Evolutionary Diversity and Function of Metacaspases in Plants: Similar to but Not Caspases
Abstract
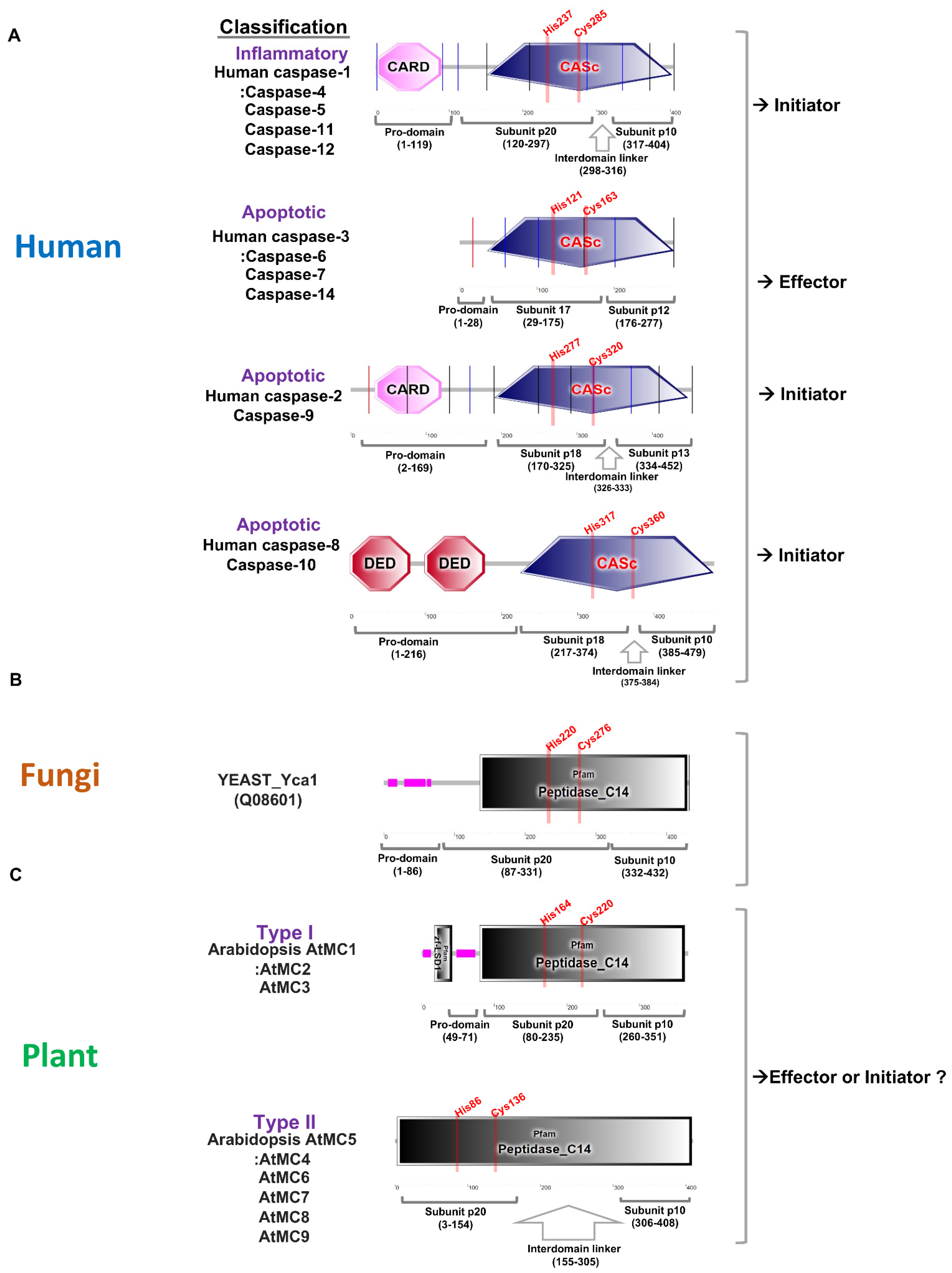
