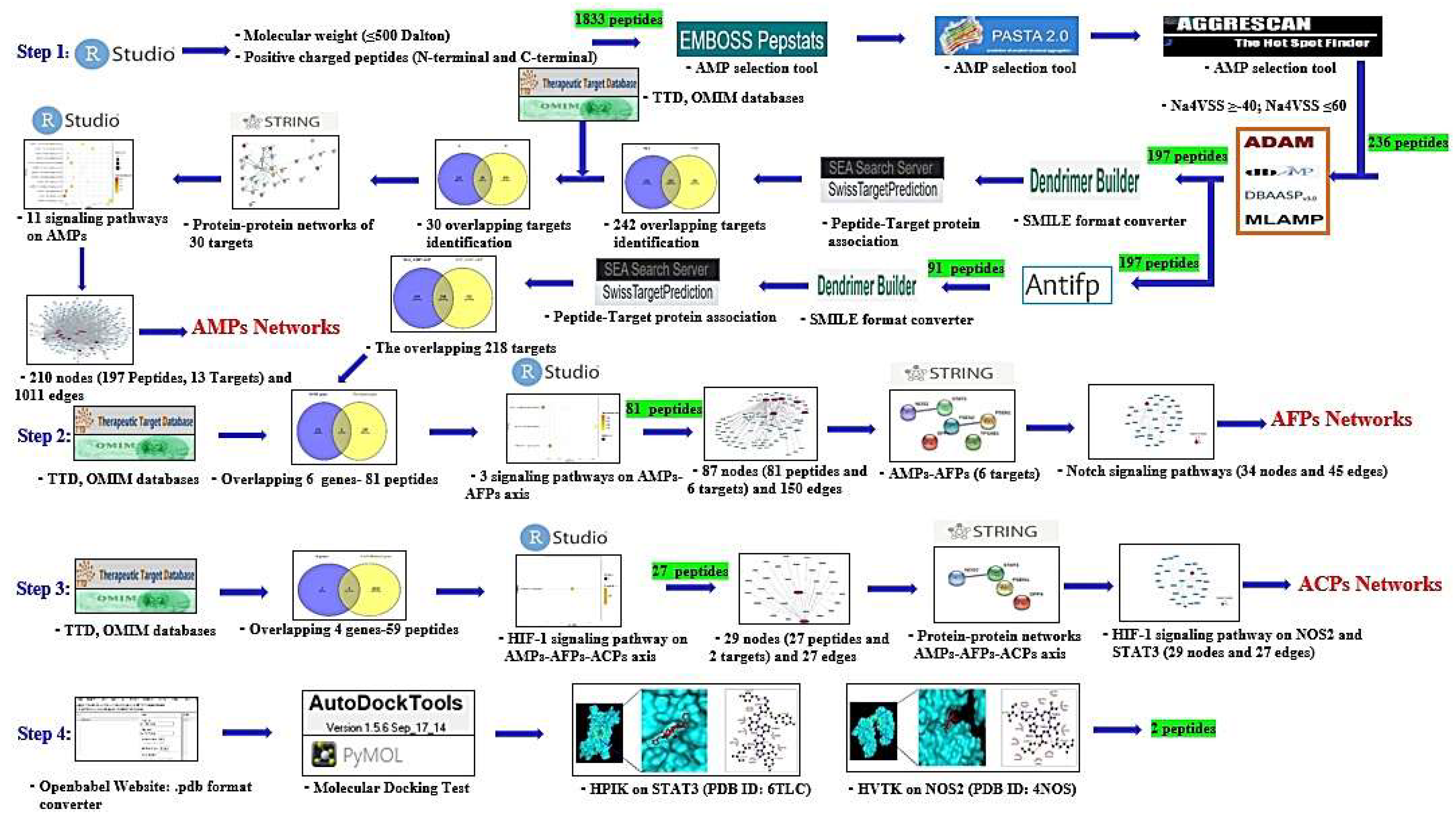
2.1. SCPs under 500 Dalton Rule
2.2. Physicochemical Refinement for AMPs
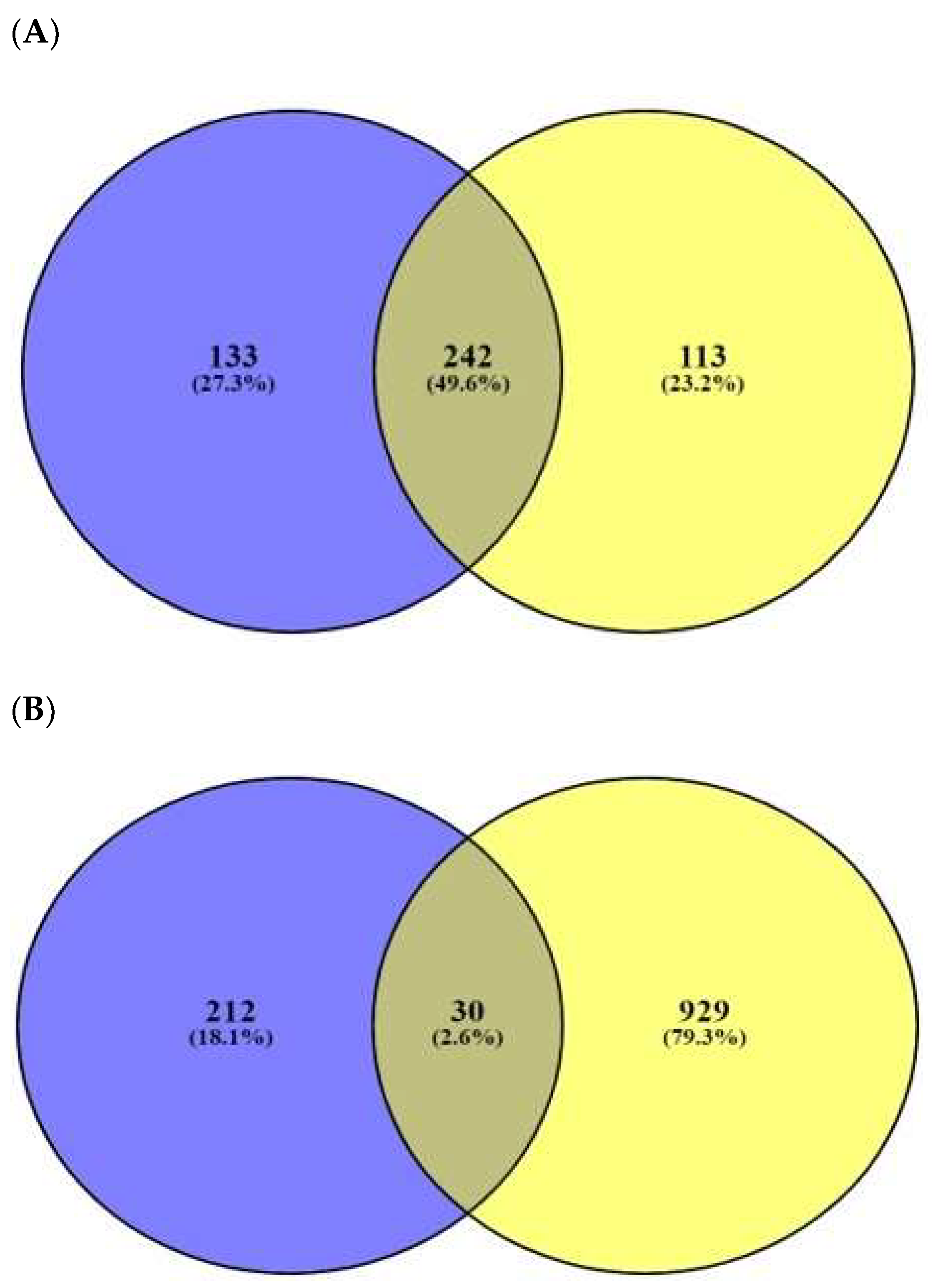
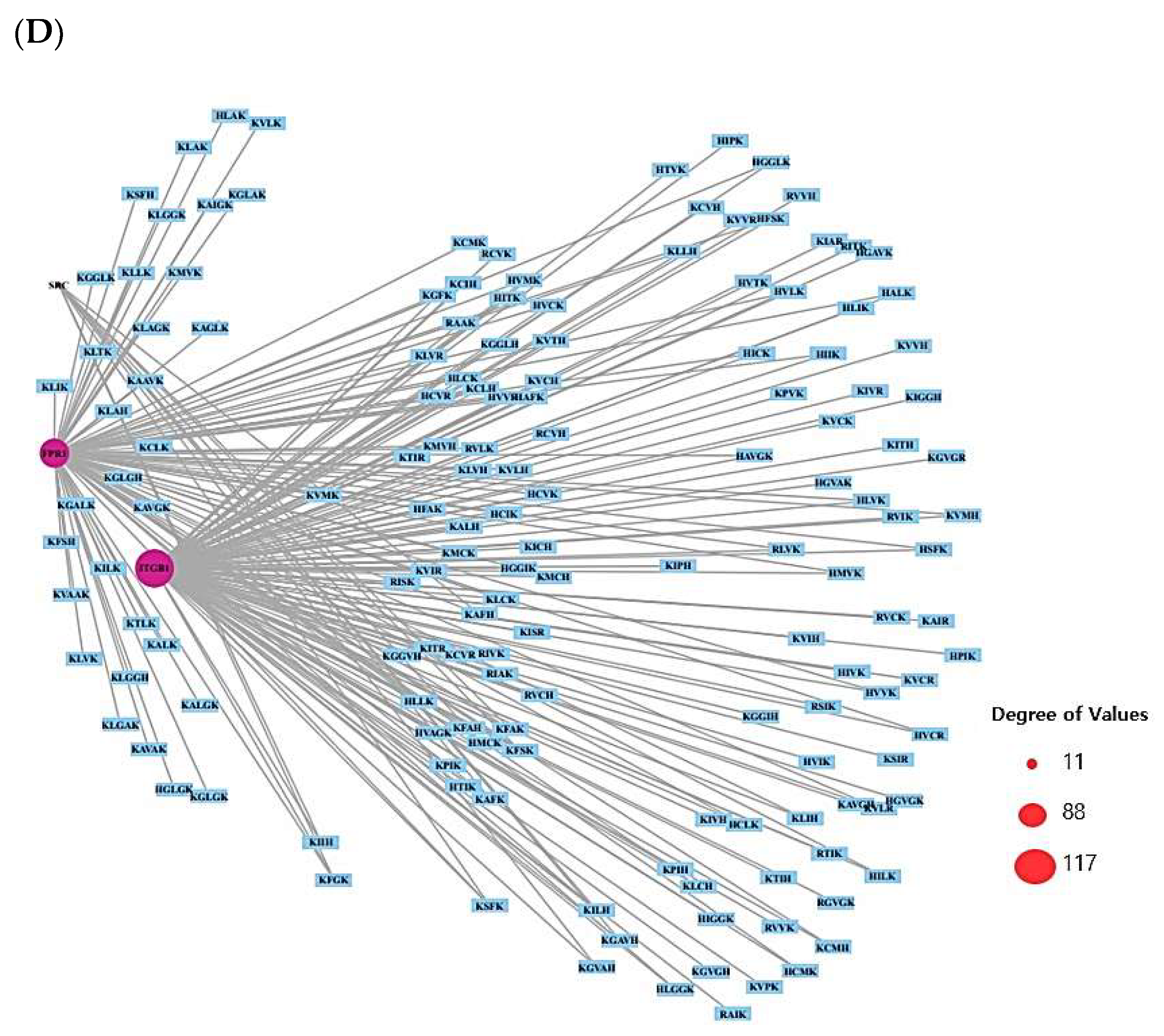
2.3. AMPs’ Targets’ Identification
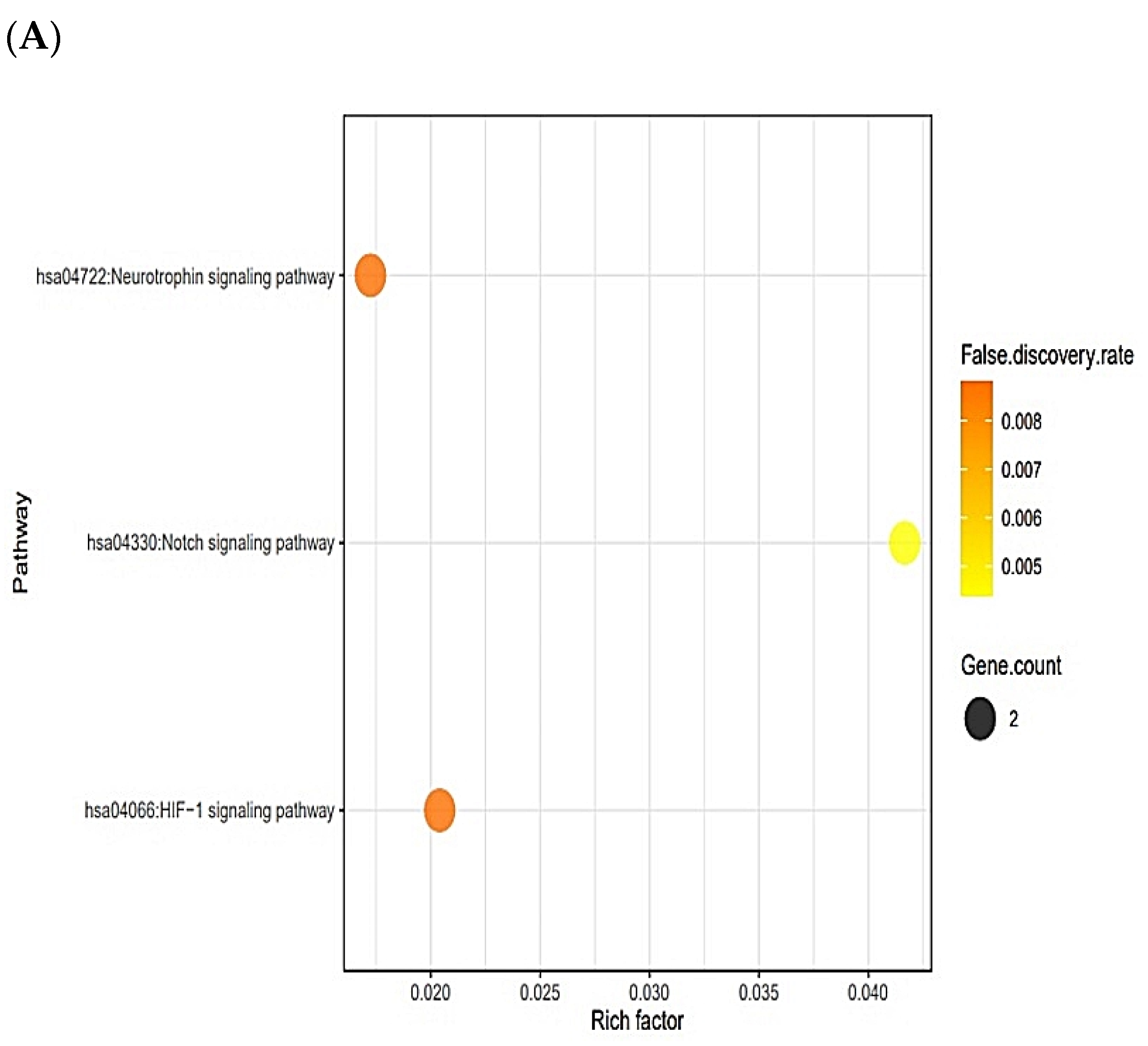
2.4. Signaling Pathways Responsive to Bacterial Infection on Human
2.5. Physicochemical Refinement for AFPs
2.6. AFPs’ Targets’ Identification
2.7. Signaling Pathways Responsive to Fungal Infection on Human
2.8. Cancer-Related Targets and ACPs’ Targets’ Identification
2.9. MDS on HIF-1 Signaling Pathway for Host Defense System
2.10. MDS of Positive Controls on HIF-1 Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. The Selection of Peptides via RStudio
4.2. AMP Evaluation and Prediction
4.3. AFP Evaluation and Prediction
4.4. The Conversion of SMILES Format
4.5. Identification of Peptide–Target Networks and Microbial-Related Targets in Database
4.6. Bubble Chart of Signaling Pathway Analysis of Overlapping Targets between Peptide–Targets and Bacterial-Responsive Targets’ Network
4.7. Identification of Peptide–Targets’ Network and Fungal-Related Targets in Database
4.8. Bubble Chart of Signaling Pathway Analysis of Overlapping Targets between Peptide–Targets and Fungal-Responsive Targets’ Network
4.9. Identification of Peptide–Targets’ Network and Cancer-Related Targets in Database
4.10. Bubble Chart of Signaling Pathway Analysis of Overlapping Targets between Peptide–Targets and Cancer-Related Targets
4.11. Preparation for Docking of Peptide Molecules
4.12. Preparation for Docking of Target Proteins and Positive Controls to Compare with Final Peptides
4.13. Peptide–Target Proteins’ Docking Test
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACPs | Anticancer peptides |
| AFPs | Antifungal peptides |
| AMPs | Antimicrobial peptides |
| AMPK | AMP-activated protein kinase |
| DEGs | Differentially Expressed Genes |
| FDA | Food and Drug Administration |
| FoxO | Forkhead box O |
| HIF-1 | Hypoxia Inducible Factor 1 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MDS | Molecular Docking Study |
| Rap1 | Repressor Activator Protein |
| SCPs | Short cationic peptides |
| SEA | Similarity Ensemble Approach |
| SMILES | Simplified Molecular Input Line Entry System |
| STP | SwissTargetPrediction |
| TCM | Traditional Chinese Medicine |
| TNF | Tumor Necrosis Factor |
| Tregs | Regulatory T cells |
References
- Uhlig, T.; Kyprianou, T.; Martinelli, F.G.; Oppici, C.A.; Heiligers, D.; Hills, D.; Calvo, X.R.; Verhaert, P. The emergence of peptides in the pharmaceutical business: From exploration to exploitation. EuPA Open Proteom. 2014, 4, 58–69. [Google Scholar] [CrossRef] [Green Version]
- Lau, J.L.; Dunn, M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef] [PubMed]
- Angell, Y.; Holford, M.; Moos, W.H. Building on Success: A Bright Future for Peptide Therapeutics. Protein Pept. Lett. 2018, 25, 1044–1050. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [Green Version]
- Bhat, Z.F.; Kumar, S.; Bhat, H.F. Bioactive peptides of animal origin: A review. J. Food Sci. Technol. 2015, 52, 5377–5392. [Google Scholar] [CrossRef] [Green Version]
