A Comparative Transcriptome and Metabolome Combined Analysis Reveals the Key Genes and Their Regulatory Model Responsible for Glucoraphasatin Accumulation in Radish Fleshy Taproots
Abstract
:1. Introduction
2. Results
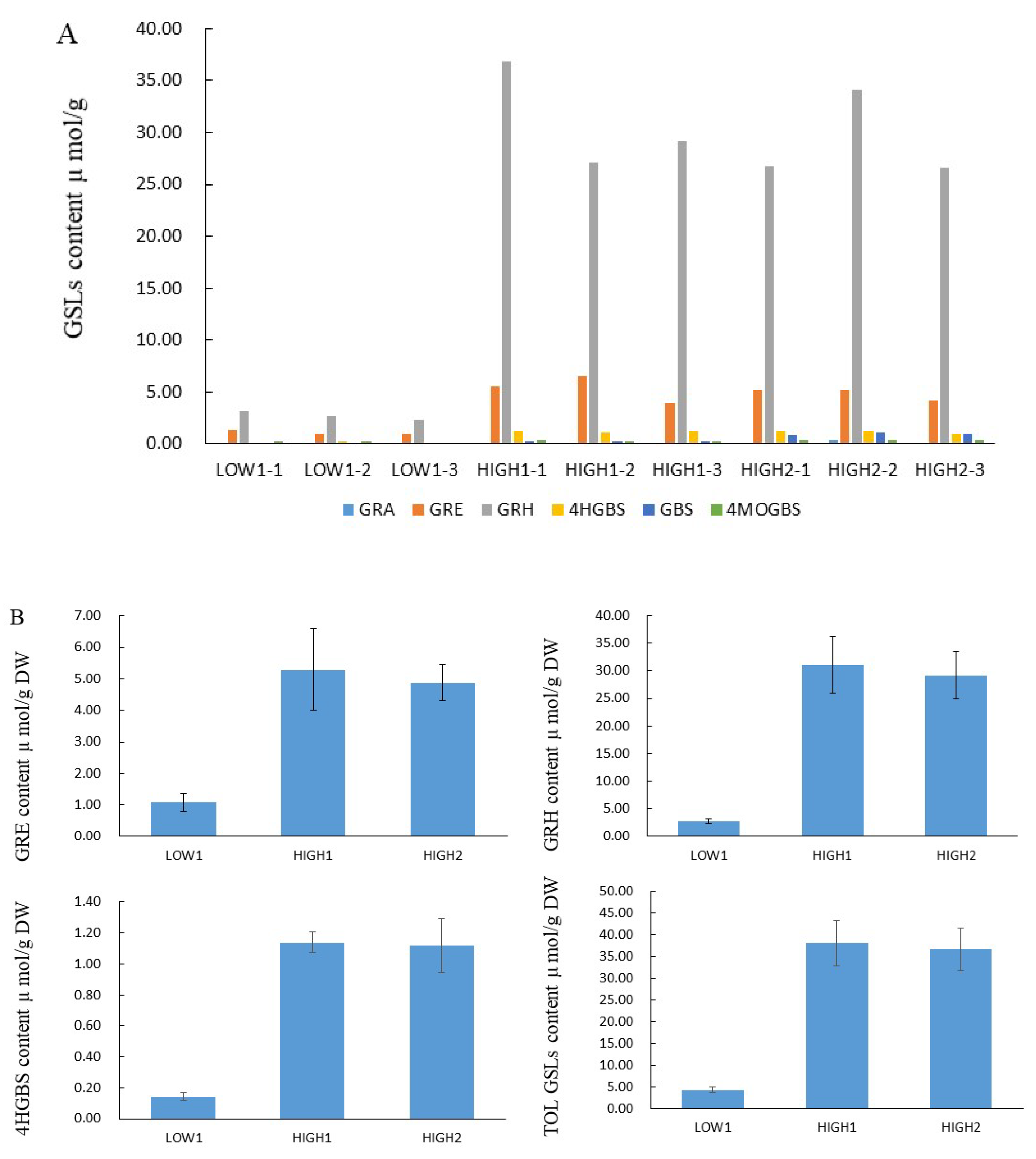
2.1. GSLs Profiles and Content of Various GSL Components in the Radish Taproots of Different Genotypes
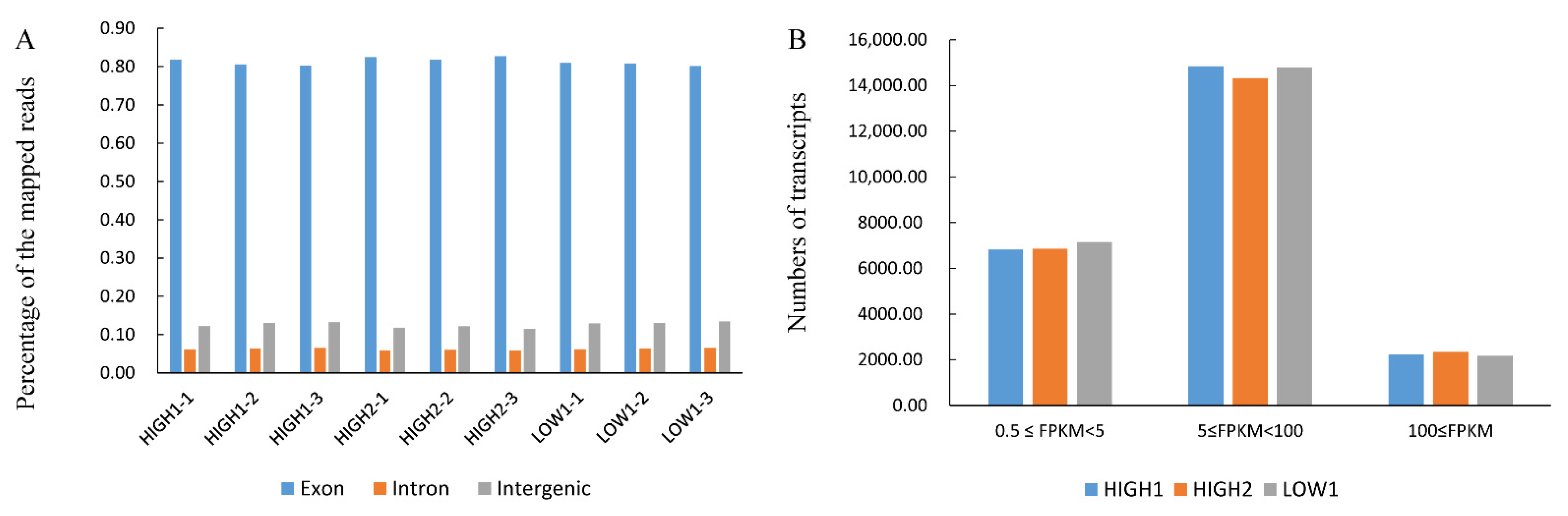
2.2. Transcriptome Analysis of Fleshy Taproots of Three Different Radish Varieties
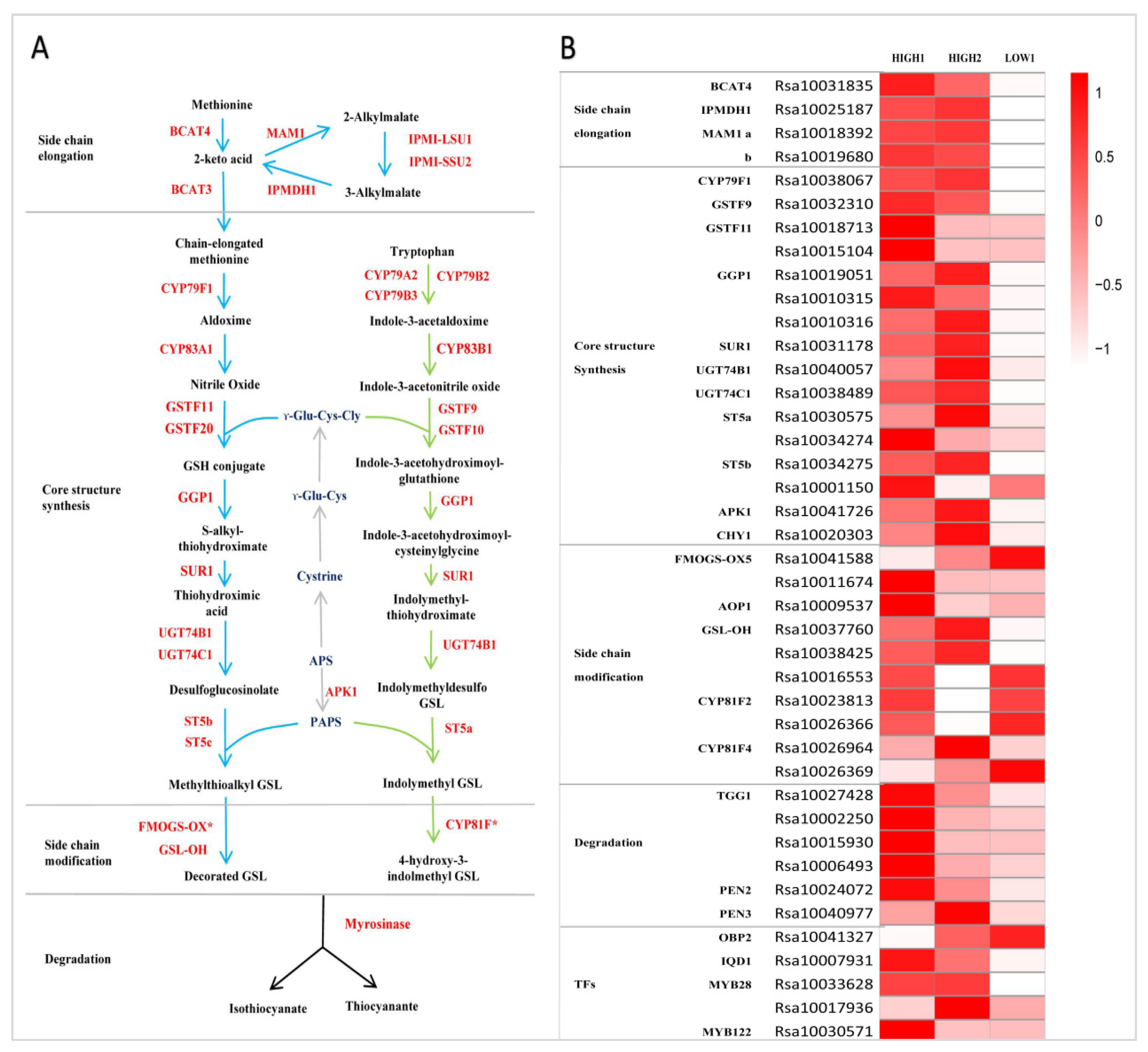
2.3. Expression of Genes Related to GSLs Biosynthesis and Degradation in Fleshy Taproots of Three Radish Varieties
2.4. Performance of Transcription Factors Related to GSLs Biosynthesis in the Fleshy Taproots of Three Radish Varieties
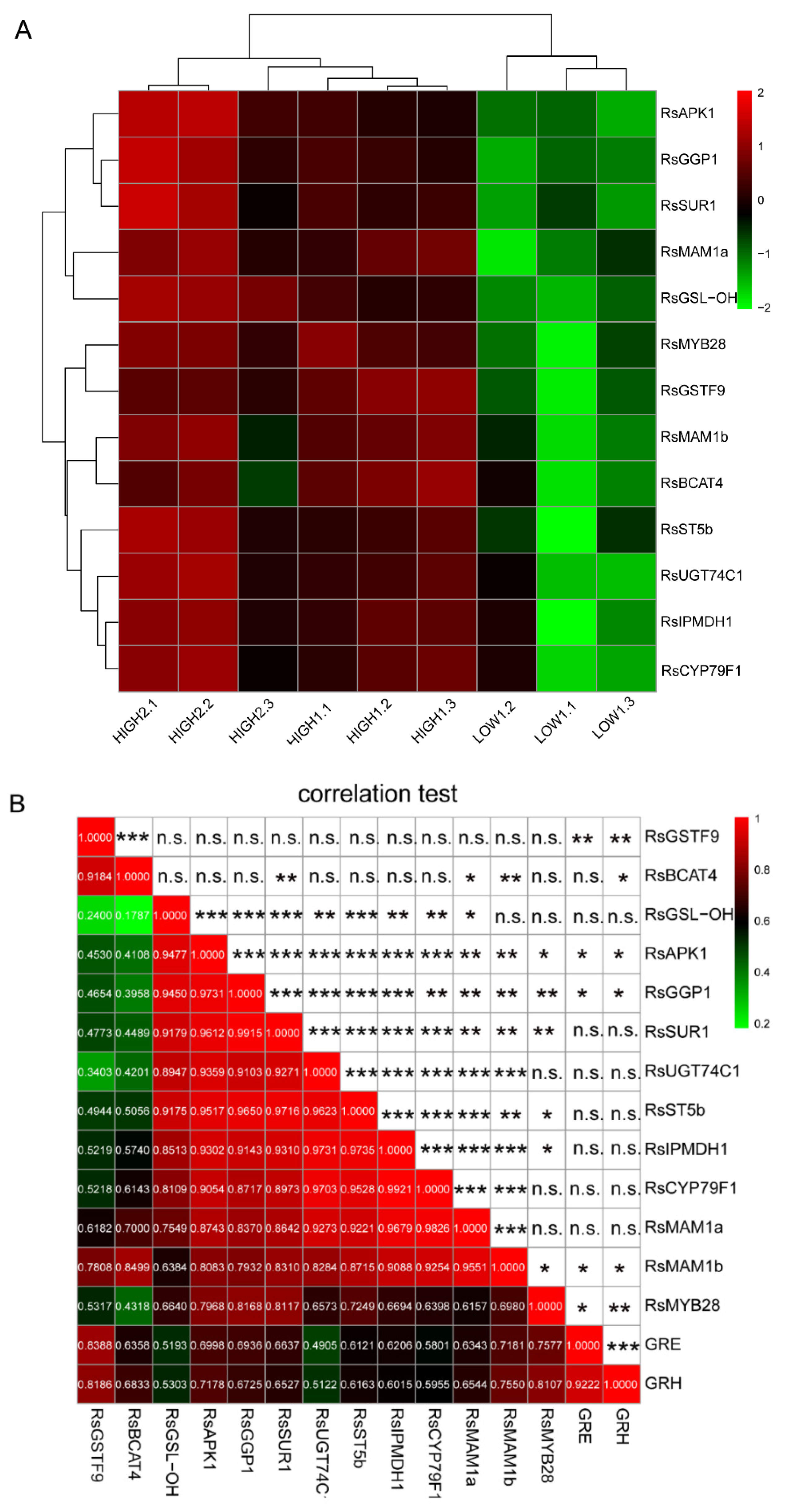
2.5. Establishment of a Co-Expression Network of the Key Genes for GSLs Biosynthesis in Radish Fleshy Taproots
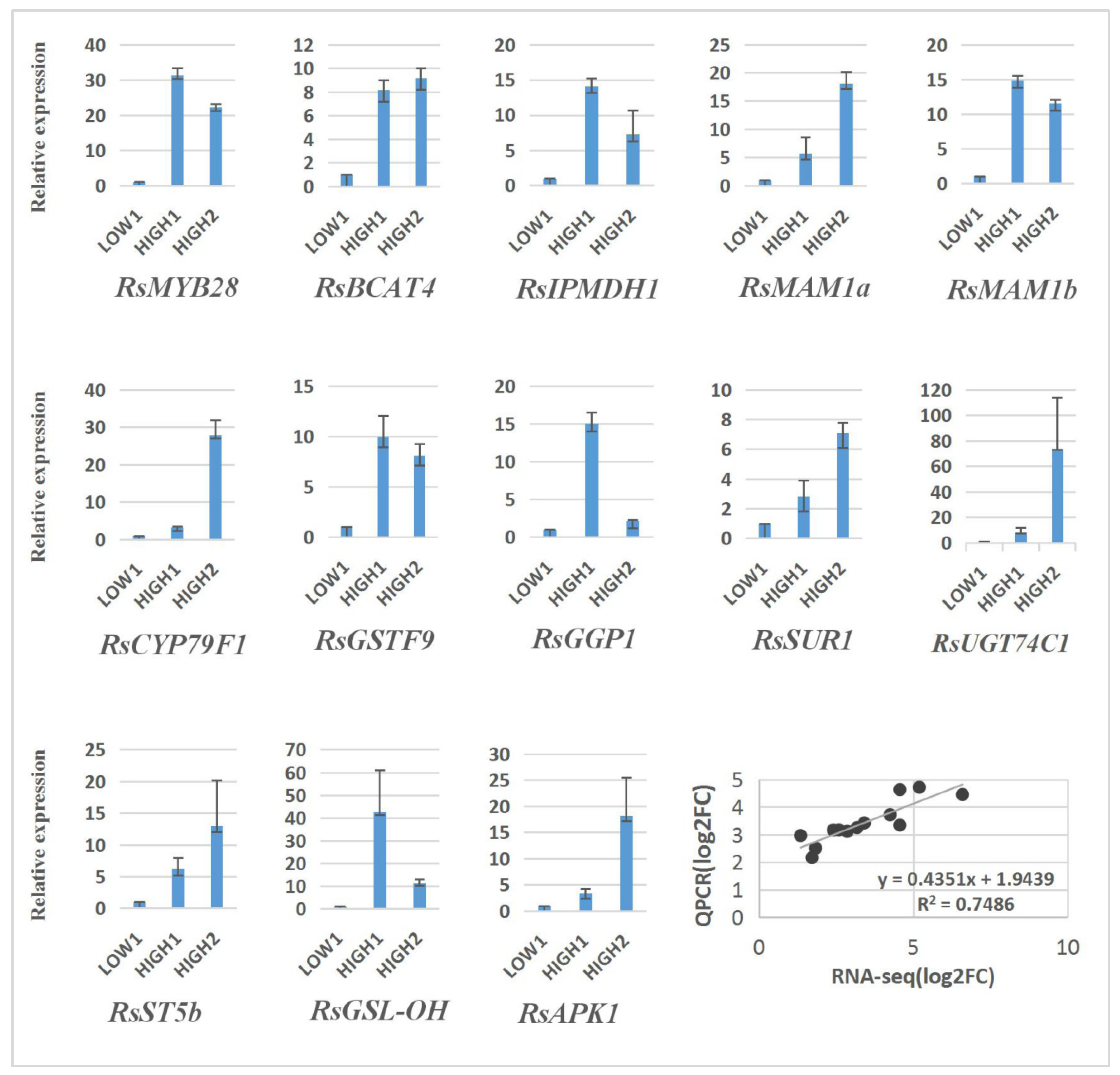
2.6. qRT–PCR Analysis of DEGs in Three Different Radish Cultivars
3. Discussion
3.1. The Content of GRH as the Main GSLs Component in Radish Taproot Is Significantly Different among Different Genotypes
3.2. RsMYB28 Regulatory Model Responsible for GSLs Accumulation in Radish Fleshy Taproots
3.3. Key Gene of GSLs Biosynthesis in Radish Fleshy Taproots
3.4. Key Genes of Indole GSLs Biosynthesis in Radish Fleshy Taproots
4. Materials and Methods
4.1. Plant Material, Cultivation, and Sampling
4.2. GSLs Extraction and Analysis
4.3. RNA Extraction, Library Construction, and RNA-Seq
4.4. Functional Annotation of Expressed Genes and Differential Expression Analysis
4.5. Co-Expression Analysis of the Key Genes for GSLs Biosynthesis and Their Relationship with GSLs Content in Radish Fleshy Taproots
4.6. Gene Expression Validation and Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Halkier, B.A.; Gershenzon, J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006, 57, 303–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sønderby, I.E.; Geu-Flores, F.; Halkier, B. Biosynthesis of glucosinolates–gene discovery and beyond. Trends Plant Sci. 2010, 15, 283–290. [Google Scholar] [CrossRef]
- Scholl, C.; Eshelman, B.D.; Barnes, D.M.; Hanlon, P.R. Raphasatin is a more potent inducer of the detoxification enzymes than its degradation products. J. Food Sci. 2011, 76, C504–C511. [Google Scholar] [CrossRef] [PubMed]
- Clarke, D.B. Glucosinolates, structures and analysis in food. Anal. Methods 2010, 2, 310–325. [Google Scholar] [CrossRef]
