Quantitative Proteomics Reveals Molecular Network Driving Stromal Cell Differentiation: Implications for Corneal Wound Healing
Abstract
:1. Introduction
2. Results
2.1. Determination of Differentiation State of Corneal Stromal Cells
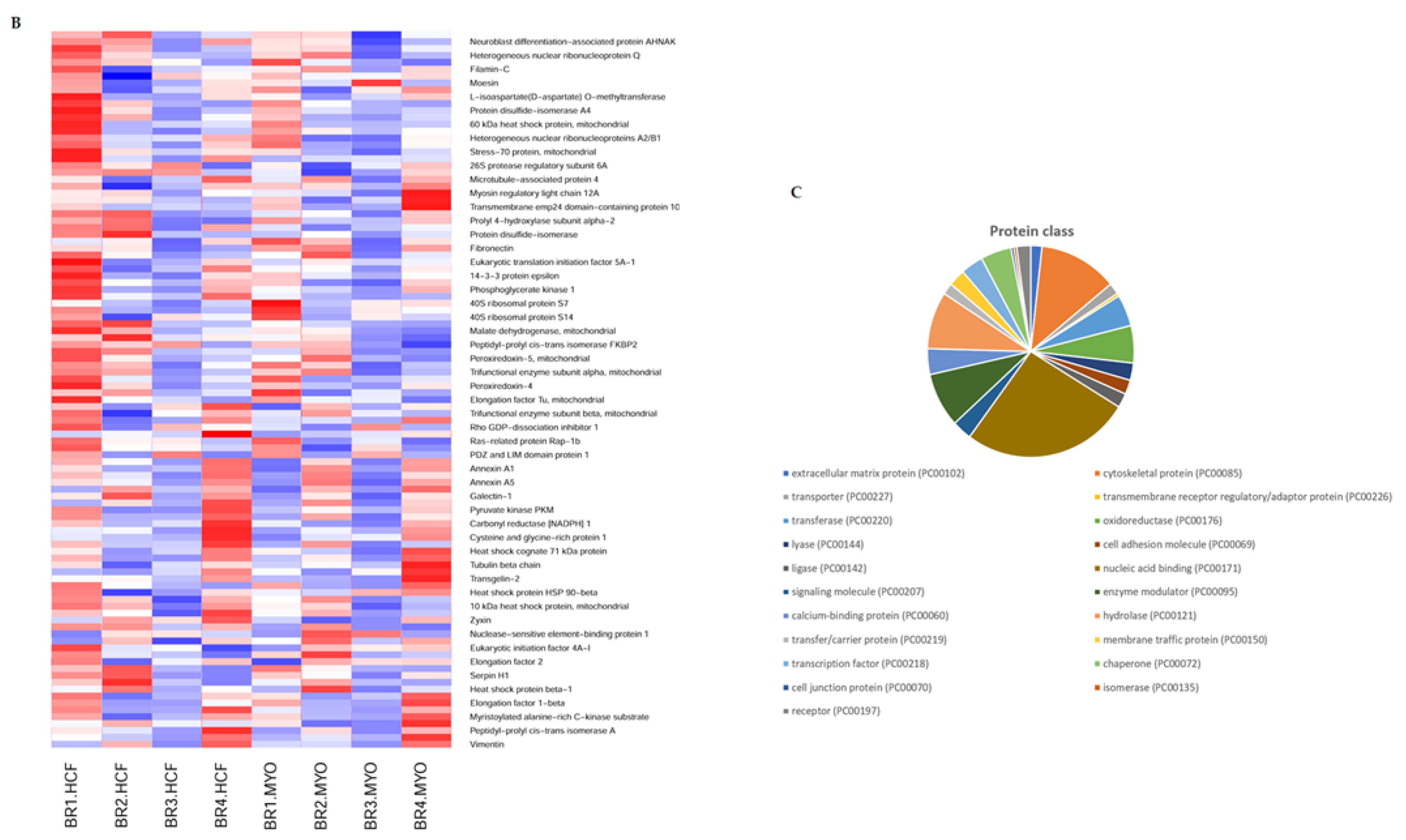
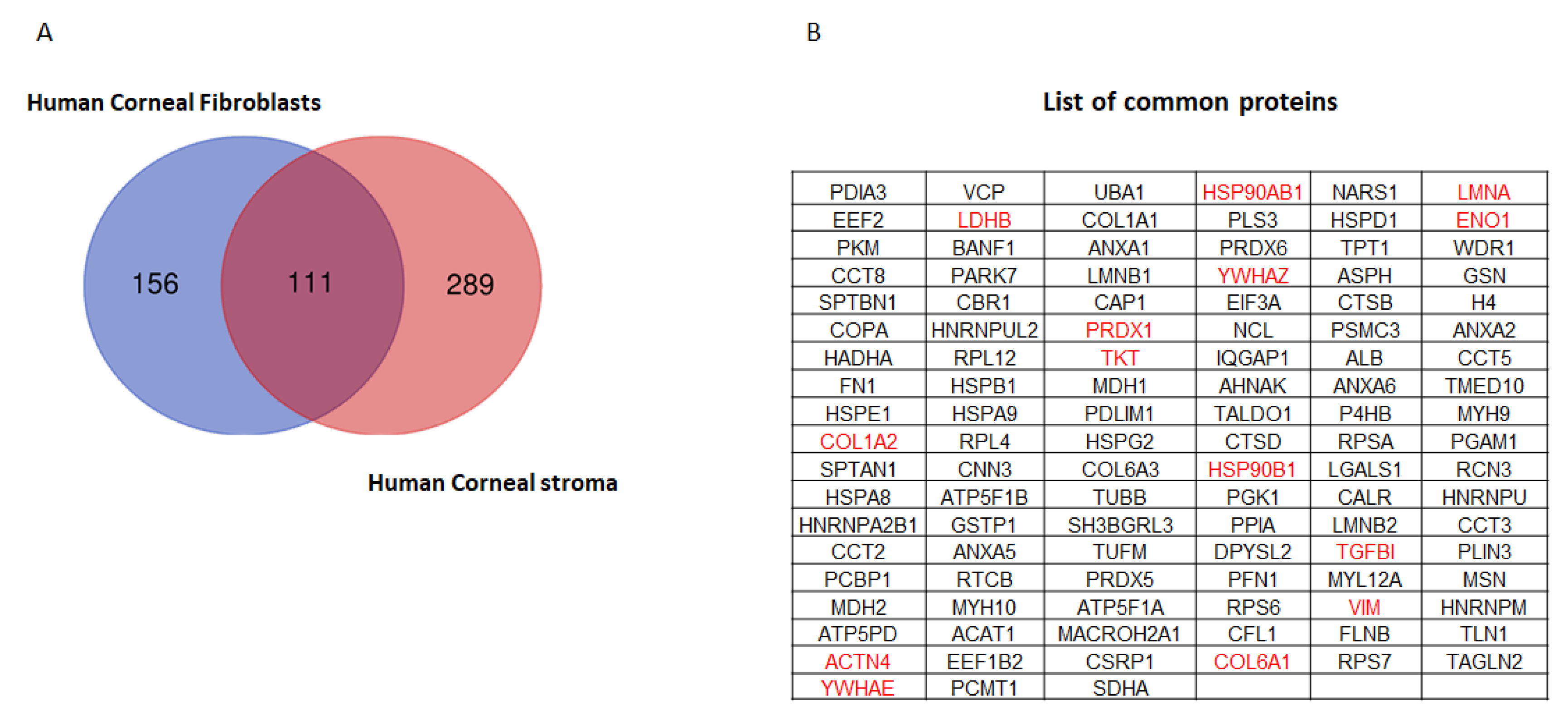
2.2. Quantitative Proteomics Reveals Enhanced Cytoskeletal Regulation and Cell Signaling
2.3. Pathway—Protein Interaction Networks
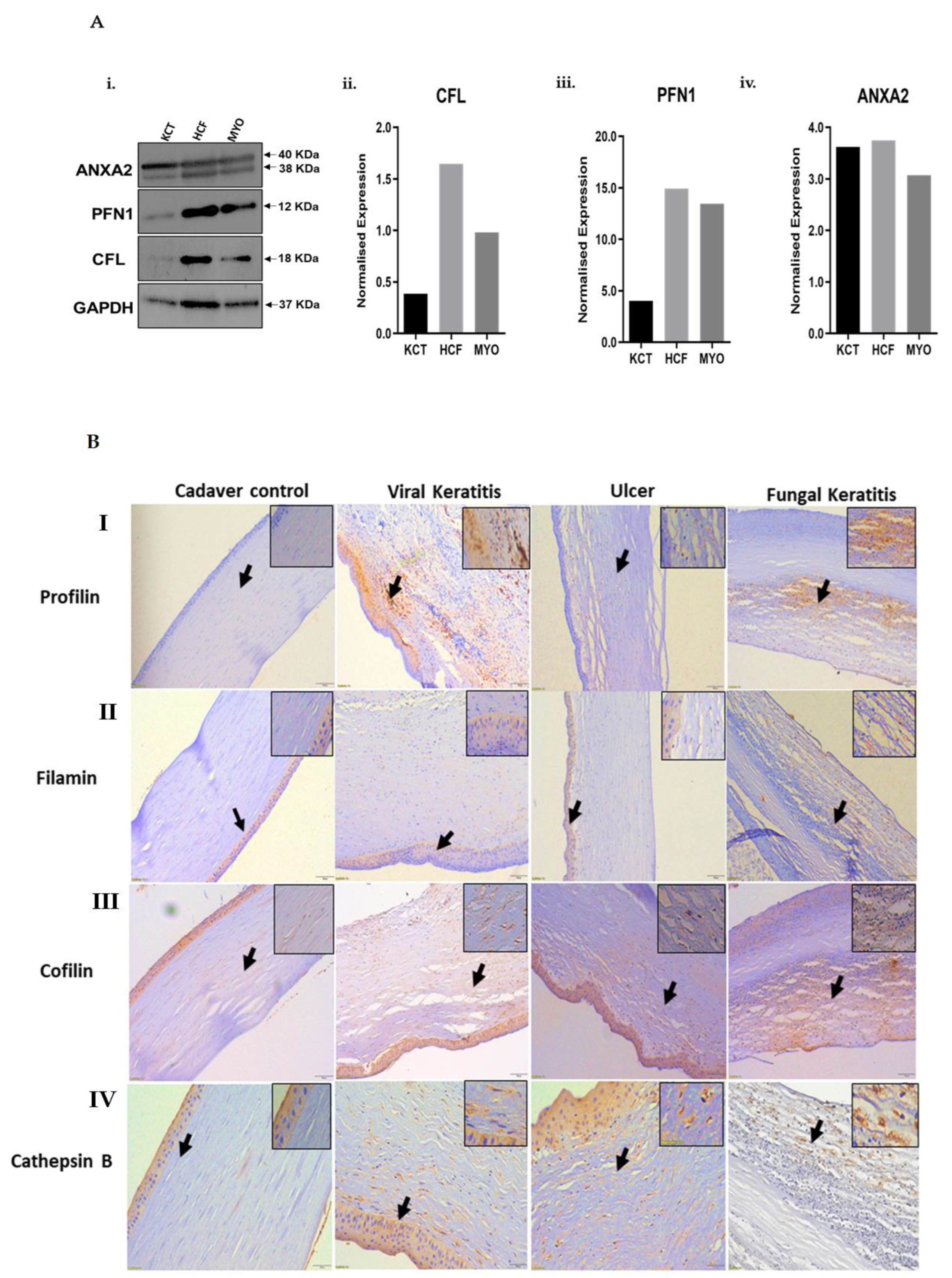
2.4. Validation by Immunofluorescence, Western Blot and Immunohistochemistry
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Differentiation of Human Corneal Fibroblasts
4.2. Gene Expression and Quantitative PCR (qPCR) Analysis
4.3. Protein Extraction from Cultured Stromal Cells and iTRAQ Labeling
4.4. Quantitative Proteomics
4.5. Statistical Analysis
4.6. Network Model Describing the Protein-Pathway Interaction
4.7. Immunofluorescence for Human Corneal Fibroblast Cells
4.8. Immunohistochemistry of Fibrotic Cornea
4.9. Immunoblotting
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilson, S.E.; Mohan, R.R.; Hong, J.W.; Lee, J.S.; Choi, R.; Mohan, R.R. The wound healing response after laser in situ keratomileusis and photorefractive keratectomy: Elusive control of biological variability and effect on custom laser vision correction. Arch. Ophthalmol. 2001, 119, 889–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spadea, L.; Giammaria, D.; Trabucco, P. Corneal wound healing after laser vision correction. Br. J. Ophthalmol. 2016, 100, 28–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ljubimov, A.V.; Saghizadeh, M. Progress in corneal wound healing. Prog. Retin. Eye Res. 2015, 49, 17–45. [Google Scholar] [CrossRef] [Green Version]
- Bernstein, A.M.; Twining, S.S.; Warejcka, D.J.; Tall, E.; Masur, S.K. Urokinase receptor cleavage, a crucial step in 14 fibroblast-to-myofibroblast differentiation. Mol. Biol. Cell 2007, 18, 2716–2727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, K.; Kurosaka, D.; Yoshino, M.; Oshima, T.; Kurosaka, H. Injured corneal epithelial cells promote myodifferentiation of corneal fibroblasts. Investig. Ophthalmol. Vis. Sci. 2002, 4, 2603–2608. [Google Scholar]