- Mishra, B.; Wang, X.; Lushnikova, T.; Zhang, Y.; Golla, R.M.; Narayana, J.L.; Wang, C.; McGuire, T.R.; Wang, G. Antibacterial, antifungal, anticancer activities and structural bioinformatics analysis of six naturally occurring temporins. Peptides 2018, 106, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Haney, E.F.; Hancock, R.E.W. Peptide design for antimicrobial and immunomodulatory applications. Biopolymers 2013, 100, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Bormann, N.; Koliszak, A.; Kasper, S.; Schoen, L.; Hilpert, K.; Volkmer, R.; Kikhney, J.; Wildemann, B. A short artificial antimicrobial peptide shows potential to prevent or treat bone infections. Sci. Rep. 2017, 7, 613–620. [Google Scholar] [CrossRef] [Green Version]
- Kuppusamy, R.; Willcox, M.; Black, D.S.C.; Kumar, N. Short cationic peptidomimetic antimicrobials. Antibiotics 2019, 8, 44. [Google Scholar] [CrossRef] [Green Version]
- Wenzel, M.; Chiriac, A.I.; Otto, A.; Zweytick, D.; May, C.; Schumacher, C.; Gust, R.; Albada, H.B.; Penkova, M.; Krämer, U.; et al. Small cationic antimicrobial peptides delocalize peripheral membrane proteins. Proc. Natl. Acad. Sci. USA 2014, 111, E1409–E1418. [Google Scholar] [CrossRef] [Green Version]
- Ziegler, A.; Nervi, P.; Dürrenberger, M.; Seelig, J. The cationic cell-penetrating peptide CPPTAT derived from the HIV-1 protein TAT is rapidly transported into living fibroblasts: Optical, biophysical, and metabolic evidence. Biochemistry 2005, 44, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Lehrer, R. Cationic peptides: A new source of antibiotics. Trends Biotechnol. 1998, 16, 82–88. [Google Scholar] [CrossRef]
- Brogden, K.A.; Ackermann, M.; McCray, P.B.; Tack, B.F. Antimicrobial peptides in animals and their role in host defences. Int. J. Antimicrob. Agents 2003, 22, 465–478. [Google Scholar] [CrossRef] [Green Version]
- Oppenheim, J.J.; Yang, D. Alarmins: Chemotactic activators of immune responses. Curr. Opin. Immunol. 2005, 17, 359–365. [Google Scholar] [CrossRef]
- Bowdish, D.; Davidson, D.; Hancock, R. A Re-evaluation of the Role of Host Defence Peptides in Mammalian Immunity. Curr. Protein Pept. Sci. 2005, 6, 35–51. [Google Scholar] [CrossRef] [Green Version]
- van Eijk, M.; Boerefijn, S.; Cen, L.; Rosa, M.; Morren, M.J.H.; van der Ent, C.K.; Kraak, B.; Dijksterhuis, J.; Valdes, I.D.; Haagsman, H.P.; et al. Cathelicidin-inspired antimicrobial peptides as novel antifungal compounds. Med. Mycol. 2020, 58, 1073–1084. [Google Scholar] [CrossRef] [Green Version]
- Netea, M.G.; Joosten, L.A.B.; Van Der Meer, J.W.M.; Kullberg, B.J.; Van De Veerdonk, F.L. Immune defence against Candida fungal infections. Nat. Rev. Immunol. 2015, 15, 630–642. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef]
- Finlay, B.B.; Hancock, R.E.W. Can innate immunity be enhanced to treat microbial infections? Nat. Rev. Microbiol. 2004, 2, 497–504. [Google Scholar] [CrossRef]
- Deslouches, B.; Peter Di, Y. Antimicrobial peptides with selective antitumor mechanisms: Prospect for anticancer applications. Oncotarget 2017, 8, 46635–46651. [Google Scholar] [CrossRef] [Green Version]