- Petersen, B.L.; Chen, S.; Hansen, C.H.; Olsen, C.E.; Halkier, B. Composition and content of glucosinolates in developing Arabidopsis thaliana. Planta 2002, 214, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; ·Quiros, C.F. Survey of glucosinolate variation in leaves of Brassica rapa crops. Genet Resour. Crop. Evol. 2010, 57, 1079–1089. [Google Scholar] [CrossRef] [Green Version]
- Sun, B.; Liu, N.; Zhao, Y.; Yan, H.; Wang, Q. Variation of glucosinolates in three edible parts of Chinese kale (Brassica alboglabra Bailey) varieties. Food Chem. 2011, 124, 941–947. [Google Scholar] [CrossRef]
- Farnham, M.W.; Stephenson, K.K.; Fahey, J.W. Glucoraphanin level in broccoli seed is largely determined by genotype. HortScience 2005, 40, 50–53. [Google Scholar] [CrossRef]
- Kakizaki, T.; Kitashiba, H.; Zou, Z.; Li, F.; Fukino, N.; Ohara, T.; Nishio, T.; Ishida, M. A 2-Oxoglutarate-Dependent Dioxygenase Mediates the Biosynthesis of Glucoraphasatin in Radish. Plant Physiol. 2017, 173, 1583–1593. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Kensler, T.W.; Cho, C.G.; Posner, G.H.; Talalay, P. Anticarcinogenic activities of sulforaphane and structurally related synthetic norbornyl isothiocyanates. Proc. Natl. Acad. Sci. USA 1994, 91, 3147–3150. [Google Scholar] [CrossRef] [Green Version]
- Razis, A.F.A.; Bagatta, M.; De Nicola, G.R.; Iori, R.; Ioannides, C. Up-regulation of cytochrome P450 and phase II enzyme systems in rat precision-cut rat lung slices by the intact glucosinolates, glucoraphanin and glucoerucin. Lung Cancer 2011, 71, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Brader, G.; Mikkelsen, M.D.; Halkier, B.A.; Tapio Palva, E. Altering glucosinolate profiles modulates disease resistance in plants. Plant J. 2006, 46, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Rodriguez, M.F.; Cymbaluk, N.; Epp, T.; Laarveld, B.; Recale, E.; Simoko, E.; Card, C. Effects of the glucosinolate sinigrin in combination with a non-iodine supplemented diet on serum iodine and thyroid hormone concentrations in non-pregnant mares. J. Equine Vet. Sci. 2020, 91, 103110. [Google Scholar] [CrossRef] [PubMed]
- Dalmazi, G.D.; Giuliani, C. Plant constituents and thyroid: A revision of the main phytochemicals that interfere with thyroid function. Food Chem. Toxicol. 2021, 152, 112158. [Google Scholar] [CrossRef]