- Jester, J.V.; Petroll, W.M.; Barry, P.A.; Cavanagh, H.D. Expression of alpha-smooth muscle (alpha-SM) actin during corneal stromal wound healing. Investig. Ophthalmol. Vis. Sci. 1995, 36, 809–819. [Google Scholar]
- Helary, C.; Ovtracht, L.; Coulomb, B.; Godeau, G.; Giraudguille, M. Dense fibrillar collagen matrices: A model to study myofibroblast behaviour during wound healing. Biomaterials 2006, 27, 4443–4452. [Google Scholar] [CrossRef]
- Mohan, R.R.; Hutcheon, A.E.; Choi, R.; Hong, J.; Lee, J.; Mohan, R.R.; Ambrósio, R.; Zieske, J.; Wilson, S. Apoptosis, necrosis, proliferation, and myofibroblast generation in the stroma following LASIK and PRK. Exp. Eye Res. 2003, 76, 71–87. [Google Scholar] [CrossRef]
- O’Brien, T.P.; Li, Q.; Ashraf, M.F.; Matteson, D.M.; Stark, W.J.; Chan, C.-C. Inflammatory Response in the Early Stages of Wound Healing After Excimer Laser Keratectomy. Arch. Ophthalmol. 1998, 116, 1470–1474. [Google Scholar] [CrossRef] [Green Version]
- Torricelli, A.A.; Santhanam, A.; Wu, J.; Singh, V.; Wilson, S.E. The corneal fibrosis response to epithelial–stromal injury. Exp. Eye Res. 2016, 142, 110–118. [Google Scholar] [CrossRef] [Green Version]
- Kymionis, G.; Liakopoulos, D.A.; Grentzelos, M.A.; Skatharoudi, C.A.; Panagopoulou, S.I. Uneventful Femtosecond Laser-assisted Flap Creation in a Patient With Postoperative PRK Corneal Haze. J. Refract. Surg. 2015, 31, 638–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, A.; Zhou, C.; Lakshman, N.; Petroll, W.M. Corneal stromal cells use both high- and low-contractility migration mechanisms in 3-D collagen matrices. Exp. Cell Res. 2012, 318, 741–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eraslan, M.; Toker, E. Mechanisms of corneal wound healing and its modulation following refractive surgery. Marmara Med. J. 2009, 22, 169–178. [Google Scholar]