- Gaspar, D.; Salomé Veiga, A.; Castanho, M.A.R.B. From antimicrobial to anticancer peptides. A review. Front. Microbiol. 2013, 4, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Digumarti, R.; Bapsy, P.P.; Suresh, A.V.; Bhattacharyya, G.S.; Dasappa, L.; Shan, J.S.; Gerber, D.E. Bavituximab plus paclitaxel and carboplatin for the treatment of advanced non-small-cell lung cancer. Lung Cancer 2014, 86, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Rodríguez, J.J.; Ochoa-Zarzosa, A.; López-Gómez, R.; López-Meza, J.E. Plant antimicrobial peptides as potential anticancer agents. BioMed Res. Int. 2015, 2015, 735087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.C.L.; Harris, J.L.; Khanna, K.K.; Hong, J.H. A comprehensive review on current advances in peptide drug development and design. Int. J. Mol. Sci. 2019, 20, 2383. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Bi, Y.; Zhang, H.; Dong, S.; Teng, L.; Lee, R.J.; Yang, Z. Cell-Penetrating Peptides in Diagnosis and Treatment of Human Diseases: From Preclinical Research to Clinical Application. Front. Pharmacol. 2020, 9, 697. [Google Scholar] [CrossRef]
- Oh, K.K.; Adnan, M.; Cho, D.H. Active ingredients and mechanisms of Phellinus linteus (grown on Rosa multiflora) for alleviation of Type 2 diabetes mellitus through network pharmacology. Gene 2020. [Google Scholar] [CrossRef]
- Oh, K.K.; Adnan, M.; Cho, D.H. Network pharmacology approach to bioactive chemical compounds identified from Lespedeza bicolor lignum methanol extract by GC–MS for amelioration of hepatitis. Gene Rep. 2020, 100851. [Google Scholar] [CrossRef]
- Oh, K.K.; Adnan, M.; Cho, D.H. Network pharmacology of bioactives from Sorghum bicolor with targets related to diabetes mellitus. PLoS ONE 2020, 15, e0240873. [Google Scholar] [CrossRef]
- Li, S.; Zhang, B. Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin. J. Nat. Med. 2013, 11, 110–120. [Google Scholar] [CrossRef]
- Pescina, S.; Ostacolo, C.; Gomez-Monterrey, I.M.; Sala, M.; Bertamino, A.; Sonvico, F.; Padula, C.; Santi, P.; Bianchera, A.; Nicoli, S. Cell penetrating peptides in ocular drug delivery: State of the art. J. Control. Release 2018, 284, 84–102. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Schultes, S.; De Graaf, C.; Haaksma, E.E.J.; De Esch, I.J.P.; Leurs, R.; Krämer, O. Ligand efficiency as a guide in fragment hit selection and optimization. Drug Discov. Today Technol. 2010, 7, e157–e162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leeson, P.D.; Springthorpe, B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat. Rev. Drug Discov. 2007, 6, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Chung, H.; Shin, Y.P.; Kim, I.W.; Natarajan, S.; Veerappan, K.; Seo, M.; Park, J.; Hwang, J.S. Transcriptome analysis of psacothea hilaris: De novo assembly and antimicrobial peptide prediction. Insects 2020, 11, 676. [Google Scholar] [CrossRef]
- Walsh, I.; Seno, F.; Tosatto, S.C.E.; Trovato, A. PASTA 2.0: An improved server for protein aggregation prediction. Nucleic Acids Res. 2014, 42, W301–W307. [Google Scholar] [CrossRef]