:1. Basic Features of Caspase and Metacaspase
2. Subcellular Localization of Caspase and Metacaspase
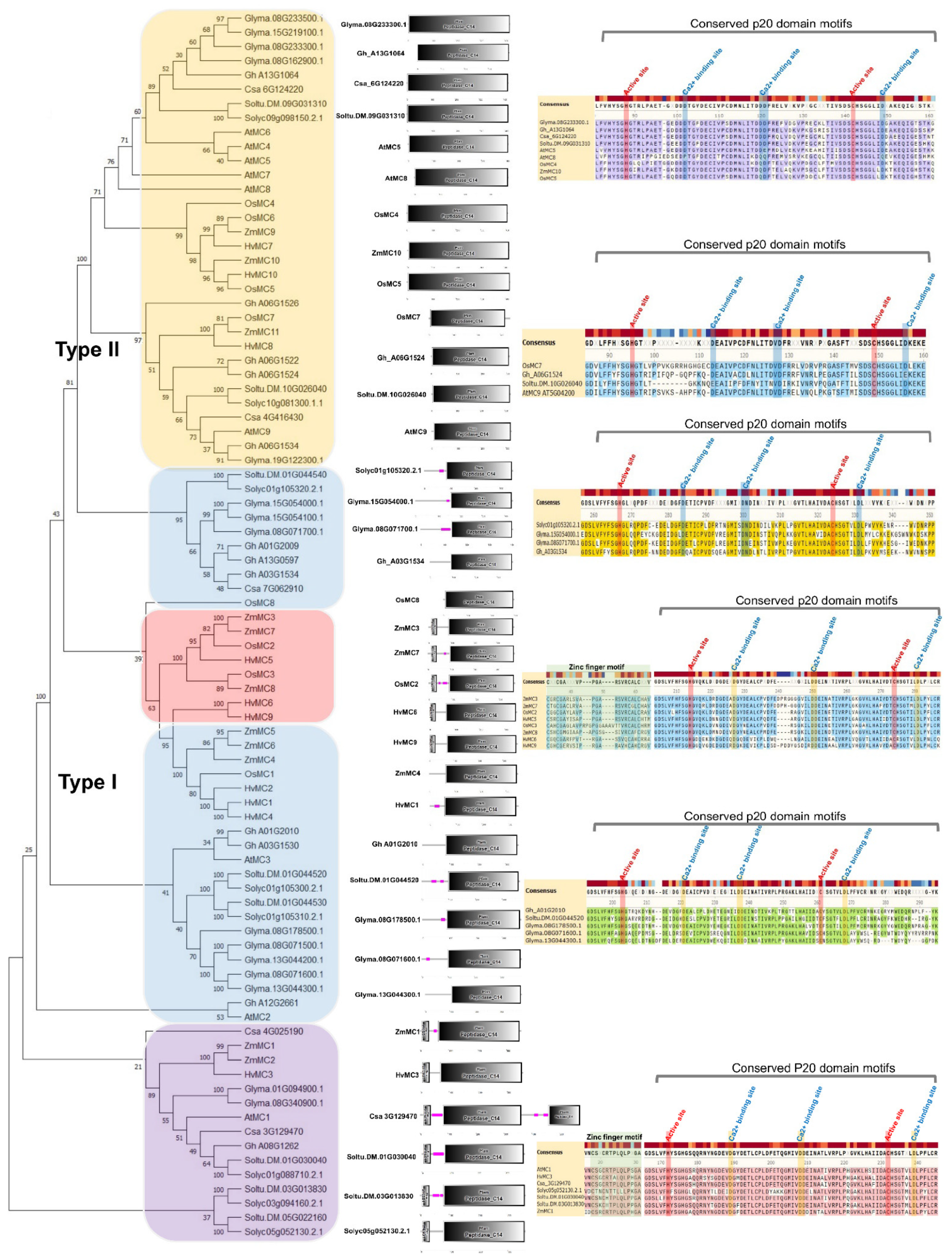
3. Diverse Metacaspase Gene