- Karamichos, D.; Hutcheon, A.E.K.; Zieske, J.D. Transforming growth factor-beta 3 regulates assembly of a non-fibrotic matrix in a 3D corneal model. J. Tissue Eng. Regen. Med. 2011, 5, 228–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrington, L.; Albon, J.; Anderson, I.; Kamma, C.; Boulton, M. Differential regulation of key stages in early corneal wound healing by TGF-beta Isoforms and their inhibitors. Investig. Ophthalmol. Vis. Sci. 2006, 47, 1886–1894. [Google Scholar] [CrossRef] [Green Version]
- Lakshman, N.; Petroll, W.M. Growth Factor Regulation of Corneal Keratocyte Mechanical Phenotypes in 3-D Collagen Matrices. Investig. Ophthalmol. Vis. Sci. 2012, 53, 1077–1086. [Google Scholar] [CrossRef] [Green Version]
- Petroll, W.; Jester, J.; Bean, J.; Cavanagh, H. Myofibroblast transformation of cat corneal endothelium by transforming growth factor-beta(1), -beta(2), and -beta(3). Investig. Ophthalmol. Vis. Sci. 1998, 39, 2018–2032. [Google Scholar]
- Funderburgh, J.; Funderburgh, M.; Mann, M.; Corpuz, L.; Roth, M. Proteoglycan expression during transforming growth factor beta-induced keratocyte-myofibroblast transdifferentiation. J. Biol. Chem. 2001, 276, 44173–44178. [Google Scholar] [CrossRef] [Green Version]
- Mulholland, B.; Tuft, S.; Khaw, P. Matrix metalloproteinase distribution during early corneal wound healing. Eye 2005, 19, 584–588. [Google Scholar] [CrossRef]
- Khamar, P.; Nishtala, K.; Shetty, R.; Panigrahi, T.; Shetty, K.; Pahuja, N.; Deshpande, V.; Ghosh, A. Early biological responses in ocular tissue after SMILE and LASIK surgery. Exp. Eye Res. 2020, 192, 107936. [Google Scholar] [CrossRef]
- Wilson, M.W.; Hungerford, J.L.; George, S.M.; Madreperla, S.A. Topical mitomycin C for the treatment of conjunctival and corneal epithelial dysplasia and neoplasia. Am. J. Ophthalmol. 1997, 124, 303–311. [Google Scholar] [CrossRef]
- Karamichos, D.; Funderburgh, M.L.; Hutcheon, A.E.K.; Zieske, J.; Du, Y.; Wu, J.; Funderburgh, J.L. A Role for Topographic Cues in the Organization of Collagenous Matrix by Corneal Fibroblasts and Stem Cells. PLoS ONE 2014, 9, e86260. [Google Scholar] [CrossRef] [Green Version]