- Scuto, A.; Kujawski, M.; Kowolik, C.; Krymskaya, L.; Wang, L.; Weiss, L.M.; DiGiusto, D.; Yu, H.; Forman, S.; Jove, R. STAT3 inhibition is a therapeutic strategy for ABC-like diffuse large B-cell lymphoma. Cancer Res. 2011, 71, 3182–3188. [Google Scholar] [CrossRef] [Green Version]
- Palumbo, P.; Lombardi, F.; Augello, F.R.; Giusti, I.; Luzzi, S.; Dolo, V.; Cifone, M.G.; Cinque, B. NOS2 inhibitor 1400 W induces autophagic flux and influences extracellular vesicle profile in human glioblastoma U87MG cell line. Int. J. Mol. Sci. 2019, 20, 3010. [Google Scholar] [CrossRef] [Green Version]
- Brown, K.L.; Hancock, R.E.W. Cationic host defense (antimicrobial) peptides. Curr. Opin. Immunol. 2006, 18, 24–30. [Google Scholar] [CrossRef]
- Tian, R.; Zou, H.; Wang, L.; Liu, L.; Song, M.; Zhang, H. Analysis of differentially expressed genes in bacterial and fungal keratitis. Indian J. Ophthalmol. 2020, 68, 39–46. [Google Scholar] [CrossRef]
- Horton, J.S.; Yamamoto, S.Y.; Bryant-Greenwood, G.D. Relaxin modulates proinflammatory cytokine secretion from human decidual macrophages. Biol. Reprod. 2011, 85, 788–797. [Google Scholar] [CrossRef] [Green Version]
- Insuela, D.B.R.; Azevedo, C.T.; Coutinho, D.S.; Magalhães, N.S.; Ferrero, M.R.; Ferreira, T.P.T.; Cascabulho, C.M.; Henriques-Pons, A.; Olsen, P.C.; Diaz, B.L.; et al. Glucagon reduces airway hyperreactivity, inflammation, and remodeling induced by ovalbumin. Sci. Rep. 2019, 9, 6478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, H.; Liao, W.; Wang, L.; Lu, Q. A Player and Coordinator: The Versatile Roles of Eosinophils in the Immune System. Transfus. Med. Hemotherapy 2016, 43, 96–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boutet, P.; Sulon, J.; Closset, R.; Detilleux, J.; Beckers, J.F.; Bureau, F.; Lekeux, P. Prolactin-induced activation of nuclear factor κB in bovine mammary epithelial cells: Role in chronic mastitis. J. Dairy Sci. 2007, 90, 155–164. [Google Scholar] [CrossRef] [Green Version]
- Yu-Lee, L.Y. Prolactin modulation of immune and inflammatory responses. Recent Prog. Horm. Res. 2002, 57, 435–455. [Google Scholar] [CrossRef] [PubMed]
- Hannan, T.J.; Hooton, T.M.; Hultgren, S.J. Estrogen and recurrent UTI: What are the facts? Sci. Transl. Med. 2013, 5, fs23–fs190. [Google Scholar] [CrossRef] [PubMed]
- Lüthje, P.; Brauner, H.; Ramos, N.L.; Övregaard, A.; Gläser, R.; Hirschberg, A.L.; Aspenström, P.; Brauner, A. Estrogen supports urothelial defense mechanisms. Sci. Transl. Med. 2013, 5, 190ra80. [Google Scholar] [CrossRef]
- Ehlers, S. Role of tumour necrosis factor (TNF) in host defence against tuberculosis: Implications for immunotherapies targeting TNF. Ann. Rheum. Dis. 2003, 62, 37ii–42. [Google Scholar] [CrossRef] [Green Version]
- Qian, Y.; Kang, Z.; Liu, C.; Li, X. IL-17 signaling in host defense and inflammatory diseases. Cell. Mol. Immunol. 2010, 7, 328–333. [Google Scholar] [CrossRef] [Green Version]
- Silwal, P.; Kim, J.K.; Yuk, J.M.; Jo, E.K. AMP-Activated protein kinase and host defense against infection. Int. J. Mol. Sci. 2018, 19, 3495. [Google Scholar] [CrossRef] [Green Version]
- Graves, D.T.; Milovanova, T.N. Mucosal Immunity and the FOXO1 Transcription Factors. Front. Immunol. 2019, 10, 2530. [Google Scholar] [CrossRef]