- Matusheski, N.V.; Jeffery, E.H. Comparison of the bioactivity of two glucoraphanin hydrolysis products found in broccoli, sulforaphane and sulforaphane nitrile. J. Agric. Food Chem. 2001, 49, 5743–5749. [Google Scholar] [CrossRef]
- Hanschen, F.S.; Herz, C.; Schlotz, N.; Kupke, F.; Rodríguez, M.M.B.; Schreiner, M.; Rohn, S.; Lamy, E. The Brassicaepithionitrile 1-cyano-2,3-epithiopropane triggers cell death in human liver cancer cellsin vitro. Mol. Nutr. Food Res. 2015, 59, 2178–2189. [Google Scholar] [CrossRef]
- Grubb, C.D.; Abel, S. Glucosinolate metabolism and its control. Trends Plant Sci. 2006, 11, 89–100. [Google Scholar] [CrossRef]
- Diebold, R.; Schuster, J.; Däschner, K.; Binder, S. The branched-chain amino acid transaminase gene family in arabidopsis encodes plastid and mitochondrial proteins. Plant Physiol. 2002, 129, 540–550. [Google Scholar] [CrossRef] [Green Version]
- Schuster, J.; Knill, T.; Reichelt, M.; Gershenzon, J.; Binder, S. Branched-chain aminotransferase4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. Plant Cell 2006, 18, 2664–2679. [Google Scholar] [CrossRef] [Green Version]
- Knill, T.; Schuster, J.; Reichelt, M.; Gershenzon, J.; Binder, S. Arabidopsis branched-chain aminotransferase 3 functions in both amino acid and glucosinolate biosynthesis. Plant Physiol. 2008, 146, 1028–1039. [Google Scholar] [CrossRef] [Green Version]
- Kroymann, J.; Textor, S.; Tokuhisa, J.G.; Falk, K.L.; Bartram, S.; Gershenzon, J.; Mitchell-Olds, T. A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol. 2001, 127, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Mawhinney, T.P.; Preuss, M.L.; Schroeder, A.C.; Chen, B.; Abraham, L.; Jez, J.M.; Chen, S. A redox-active isopropylmalate dehydrogenase functions in the biosynthesis of glucosinolates and leucine in Arabidopsis. Plant J. 2009, 60, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.H.; Wittstock, U.; Olsen, C.E.; Hick, A.J.; Pickett, J.A.; Halkier, B. Cytochrome P450 CYP79F1 from Arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J. Biol. Chem. 2001, 276, 11078–11085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Glawischnig, E.; Jørgensen, K.; Naur, P.; Jørgensen, B.; Olsen, C.E.; Hansen, C.H.; Rasmussen, H.; Pickett, J.A.; Halkier, B.A. CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J. 2003, 33, 923–937. [Google Scholar] [CrossRef] [Green Version]
- Hemm, M.R.; Ruegger, M.O.; Chapple, C. The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 2003, 15, 179–194. [Google Scholar] [CrossRef] [Green Version]