- Pei, Y.; Sherry, D.M.; McDermott, A.M. Thy-1 distinguishes human corneal fibroblasts and myofibroblasts from keratocytes. Exp. Eye Res. 2004, 79, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Jester, J.V.; Barry-Lane, P.A.; Cavanagh, H.D.; Petroll, W.M. Induction of alpha-smooth muscle actin expression and myofibroblast transformation in cultured corneal keratocytes. Cornea 1996, 15, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Schure, R.; Costa, K.D.; Rezaei, R.; Lee, W.; Laschinger, C.; Tenenbaum, H.C.; McCulloch, C.A. Impact of matrix metalloproteinases on inhibition of mineralization by fetuin. J. Periodontal Res. 2013, 48, 357–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myrna, K.E.; Pot, S.A.; Murphy, C.J. Meet the corneal myofibroblast: The role of myofibroblast transformation in corneal wound healing and pathology. Vet.-Ophthalmol. 2009, 12, 25–27. [Google Scholar] [CrossRef]
- Honda, E.; Park, A.-M.; Yoshida, K.; Tabuchi, M.; Munakata, H. Myofibroblasts: Biochemical and Proteomic Approaches to Fibrosis. Tohoku J. Exp. Med. 2013, 230, 67–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerke, V.; Moss, S.E. Annexins: From Structure to Function. Physiol. Rev. 2002, 82, 331–371. [Google Scholar] [CrossRef] [PubMed]
- Desmoulière, A.; Geinoz, A.; Gabbiani, F. Transforming growth factor-beta 1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J. Cell Biol. 1993, 122, 103–111. [Google Scholar] [CrossRef] [Green Version]
- Karring, H.; Thøgersen, I.B.; Klintworth, G.K.; Enghild, J.J.; Møller-Pedersen, T. Proteomic Analysis of the Soluble Fraction from Human Corneal Fibroblasts with Reference to Ocular Transparency. Mol. Cell. Proteom. 2004, 3, 660–674. [Google Scholar] [CrossRef] [Green Version]
- Jonathan, D.; Winkelman; Colleen, G.; Peifer, B.M.; Kovar, D.R. Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. Proc. Natl. Acad. Sci. USA 2014, 111, 4121–4126. [Google Scholar]