- Palazon, A.; Goldrath, A.W.; Nizet, V.; Johnson, R.S. HIF Transcription Factors, Inflammation, and Immunity. Immunity 2014, 41, 518–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mor, A.; Haklai, R.; Ben-Moshe, O.; Mekori, Y.A.; Kloog, Y. Inhibition of contact sensitivity by farnesylthiosalicylic acid-amide, a potential rap1 inhibitor. J. Investig. Dermatol. 2011, 131, 2040–2048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heese, K.; Hock, C.; Otten, U. Inflammatory signals induce neurotrophin expression in human microglial cells. J. Neurochem. 1998, 70, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Mannion, R.J.; Costigan, M.; Decosterd, I.; Amaya, F.; Ma, Q.P.; Holstege, J.C.; Ji, R.R.; Acheson, A.; Lindsay, R.M.; Wilkinson, G.A.; et al. Neurotrophins: Peripherally and centrally acting modulators of tactile stimulus-induced inflammatory pain hypersensitivity. Proc. Natl. Acad. Sci. USA 1999, 96, 9385–9390. [Google Scholar] [CrossRef] [Green Version]
- Grahl, N.; Shepardson, K.M.; Chung, D.; Cramer, R.A. Hypoxia and fungal pathogenesis: To air or not to air? Eukaryotic Cell 2012, 11, 560–570. [Google Scholar] [CrossRef] [Green Version]
- Ito, T.; Connett, J.M.; Kunkel, S.L.; Matsukawa, A. Notch system in the linkage of innate and adaptive immunity. J. Leukoc. Biol. 2012, 92, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Burroughs, S.K.; Kaluz, S.; Wang, D.; Wang, K.; Van Meir, E.G.; Wang, B. Hypoxia inducible factor pathway inhibitors as anticancer therapeutics. Future Med. Chem. 2013, 5, 553–572. [Google Scholar] [CrossRef] [Green Version]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 2003, 3, 721–732. [Google Scholar] [CrossRef]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 2019, 18, 157. [Google Scholar] [CrossRef] [Green Version]
- Rice, P.; Longden, L.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Lee, H.T.; Lee, C.C.; Yang, J.R.; Lai, J.Z.C.; Chang, K.Y.; Ray, O. A large-scale structural classification of Antimicrobial peptides. BioMed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Jhong, J.H.; Chi, Y.H.; Li, W.C.; Lin, T.H.; Huang, K.Y.; Lee, T.Y. dbAMP: An integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data. Nucleic Acids Res. 2019, 47, D285–D297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 2021, 49, D288–D297. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Xu, D. Imbalanced multi-label learning for identifying antimicrobial peptides and their functional types. Bioinformatics 2016, 32, 3745–3752. [Google Scholar] [CrossRef]
- Agrawal, P.; Bhalla, S.; Chaudhary, K.; Kumar, R.; Sharma, M.; Raghava, G.P.S. In Silico Approach for Prediction of Antifungal Peptides. Front. Microbiol. 2018, 9, 323. [Google Scholar] [CrossRef] [Green Version]
- Maingi, V.; Jain, V.; Bharatam, P.V.; Maiti, P.K. Dendrimer building toolkit: Model building and characterization of various dendrimer architectures. J. Comput. Chem. 2012, 33, 1997–2011. [Google Scholar] [CrossRef]
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W3664. [Google Scholar] [CrossRef] [Green Version]