- Mikkelsen, M.D.; Naur, P.; Halkier, B.A. Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 2004, 37, 770–777. [Google Scholar] [CrossRef]
- Gachon, C.; Langlois-Meurinne, M.; Henry, Y.; Saindrenan, P. Transcriptional co-regulation of secondary metabolism enzymes in Arabidopsis: Functional and evolutionary implications. Plant Mol. Biol. 2005, 58, 229–245. [Google Scholar] [CrossRef]
- Piotrowski, M.; Schemenewitz, A.; Lopukhina, A.; Müller, A.; Janowitz, T.; Weiler, E.W.; Oecking, C. Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J. Biol. Chem. 2004, 279, 50717–50725. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Qiu, Y.; Wang, X.; Yue, Z.; Yang, X.; Chen, X.; Zhang, X.; Shen, D.; Wang, H.; Song, J.; et al. Insights into the species-specific metabolic engineering of glucosinolates in radish (Raphanus sativus L.) based on comparative genomic analysis. Sci. Rep. 2017, 7, 16040. [Google Scholar] [CrossRef] [Green Version]
- Gigolashvili, T.; Yatusevich, R.; Berger, B.; Müller, C.; Flügge, U.-I. The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007, 51, 247–261. [Google Scholar] [CrossRef]
- Celenza, J.L.; Quiel, J.A.; Smolen, G.A.; Merrikh, H.; Silvestro, A.R.; Normanly, J.; Bender, J. The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 2005, 137, 253–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gigolashvili, T.; Yatusevich, R.; Rollwitz, I.; Humphry, M.; Gershenzon, J.; Flügge, U.-I. The plastidic bile acid transporter 5 is required for the biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana. Plant Cell 2009, 21, 1813–1829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sønderby, I.E.; Burow, M.; Rowe, H.C.; Kliebenstein, D.; Halkier, B.A. A complex interplay of three R2R3 MYB transcription factors determines the profile of aliphatic glucosinolates in Arabidopsis. Plant Physiol. 2010, 153, 348–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, L.; Chen, H.; Cao, B.; Lei, J.; Chen, G. Molecular characterization of MYB28 involved in Aliphatic glucosinolate biosynthesis in Chinese kale (Brassica oleracea var. alboglabra Bailey). Front. Plant Sci. 2017, 8, 1083. [Google Scholar] [CrossRef] [Green Version]
- Augustine, R.; Mukhopadhyay, A.; Bisht, N.C. Targeted silencing of BjMYB28 transcription factor gene directs development of low glucosinolate lines in oilseed Brassica juncea. Plant Biotechnol. J. 2013, 11, 855–866. [Google Scholar] [CrossRef]