- Bear, J.E.; Gertler, F.B. Ena/VASP: Towards resolving a pointed controversy at the barbed end. J. Cell Sci. 2009, 122, 1947–1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bashaw, G.J.; Kidd, T.; Murray, D.; Pawson, T.; Goodman, C.S. Repulsive Axon Guidance: Abelson and Enabled Play Opposing Roles Downstream of the Roundabout Receptor. Cell 2000, 101, 703–715. [Google Scholar] [CrossRef] [Green Version]
- Dickinson, R.E.; Duncan, W.C. The SLIT–ROBO pathway: A regulator of cell function with implications for the reproductive system. Reproduction 2010, 139, 697–704. [Google Scholar] [CrossRef] [Green Version]
- Blockus, H.; Chédotal, A. Slit-Robo signaling. Development 2016, 143, 3037–3044. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-H. Pull and push: Talin activation for integrin signaling. Cell Res. 2012, 22, 1512–1514. [Google Scholar] [CrossRef] [Green Version]
- Das, M.; Ithychanda, S.S.; Qin, J.; Plow, E.F. Mechanisms of talin-dependent integrin signaling and crosstalk. Biochim. Biophys. Acta (BBA) Biomembr. 2014, 1838, 579–588. [Google Scholar] [CrossRef] [Green Version]
- Cintron, C.; Covington, H.; Kublin, C.L. Morphogenesis of rabbit corneal stroma. Investig. Ophthalmol. Vis. Sci. 1983, 24, 543–556. [Google Scholar] [CrossRef]
- Cornuet, P.K.; Blochberger, C.; Hassell, J.R. Molecular polymorphism of lumican during corneal development. Investig. Ophthalmol. Vis. Sci. 1994, 35, 870–877. [Google Scholar]
- Funderburgh, J.L.; Caterson, B.; Conrad, G.W. Keratan sulfate proteoglycan during embryonic development of the chicken cornea. Dev. Biol. 1986, 116, 267–277. [Google Scholar] [CrossRef]
- Young, R.D.; Gealy, E.C.; Liles, M.; Caterson, B.; Ralphs, J.R.; Quantock, A.J. Keratan Sulfate Glycosaminoglycan and the Association with Collagen Fibrils in Rudimentary Lamellae in the Developing Avian Cornea. Investig. Ophthalmol. Vis. Sci. 2007, 48, 3083–3088. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Nagata, K. Biology of Hsp47 (Serpin H1), a collagen-specific molecular chaperone. Semin. Cell Dev. Biol. 2017, 62, 142–151. [Google Scholar] [CrossRef]