- Lin, G.; Chai, J.; Yuan, S.; Mai, C.; Cai, L.; Murphy, R.W.; Zhou, W.; Luo, J. VennPainter: A Tool for the Comparison and Identification of Candidate Genes Based on Venn Diagrams. PLoS ONE 2016, 11, e0154315. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, S.; Li, F.; Zhou, Y.; Zhang, Y.; Wang, Z.; Zhang, R.; Zhu, J.; Ren, Y.; Tan, Y.; et al. Therapeutic target database 2020: Enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020, 48, D1031–D1041. [Google Scholar] [CrossRef] [Green Version]
- Amberger, J.S.; Bocchini, C.A.; Scott, A.F.; Hamosh, A. OMIM.org: Leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019, 47, D1038–D1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open chemical toolbox. J. Cheminform. 2011, 3, e00950-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software news and updates AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khanal, P.; Patil, B.M.; Chand, J.; Naaz, Y. Anthraquinone Derivatives as an Immune Booster and their Therapeutic Option Against COVID-19. Nat. Prod. Bioprospect. 2020, 10, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]

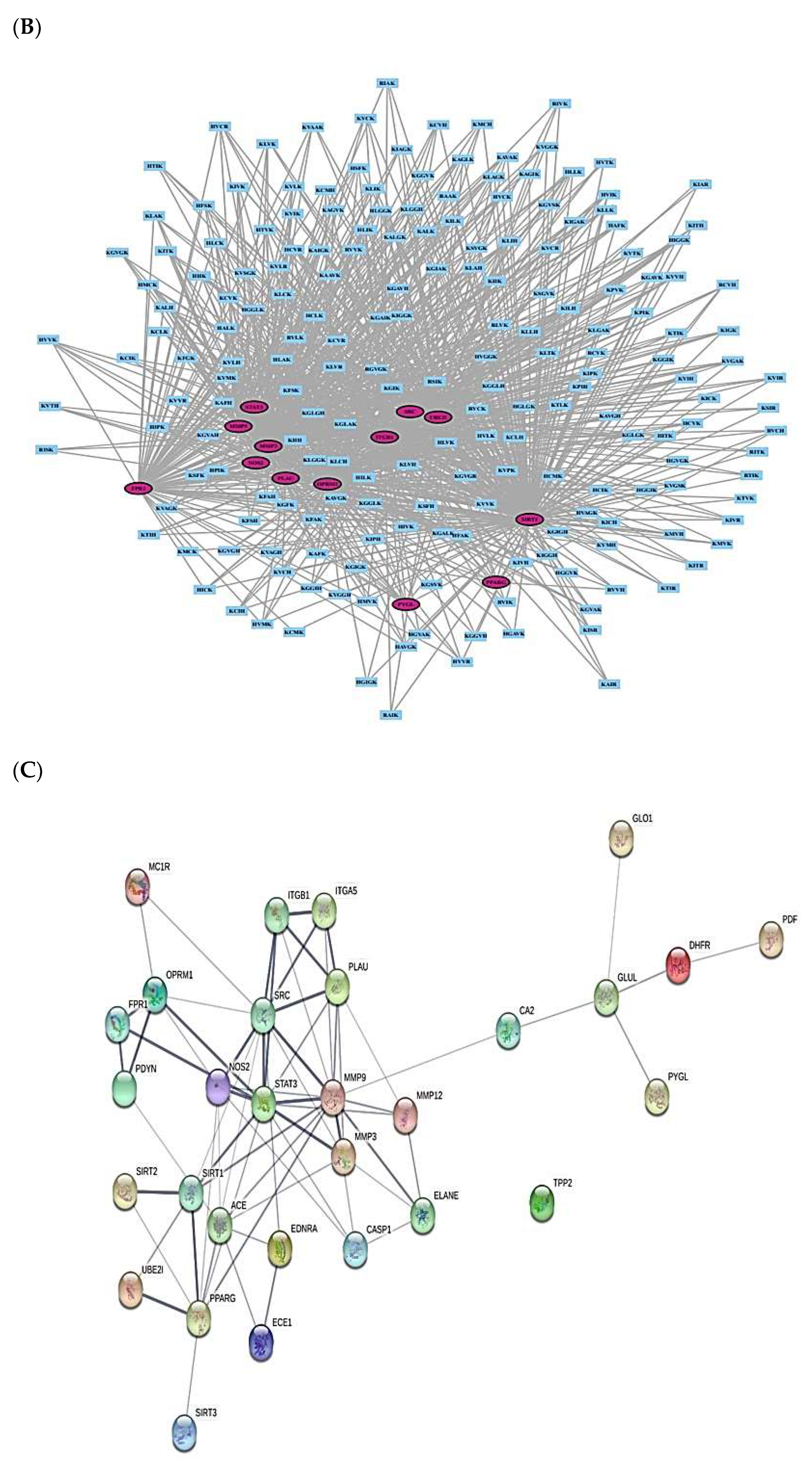
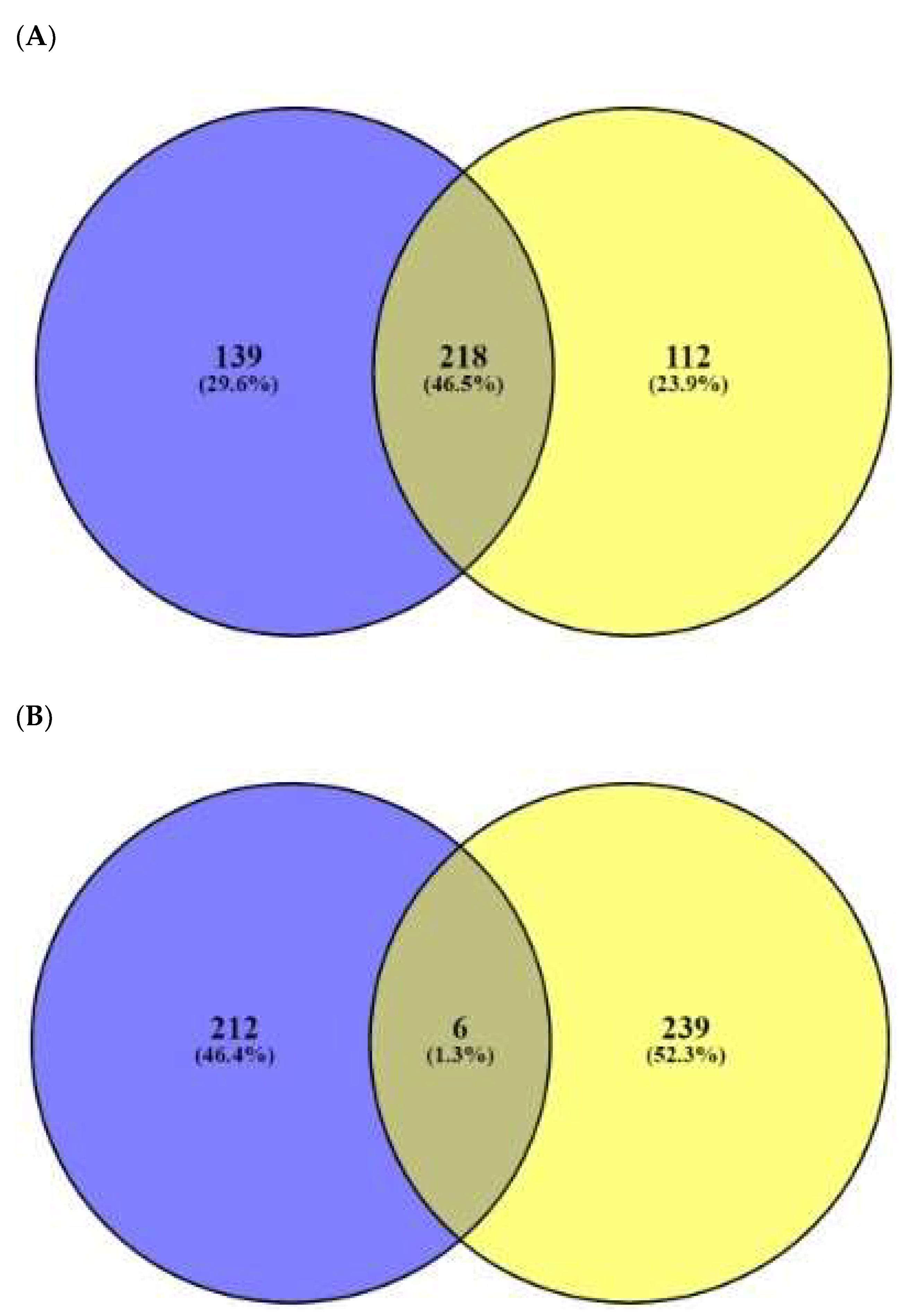
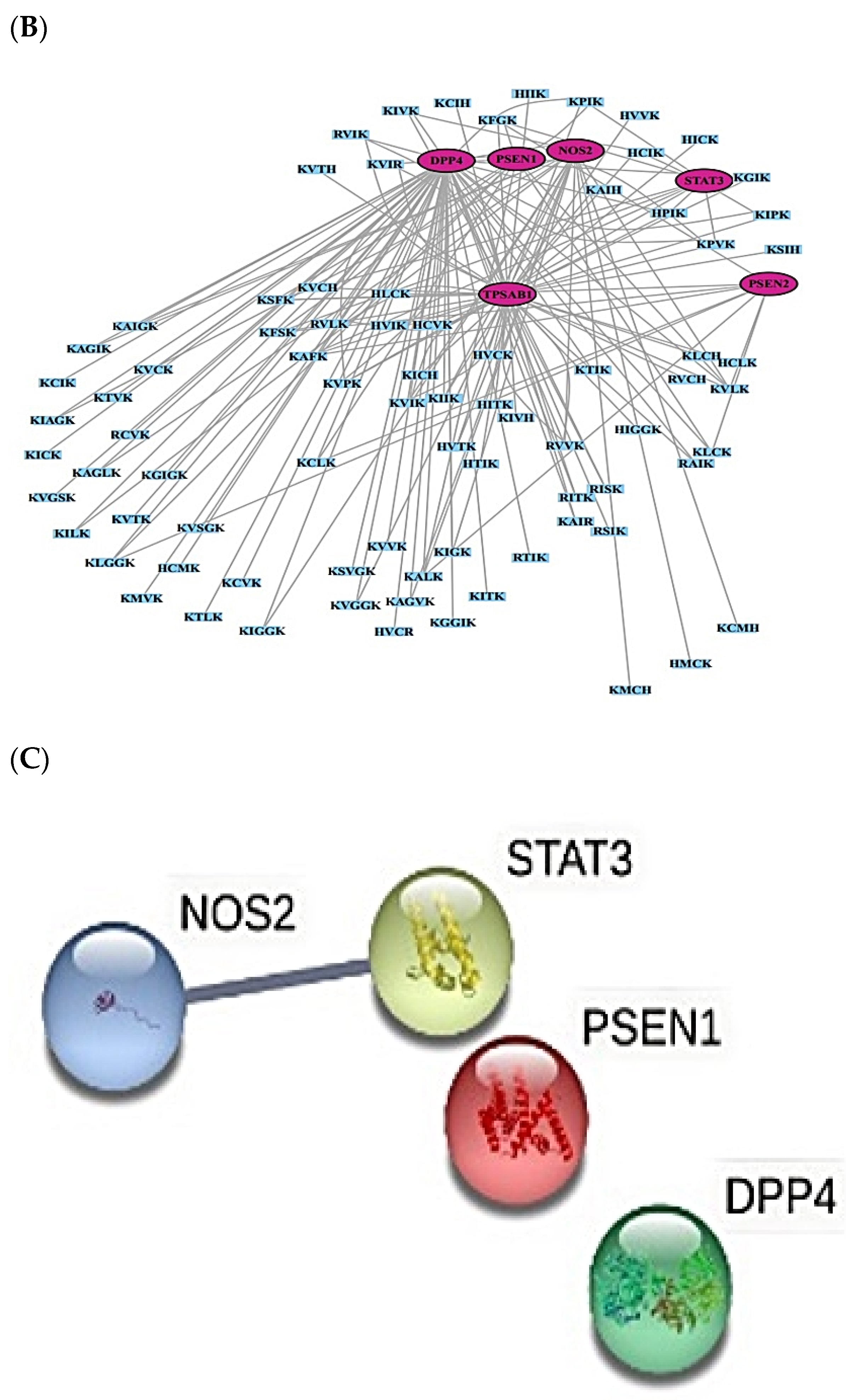


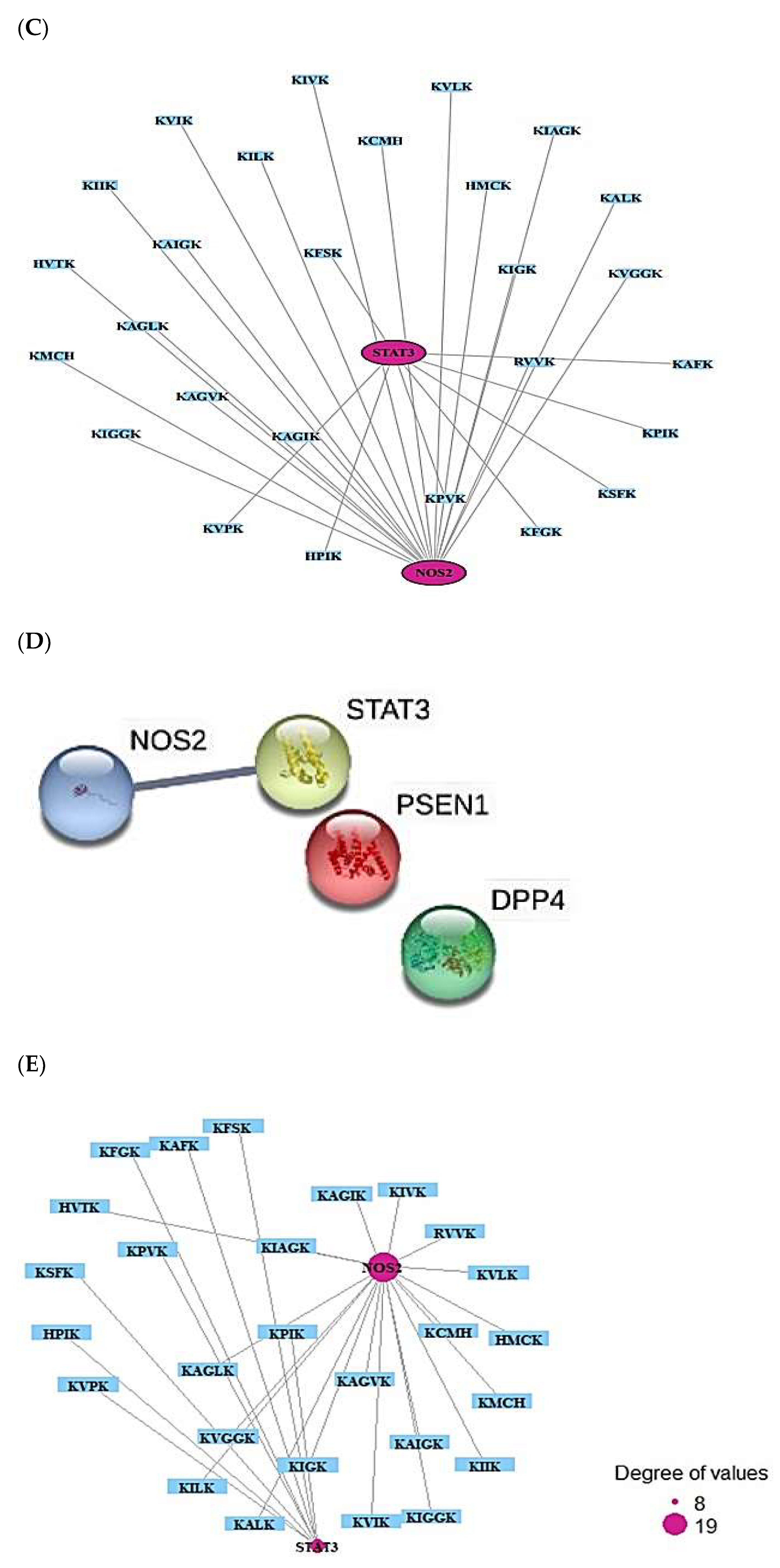


| No. | Targets | No. | Targets |
|---|---|---|---|
| 1 | ACE | 16 | CA2 |
| 2 | ECE1 | 17 | ITGB1 |
| 3 | EDNRA | 18 | GLO1 |
| 4 | MMP3 | 19 | MC1R |
| 5 | SIRT1 | 20 | OPRM1 |
| 6 | SIRT2 | 21 | PPARG |
| 7 | TPP2 | 22 | PYGL |
| 8 | UBE2I | 23 | SRC |
| 9 | CASP1 | 24 | PLAU |
| 10 | FPR1 | 25 | ELANE |
| 11 | MMP9 | 26 | STAT3 |
| 12 | PDYN | 27 | NOS2 |
| 13 | MMP12 | 28 | GLUL |
| 14 | SIRT3 | 29 | DHFR |
| 15 | 30 | ITGA5 |
| KEGG ID | Description | Targets | False Discovery Rate |
|---|---|---|---|
| hsa04917 | Prolactin signaling pathway | SRC, STAT3 | 0.0283 |
| hsa04926 | Relaxin signaling pathway | SRC, NOS2, MMP9 | 0.0093 |
| hsa04915 | Estrogen signaling pathway | SRC, OPRM1, MMP9 | 0.0093 |
| hsa04657 | IL-17 signaling pathway | MMP3, MMP9 | 0.0359 |
| hsa04064 | NF-kappa B signaling pathway | PLAU, UBE2I | 0.0359 |
| hsa04066 | HIF-1 signaling pathway | STAT3, NOS2 | 0.0359 |
| hsa04922 | Glucagon signaling pathway | SIRT1, PYGL | 0.0360 |
| hsa04668 | TNF signaling pathway | MMP3, MMP9 | 0.0389 |
| hsa04152 | AMPK signaling pathway | SIRT1, PPARG | 0.0448 |
| hsa04068 | FoxO signaling pathway | STAT3, SIRT1 | 0.0496 |
| hsa04015 | Rap1 signaling pathway | SRC, ITGB1, FPR1 | 0.0243 |
| KEGG ID | Description | Targets | False Discovery Rate |
|---|---|---|---|
| hsa04330 | Notch signaling pathway | PSEN1, PSEN2 | 0.0044 |
| hsa04066 | HIF-1 signaling pathway | NOS2, STAT3 | 0.0088 |
| hsa04722 | Neurotrophin signaling pathway | PSEN1, PSEN2 | 0.0088 |
| KEGG ID | Description | Targets | False Discovery Rate |
|---|---|---|---|
| hsa04066 | HIF-1 signaling pathway | NOS2, STAT3 | 0.0071 |
| No. | Peptide Sequence | Residue Mass | Targets | Charge | Isoelectric Point | Aggregation Propensity |
|---|---|---|---|---|---|---|
| (Da) | (>0) | (8≤; ≥12) | (Na4VSS ≥ −40; Na4VSS ≤ 60) | |||
| 1 | KPIK | 466.65 | NOS2 | 2 | 10.8 | −34.6 |
| 2 | KPVK | 452.62 | NOS2 | 2 | 10.8 | −39.1 |