- Augustine, R.; Majee, M.; Gershenzon, J.; Bisht, N.C. Four genes encoding MYB28, a major transcriptional regulator of the aliphatic glucosinolate pathway, are differentially expressed in the allopolyploid Brassica juncea. J. Exp. Bot. 2013, 64, 4907–4921. [Google Scholar] [CrossRef] [PubMed]
- Araki, R.; Hasumi, A.; Nishizawa, O.I.; Sasaki, K.; Kuwahara, A.; Sawada, Y.; Totoki, Y.; Toyoda, A.; Sakaki, Y.; Li, Y. Novel bioresources for studies of B rassica oleracea: Identification of a kale MYB transcription factor responsible for glucosinolate production. Plant Biotechnol. J. 2013, 11, 1017–1027. [Google Scholar] [CrossRef]
- Wang, B.; Wu, Z.; Li, Z.; Zhang, Q.; Hu, J.; Xiao, Y.; Cai, D.; Wu, J.; King, G.J.; Li, H.; et al. Dissection of the genetic architecture of three seed-quality traits and consequences for breeding in Brassica napus. Plant Biotechnol. J. 2018, 16, 1336–1348. [Google Scholar] [CrossRef] [Green Version]
- Levy, M.; Wang, Q.; Kaspi, R.; Parrella, M.P.; Abel, S. Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 2005, 43, 79–96. [Google Scholar] [CrossRef]
- Skirycz, A.; Reichelt, M.; Burow, M.; Birkemeyer, C.; Rolcik, J.; Kopka, J.; Zanor, M.I.; Gershenzon, J.; Strnad, M.; Szopa, J.; et al. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 2006, 47, 10–24. [Google Scholar] [CrossRef]
- Crisp, P. Radish, Raphanus sativus (Cruciferae). In Evolution of Crop Plants, 2nd ed; Smartt, J., Simmonds, N.W., Eds.; Longman Scientific & Technical: Harlow, UK, 1995; pp. 86–89. [Google Scholar]
- Maldini, M.; Foddai, M.; Natella, F.; Petretto, G.L.; Rourke, J.P.; Chessa, M.; Pintore, G. Identification and quantification of glucosinolates in different tissues of Raphanus raphanistrum by liquid chromatography tandem-mass spectrometry. J. Food Compos. Anal. 2017, 61, 20–27. [Google Scholar] [CrossRef]
- Barillari, J.; Cervellati, R.; Paolini, M.; Tatibout, A.; Rollin, P.; Iori, R. Isolation of 4-Methylthio-3-butenyl Glucosinolate from Raphanus sativus Sprouts (Kaiware Daikon) and Its Redox Properties. J. Agric. Food Chem. 2005, 53, 9890–9896. [Google Scholar] [CrossRef] [PubMed]
- Ishida, M.; Nagata, M.; Ohara, T.; Kakizaki, T.; Hatakeyama, K.; Nishio, T. Small variation of glucosinolate composition in Japanese cultivars of radish (Raphanus sativus L.) requires simple quantitative analysis for breeding of glucosinolate component. Breed. Sci. 2012, 62, 63–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, G.; Lim, S.; Chae, W.B.; Park, J.E.; Park, H.R.; Lee, E.J.; Huh, J.H. Root Glucosinolate Profiles for screening of radish (Raphanus sativus L.) genetic resources. J. Agric. Food Chem. 2016, 64, 61–70. [Google Scholar] [CrossRef]