- Ishida, Y.; Kubota, H.; Yamamoto, A.; Kitamura, A.; Bächinger, H.P.; Nagata, K. Type I Collagen in Hsp47-null Cells Is Aggregated in Endoplasmic Reticulum and Deficient in N-Propeptide Processing and Fibrillogenesis. Mol. Biol. Cell 2006, 17, 2346–2355. [Google Scholar] [CrossRef]
- Tremble, P.M.; Lane, T.F.; Sage, E.H.; Werb, Z. SPARC, a secreted protein associated with morphogenesis and tissue remodeling, induces expression of metalloproteinases in fibroblasts through a novel extracellular matrix-dependent pathway. J. Cell Biol. 1993, 121, 1433–1444. [Google Scholar] [CrossRef] [Green Version]
- Barnes, A.; Chang, W.; Morello, R.; Cabral, W.A.; Weis, M.; Eyre, D.R.; Leikin, S.; Makareeva, E.; Kuznetsova, N.; Uveges, T.E.; et al. Deficiency of Cartilage-Associated Protein in Recessive Lethal Osteogenesis Imperfecta. New Engl. J. Med. 2006, 355, 2757–2764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malmstrom, J.; Lindberg, H.; Lindberg, C.; Bratt, C.; Wieslander, E.; Delander, E.L.; Särnstrand, B.; Burns, J.S.; Mose-Larsen, P.; Fey, S.; et al. Transforming growth factor-beta 1 specifically induce proteins involved in the myofibroblast contractile apparatus. Mol. Cell Proteom. 2004, 3, 466–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, H.; Hajjar, K. Annexin A2 in Fibrinolysis, Inflammation and Fibrosis. Int. J. Mol. Sci. 2021, 22, 6836. [Google Scholar] [CrossRef]
- Buss, D.G.; Giuliano, E.A.; Sharma, A.; Mohan, R.R. Isolation and cultivation of equine corneal keratocytes, fibroblasts and myofibroblasts. Vet.-Ophthalmol. 2010, 13, 37–42. [Google Scholar] [CrossRef] [Green Version]
- Beales, M.P.; Funderburgh, J.L.; Jester, J.; Hassell, J.R. Proteoglycan synthesis by bovine keratocytes and corneal fibroblasts: Maintenance of the keratocyte phenotype in culture. Investig. Ophthalmol. Vis. Sci. 1999, 40, 1658–1663. [Google Scholar]