| 3 | KVPK | 452.62 | NOS2 | 2 | 10.8 | −39.1 |
| 4 | HPIK | 475.62 | NOS2 | 1.5 | 9.2 | −36.6 |
| 5 | KAFK | 474.62 | NOS2 | 2 | 10.8 | −30.0 |
| 6 | KFGK | 460.60 | NOS2 | 2 | 10.8 | −40.0 |
| 7 | KSFK | 490.62 | NOS2 | 2 | 10.8 | −35.1 |
| 8 | KFSK | 490.62 | NOS2 | 2 | 10.8 | −35.1 |
| 9 | RVVK | 482.66 | STAT3 | 2 | 11.7 | −6.7 |
| 10 | HMCK | 499.68 | STAT3 | 1.5 | 8.0 | −36.1 |
| 11 | KMCH | 499.68 | STAT3 | 1.5 | 8.0 | −36.1 |
| 12 | HVTK | 465.58 | STAT3 | 1.5 | 9.2 | −37.7 |
| 13 | KCMH | 499.68 | STAT3 | 1.5 | 8.0 | −36.1 |
| 14 | KIIK | 482.70 | STAT3 | 2 | 10.8 | 8.6 |
| 15 | KVIK | 468.67 | STAT3 | 2 | 10.8 | 4.0 |
| 16 | KILK | 482.70 | STAT3 | 2 | 10.8 | −0.3 |
| 17 | KVLK | 468.67 | STAT3 | 2 | 10.8 | −4.8 |
| 18 | KALK | 440.61 | STAT3 | 2 | 10.8 | −37.4 |
| 19 | KIVK | 468.67 | STAT3 | 2 | 10.8 | 4.0 |
| 20 | KIGK | 426.59 | STAT3 | 2 | 10.8 | −38.6 |
| 21 | KAIGK | 497.57 | STAT3 | 2 | 10.8 | −19.0 |
| 22 | KIAGK | 497.57 | STAT3 | 2 | 10.8 | −19.0 |
| 23 | KAGVK | 483.64 | STAT3 | 2 | 10.8 | −23.6 |
| 24 | KAGIK | 497.67 | STAT3 | 2 | 10.8 | −19.0 |
| 25 | KAGLK | 497.67 | STAT3 | 2 | 10.8 | −27.8 |
| 26 | KIGGK | 483.65 | STAT3 | 2 | 10.8 | −29.0 |
| 27 | KVGGK | 469.62 | STAT3 | 2 | 10.8 | −33.5 |
| Peptide Sequence | Binding Energy (kcal/mol) | Hydrogen Bond Interactions | Hydrophobic Interactions |
| Amino Acid Residue | Amino Acid Residue | ||
| HPIK | −7.3 | Gln361, Tyr446 | Gln448, Glu444, Leu358 |
| Glu357, His447, Gln448 | |||
| KAFK | −7.1 | Lys363, Thr443, Tyr446, | Gln448, Glu357, His447 |
| Glu357, Gly449, Gln448, | Val445, Glu444 | ||
| Gln361 | |||
| KPIK | −7.0 | Gln361, Tyr446, Gln361 | Glu444, Val445, Gly449, |
| Gln448, Glu357, Gln448, | |||
| Leu362 | |||
| KPVK | −6.8 | Glu444, Tyr446, Gln361, | Gln448, Glu357, Leu358, |
| Tyr446 | Gly449, Val445 | ||
| KVPK | −6.8 | Gln361 | His447 Glu357, Gln448, |
| Tyr446, Gly449, Val445 | |||
| KFGK | −6.8 | Glu306, Arg278, Lys282, | Ile309, Tyr360, Lys283, |
| Gln361, Gln448 | Gln279, Glu286, Leu362, | ||
| Lys363, Leu450, Gly449, | |||
| Val310 | |||
| KSFK | −6.7 | Lys363, Gly449, Gln448 | Glu444, His447, Tyr446, |
| Tyr446, Gln361, Thr443 | His447, Glu357, Gln448 | ||
| Val445, Glu357, Leu358 | |||
| KFSK | −6.4 | Tyr446, Gly449, Gln361 | Gln448, Glu444, Val445 |
| Gln448 | His447, Glu357, His447, | ||
| Glu357 | |||
| Compound (PubChem ID) | Binding energy (kcal/mol) | Hydrogen Bond Interactions | Hydrophobic Interactions |
| Amino Acid Residue | Amino Acid Residue | ||
| Stattic (2779853) | −6.1 | Gly449, Tyr446, Gln361 | Gln448, Tyr446, Glu357, |
| His447 |
| Peptide Sequence | Binding Energy (kcal/mol) | Hydrogen Bond Interactions | Hydrophobic Interactions |
| Amino Acid Residue | Amino Acid Residue | ||
| HVTK | −6.6 | Gln149, Ser486, Ser453 | Asn148, Glu145, Lys103 |
| Leu485, An196, Arg195, | |||
| Gln192, Arg454, Ser153, | |||
| Gly152 | |||
| KILK | −6.4 | Ser486, Gln149, Glu145 | Lys105, ALa104, Leu485, |
| Lys103, Gln192, Gly152, | |||
| Leu100, Ser153, Pro273 | |||
| KAGVK | −6.1 | Ser153, Gln192, Asn196, | Gly152, Arg195, Gln149, |
| Arg454, Ser453 | Ser486, Leu485, Glu145, | ||
| Lys103, Leu100 | |||