- Kim, S.-J.; Uddin, M.R.; Park, S.U. Glucosinolate accumulation in three important radish (Raphanus sativus) cultivars. Australian J. Crop Sci. 2013, 7, 1843–1847. [Google Scholar]
- Carlson, D.G.; Daxenbichler, M.E.; Vanetten, C.H.; Hill, C.B.; Williams, P.H. Glucosinolates in radish cultivars. J. Am. Soc. Hortic. Sci. 1985, 110, 634–638. [Google Scholar]
- Rampal, G.; Thind, T.S.; Vig, A.P.; Arora, S. Antimutagenic Potential of Glucosinolate-Rich Seed Extracts of Broccoli (Brassica oleracea L. var Italica plenck). Int. J. Toxicol. 2010, 29, 616–624. [Google Scholar] [CrossRef]
- Voa, Q.V.; Trenerryb, C.; Rochfortc, S.; Wadesonc, J.; Hughes, A.B. Synthesis and anti-inflammatory activity of aromatic glucosinolates. Bioorganic Med. Chem. 2013, 21, 5945–5954. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Kostov, R.V. Glucosinolates and isothiocyanates in health and disease. Trends Mol. Med. 2012, 18, 337–347. [Google Scholar] [CrossRef]
- Zou, Z.; Ishida, M.; Li, F.; Kakizaki, T.; Suzuki, S.; Kitashiba, H.; Nishio, T. QTL analysis using SNP markers developed by next-generation sequencing for identification of candidate genes controlling 4-Methylthio-3-Butenyl glucosinolate contents in roots of radish, Raphanus sativus L. PLoS ONE 2013, 8, e53541. [Google Scholar] [CrossRef]
- Kuang, P.; Song, D.; Yuan, Q.; Lv, X.; Zhao, D.; Liang, H. Preparative separation and purification of sulforaphene from radish seeds by high-speed countercurrent chromatography. Food Chem. 2013, 136, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-N.; Won, S.Y.; Seo, M.-S.; Lee, J.; Lee, S.M.; Kwon, S.-J.; Kim, J.S. Induction of glucoraphasatin biosynthesis genes by MYB29 in radish (Raphanus sativus L.) roots. Int. J. Mol. Sci. 2020, 21, 5721. [Google Scholar] [CrossRef] [PubMed]
- Hua, B. Analysis and Evaluation on Glucosinolate Composition and Sulforaphane Content of Radish (Raphanus sativus L.) Germplasm Resources; Chinese Academy of Agricultural Science: Beijing, China, 2013. [Google Scholar]
- Zhang, X.; Yue, Z.; Mei, S.; Qiu, Y.; Yang, X.; Chen, X.; Cheng, F.; Wu, Z.; Sun, Y.; Jing, Y. A de novo genome of a Chinese radish cultivar. Hortic. Plant J. 2015, 1, 155–164. [Google Scholar]