- Masur, S.K.; Dewal, H.S.; Dinh, T.T.; Erenburg, I.; Petridou, S. Myofibroblasts differentiate from fibroblasts when plated at low density. Proc. Natl. Acad. Sci. USA 1996, 93, 4219–4223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shetty, R.; Ghosh, A.; Lim, R.R.; Subramani, M.; Mihir, K.; Reshma, A.R.; Ranganath, A.; Nagaraj, S.; Nuijts, R.M.; Beuerman, R.; et al. Elevated expression of matrix metalloproteinase-9 and inflammatory cytokines in keratoconus patients is inhibited by cyclosporine A. Investig. Ophthalmol. Vis. Sci. 2015, 56, 738–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidiropoulos, K.; Viteri, G.; Sevilla, C.; Jupe, S.; Webber, M.; Orlic-Milacic, M.; Jassal, B.; May, B.; Shamovsky, V.; Duenas, C.; et al. Reactome enhanced pathway visualization. Bioinformatics 2017, 33, 3461–3467. [Google Scholar] [CrossRef] [PubMed] [Green Version]



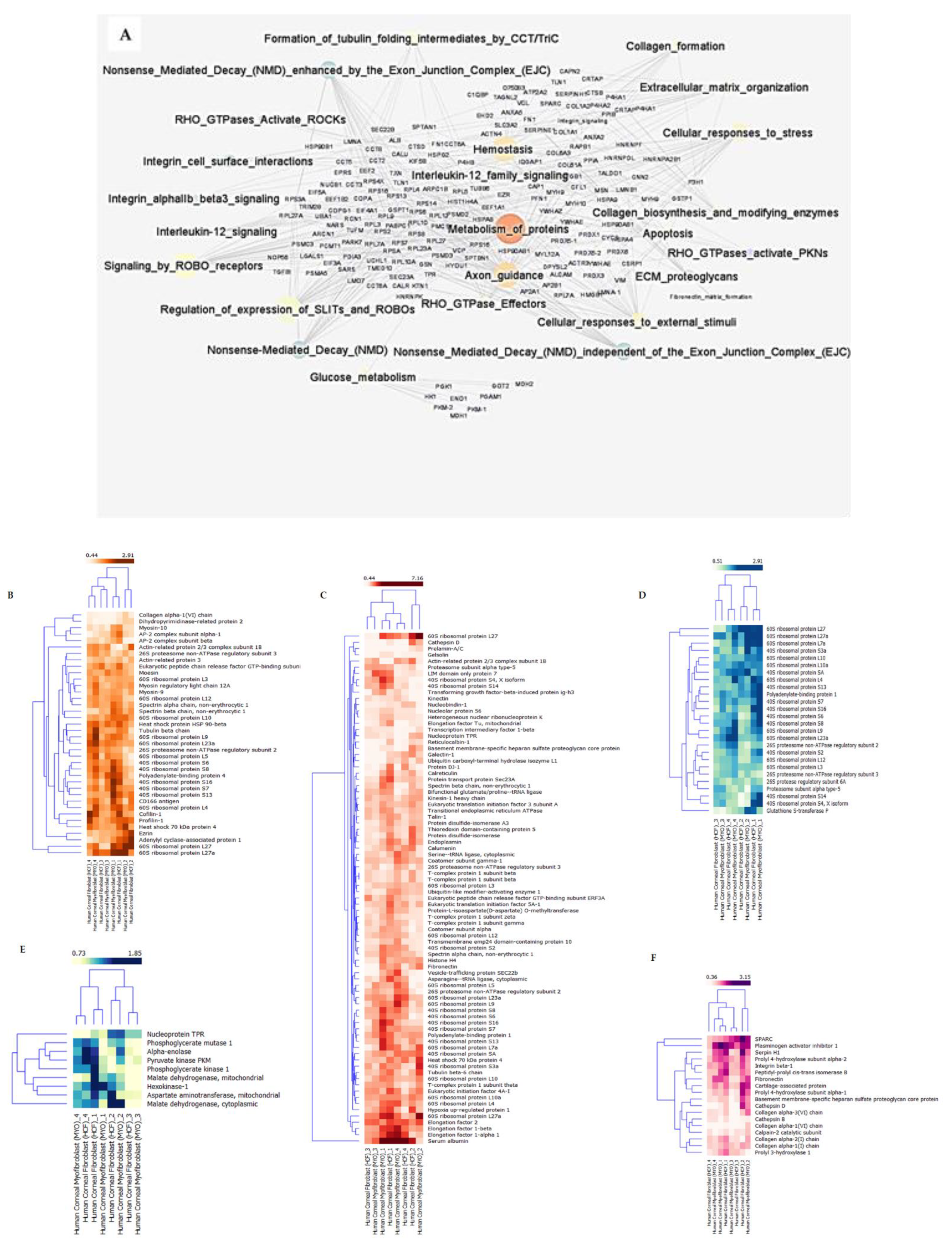

| Pathway Identifier | Pathway Name | Proteins Identified | Total Proteins | Entities p Value | Entities FDR |
|---|---|---|---|---|---|
| R-HSA-422475 | Axon guidance | 52 | 557 | 1.11 × 10−16 | 2.95 × 10−14 |
| R-HSA-975956 | Nonsense-mediated decay (NMD) independent of the exon junction complex (EJC) | 23 | 96 | 6.66 × 10−16 | 1.06 × 10−13 |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 28 | 172 | 6.11 × 10−15 | 5.36 × 10−13 |
| R-HSA-975957 | Nonsense-mediated decay (NMD) enhanced by the exon junction complex (EJC) | 23 | 117 | 3.94 × 10−14 | 2.09 × 10−12 |
| R-HSA-927802 | Nonsense-mediated decay (NMD) | 23 | 117 | 3.94 × 10−14 | 2.09 × 10−12 |
| R-HSA-376176 | Signaling by ROBO receptors | 30 | 218 | 4.71 × 10−14 | 2.17 × 10−12 |
| R-HSA-9020591 | Interleukin-12 signaling | 13 | 46 | 2.09 × 10−10 | 6.08 × 10−9 |
| R-HSA-447115 | Interleukin-12 family signaling | 13 | 56 | 2.18 × 10−9 | 5.66 × 10−8 |
| R-HSA-392499 | Metabolism of proteins | 85 | 2010 | 1.14 × 10−7 | 2.29 × 10−6 |
| R-HSA-1650814 | Collagen biosynthesis and modifying enzymes | 11 | 67 | 1.13 × 10−6 | 2.03 × 10−5 |
| R-HSA-1474290 | Collagen formation | 12 | 90 | 3.09 × 10−6 | 4.95 × 10−5 |
| R-HSA-389960 | Formation of tubulin folding intermediates by CCT/TriC | 7 | 26 | 4.54 × 10−6 | 7.27 × 10−5 |
| R-HSA-2262752 | Cellular responses to stress | 26 | 408 | 1.08 × 10−5 | 1.62 × 10−4 |
| R-HSA-3000178 | ECM proteoglycans | 9 | 76 | 1.29 × 10−4 | 0.00168208 |