| KIGGK | −6.0 | Glu450, Arg454, Asn196, | Ser453, Trp206, Leu100, |
| Lys103, Gln192, Arg195 | Leu485, Ser486, Gly152, | ||
| Gln149, Ser153 | |||
| KAGLK | −5.8 | Gln149, Gln192, Asn148, | Arg454, Arg195, Gly275, |
| Glu145 | Asp274, Pro273, Gly152, | ||
| Ser486, Lys103, Phe188, | |||
| Leu485, Leu100 | |||
| KAIGK | −5.6 | Arg454, Ser486, Gln192 | Gln149, Lys103, Leu485, |
| Leu100, Glu145, Asn148, | |||
| Gly152 | |||
| HMCK | −5.5 | Gln192, Ser486, Glu245 | Ser153, Gly152, Arg195, |
| Asn196, Arg454 | Lys103, Leu485, Gln149, | ||
| Leu100, Ser453 | |||
| KIAGK | −5.5 | Glu145, Gln192, Arg195 | Lys103, Leu100, Leu485, |
| Arg454, Gln149, Asn148 | |||
| KVIK | −5.5 | Glu145, Asn196, Gln192, | Ser486, Leu485, Gln149, |
| Arg454 | Asn148, Pro273, Lys103 | ||
| KALK | −5.5 | Ser486, Gln149, Asp274 | Leu100, Gly152, Pro273, |
| Asn148, Glu145, Lys103, | |||
| Leu485 | |||
| RVVK | −5.4 | Gln192 | Ser153, Arg454, Gln149, |
| Gly152, Asp274, Asn148, | |||
| Lys103, Leu485, Leu100 | |||
| KIIK | −5.4 | Lys103, Gln192, Arg195 | Leu485, Gln149, Leu100, |
| Ser453, Ser153, Gly152, | |||
| Glu145, Ser486 | |||
| KIVK | −5.4 | Pro273, An148, Glu145 | Ser486, Lys103, Leu485, |
| Asn196, Leu100, Arg454, | |||
| Gln149, Gly152 | |||
| KMCH | −5.3 | Arg195, Gln192, Ser486, | Ser153, Asp274, Asn148, |
| Gln149, Lys103 | Gly275, Pro273, Glu145, | ||
| Leu100, Arg454 | |||
| KVGGK | −5.3 | Thr121, Arg86, Thr126 | Trp90, Glu479, Ile119, |
| Val85, Arg83, His84, | |||
| Leu116, Thr109, Pro122, | |||
| Lys123 | |||
| KIGK | −5.3 | Gly152, Lys103, Glu145, | Asn148, Leu485, Ser486, |
| Asn196, Arg454 | Leu100, Gln149, Ser453 | ||
| Ser153 | |||
| KCMH | −5.1 | Lys103, Gln149, Gln192 | Ser453, Gly152, Leu100, |
| Leu485, Ser486, Ser153 | |||
| KVLK | −5.1 | Ile277, Asn390, Gly279 | Arg278, Ser276, Leu344, |
| Arg301, Ile391, Pro281, | |||
| Tyr389, Arg388 | |||
| KAGIK | −5.0 | Asn196 | Arg454, Arg195, Ser153, |
| Leu100, Lys103, Ser486, | |||
| Leu485, Glu145, Asn148, | |||
| Gln149, Gly152, Gln192 | |||
| Compound (PubChem ID) | Binding energy (kcal/mol) | Hydrogen Bond Interactions | Hydrophobic Interactions |
| Amino Acid Residue | Amino Acid Residue | ||
| 1400 W(1433) | −5.2 | Gln97 | Gly455, Arg452, Tyr451, |
| Met94, Gln448, Thr95, | |||
| Phe96 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, K.-K.; Adnan, M.; Cho, D.-H. Uncovering a Hub Signaling Pathway of Antimicrobial-Antifungal-Anticancer Peptides’ Axis on Short Cationic Peptides via Network Pharmacology Study. Int. J. Mol. Sci. 2022, 23, 2055. https://doi.org/10.3390/ijms23042055
Oh K-K, Adnan M, Cho D-H. Uncovering a Hub Signaling Pathway of Antimicrobial-Antifungal-Anticancer Peptides’ Axis on Short Cationic Peptides via Network Pharmacology Study. International Journal of Molecular Sciences. 2022; 23(4):2055. https://doi.org/10.3390/ijms23042055
Chicago/Turabian StyleOh, Ki-Kwang, Md. Adnan, and Dong-Ha Cho. 2022. "Uncovering a Hub Signaling Pathway of Antimicrobial-Antifungal-Anticancer Peptides’ Axis on Short Cationic Peptides via Network Pharmacology Study" International Journal of Molecular Sciences 23, no. 4: 2055. https://doi.org/10.3390/ijms23042055