- Mithen, R.; Clarke, J.; Lister, C.; Dean, C. Genetics of aliphatic glucosinolates. III. Side chain structure of aliphatic glucosinolates in Arabidopsis thaliana. Heredity 1995, 74, 210–215. [Google Scholar] [CrossRef]
- Barth, C.; Jander, G. Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 2006, 46, 549–562. [Google Scholar] [CrossRef]
- Gigolashvili, T.; Engqvist, M.; Yatusevich, R.; Müller, C.; Flügge, U.I. HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 2008, 177, 627–642. [Google Scholar] [CrossRef]
- Gigolashvili, T.; Berger, B.; Mock, H.P.; Müller, C.; Weisshaar, B.; Flügge, U.I. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007, 50, 886–901. [Google Scholar] [CrossRef] [Green Version]
- Farnham, M. A comprehensive program to enhance glucoraphanin content of broccoli heads and seed. In Proceedings of the International Symposium on Human Health Effects of Fruits and Vegetables, Québec City, QC, Canada, 17–20 August 2005; p. 30. [Google Scholar]
- Hirai, M.; Sugiyama, K.; Sawada, Y.; Tohge, T.; Obayashi, T.; Suzuki, A.; Araki, R.; Sakurai, N.; Suzuki, H.; Aoki, K.; et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 6478–6483. [Google Scholar] [CrossRef] [Green Version]
- Frerigmann, H. Glucosinolate regulation in a complex relationship–MYC and MYB–no one can act without each other. Adv. Bot. Res. 2016, 80, 57–97. [Google Scholar]
- Seo, M.-S.; Jin, M.; Chun, J.-H.; Kim, S.-J.; Park, B.-S.; Shon, S.-H.; Kim, J.S. Functional analysis of three BrMYB28 transcription factors controlling the biosynthesis of glucosinolates in Brassica rapa. Plant Mol. Biol. 2016, 90, 503–516. [Google Scholar] [CrossRef] [Green Version]
- Zang, Y.-X.; Kim, J.-H.; Park, Y.-D.; Kim, D.-H.; Hong, S.-B. Metabolic engineering of aliphatic glucosinolates in Chinese cabbage plants expressing Arabidopsis MAM1, CYP79F1, and CYP83A1. BMB Rep. 2008, 41, 472–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, M.; Mukhopadhyay, A.; Gupta, V.; Pental, D.; Pradhan, A.K. BjuB.CYP79F1 regulates synthesis of propyl fraction of aliphatic glucosinolates in oilseed mustard Brassica juncea: Functional validation through genetic and transgenic approaches. PLoS ONE 2016, 11, e0150060. [Google Scholar] [CrossRef]