| R-HSA-8953897 | Cellular responses to external stimuli | 27 | 505 | 1.42 × 10−4 | 0.001704805 |
| R-HSA-109581 | Apoptosis | 14 | 178 | 1.56 × 10−4 | 0.001872861 |
| R-HSA-70326 | Glucose metabolism | 10 | 98 | 1.83 × 10−4 | 0.002199433 |
| R-HSA-1474244 | Extracellular matrix organization | 19 | 301 | 1.93 × 10−4 | 0.002310664 |
| R-HSA-216083 | Integrin cell surface interactions | 7 | 85 | 0.005395875 | 0.032375248 |
| R-HSA-195258 | RHO GTPase effectors | 15 | 295 | 0.006736196 | 0.034468636 |
| R-HSA-1566977 | Fibronectin matrix formation | 2 | 6 | 0.009769066 | 0.047246897 |
| R-HSA-109582 | Hemostasis | 28 | 722 | 0.011423688 | 0.047246897 |
| R-HSA-5627117 | RHO GTPases activate ROCKs | 3 | 19 | 0.011811724 | 0.047246897 |
| R-HSA-5625740 | RHO GTPases activate PKNs | 5 | 63 | 0.020332384 | 0.064732946 |
| R-HSA-354192 | Integrin alpha IIb beta3 signaling | 3 | 28 | 0.032248972 | 0.08304909 |
| R-HSA-9006921 | Integrin signaling | 3 | 28 | 0.032248972 | 0.08304909 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishtala, K.; Panigrahi, T.; Shetty, R.; Kumar, D.; Khamar, P.; Mohan, R.R.; Deshpande, V.; Ghosh, A. Quantitative Proteomics Reveals Molecular Network Driving Stromal Cell Differentiation: Implications for Corneal Wound Healing. Int. J. Mol. Sci. 2022, 23, 2572. https://doi.org/10.3390/ijms23052572
Nishtala K, Panigrahi T, Shetty R, Kumar D, Khamar P, Mohan RR, Deshpande V, Ghosh A. Quantitative Proteomics Reveals Molecular Network Driving Stromal Cell Differentiation: Implications for Corneal Wound Healing. International Journal of Molecular Sciences. 2022; 23(5):2572. https://doi.org/10.3390/ijms23052572
Chicago/Turabian StyleNishtala, Krishnatej, Trailokyanath Panigrahi, Rohit Shetty, Dhanananajay Kumar, Pooja Khamar, Rajiv R. Mohan, Vrushali Deshpande, and Arkasubhra Ghosh. 2022. "Quantitative Proteomics Reveals Molecular Network Driving Stromal Cell Differentiation: Implications for Corneal Wound Healing" International Journal of Molecular Sciences 23, no. 5: 2572. https://doi.org/10.3390/ijms23052572