- Magrath, R.; Bano, F.; Morgner, M.; Parkin, I.; Sharpe, A.; Lister, C.; Dean, C.; Turner, J.; Lydiate, D.; Mithen, R. Genetics of aliphatic glucosinolates. I. Side chain elongation in Brassica napus and Arabidopsis thaliana. Heredity 1994, 72, 290–299. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Gao, M.; Yang, B.; Quiros, C.F. Gene for gene alignment between the Brassica and Arabidopsis genomes by direct transcriptome mapping. Theor. Appl. Genet. 2003, 107, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Skipsey, M.; Edwards, R. Roles for glutathione transferases in plant secondary metabolism. Phytochemistry 2010, 71, 338–350. [Google Scholar] [CrossRef]
- Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences. Chinese Vegetable Cultivation; China Agriculture Press: Beijing, China, 2009; pp. 276–291. [Google Scholar]
- He, H.; Chen, H.; Schnitzler, W.H. Glucosinolate composition and contents in Brassica vegetables. Sci. Agric. Sin. 2002, 35, 192–197. [Google Scholar]
- Møller, P.; Olsen, O.; Plöger, A.; Rasmussen, K.; Sørensen, H. Quantitative analysis of individual glucosinolates. In Advances in the Production and Utilization of Cruciferous Crops; Springer: Dordrecht, The Netherlands, 1985; pp. 111–126. [Google Scholar]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly, differential expression, and differential regulation for RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Nat. Preced. 2010, 11, R106. [Google Scholar]
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Mering, V.C. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wang, P.; Wang, J.; Wang, H.; Liu, T.; Zhang, X.; Song, J.; Yang, W.; Wu, C.; Yang, H.; et al. A Comparative Transcriptome and Metabolome Combined Analysis Reveals the Key Genes and Their Regulatory Model Responsible for Glucoraphasatin Accumulation in Radish Fleshy Taproots. Int. J. Mol. Sci. 2022, 23, 2953. https://doi.org/10.3390/ijms23062953
Li X, Wang P, Wang J, Wang H, Liu T, Zhang X, Song J, Yang W, Wu C, Yang H, et al. A Comparative Transcriptome and Metabolome Combined Analysis Reveals the Key Genes and Their Regulatory Model Responsible for Glucoraphasatin Accumulation in Radish Fleshy Taproots. International Journal of Molecular Sciences. 2022; 23(6):2953. https://doi.org/10.3390/ijms23062953
Chicago/Turabian StyleLi, Xiaoman, Peng Wang, Jinglei Wang, Haiping Wang, Tongjin Liu, Xiaohui Zhang, Jiangping Song, Wenlong Yang, Chunhui Wu, Haohui Yang, and et al. 2022. "A Comparative Transcriptome and Metabolome Combined Analysis Reveals the Key Genes and Their Regulatory Model Responsible for Glucoraphasatin Accumulation in Radish Fleshy Taproots" International Journal of Molecular Sciences 23, no. 6: 2953. https://doi.org/10.3390/ijms23062953