Duplications and Conserved Cysteine Protease Features
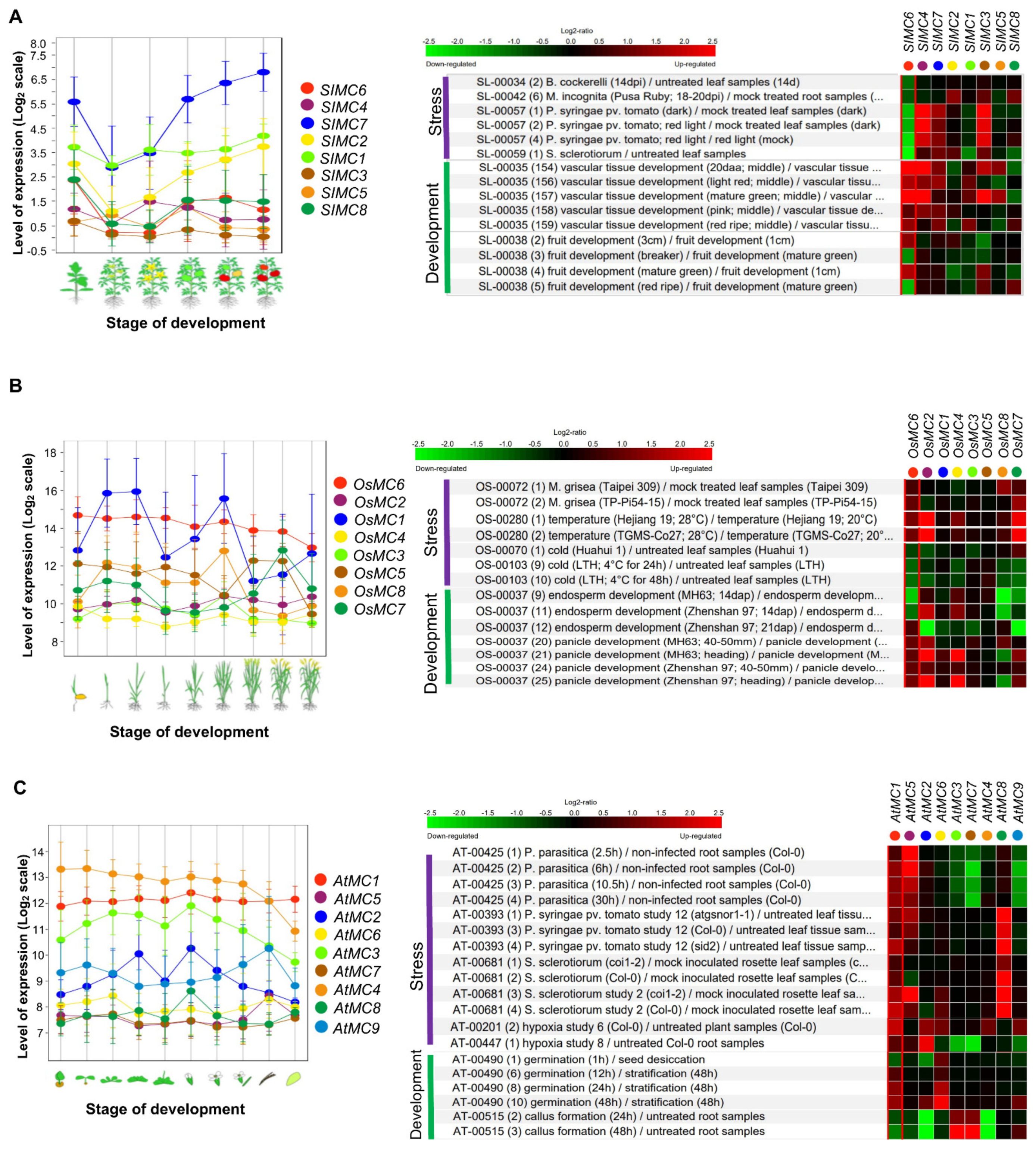
4. Metacaspase Functions in Development, Biotic, and Abiotic Stresses
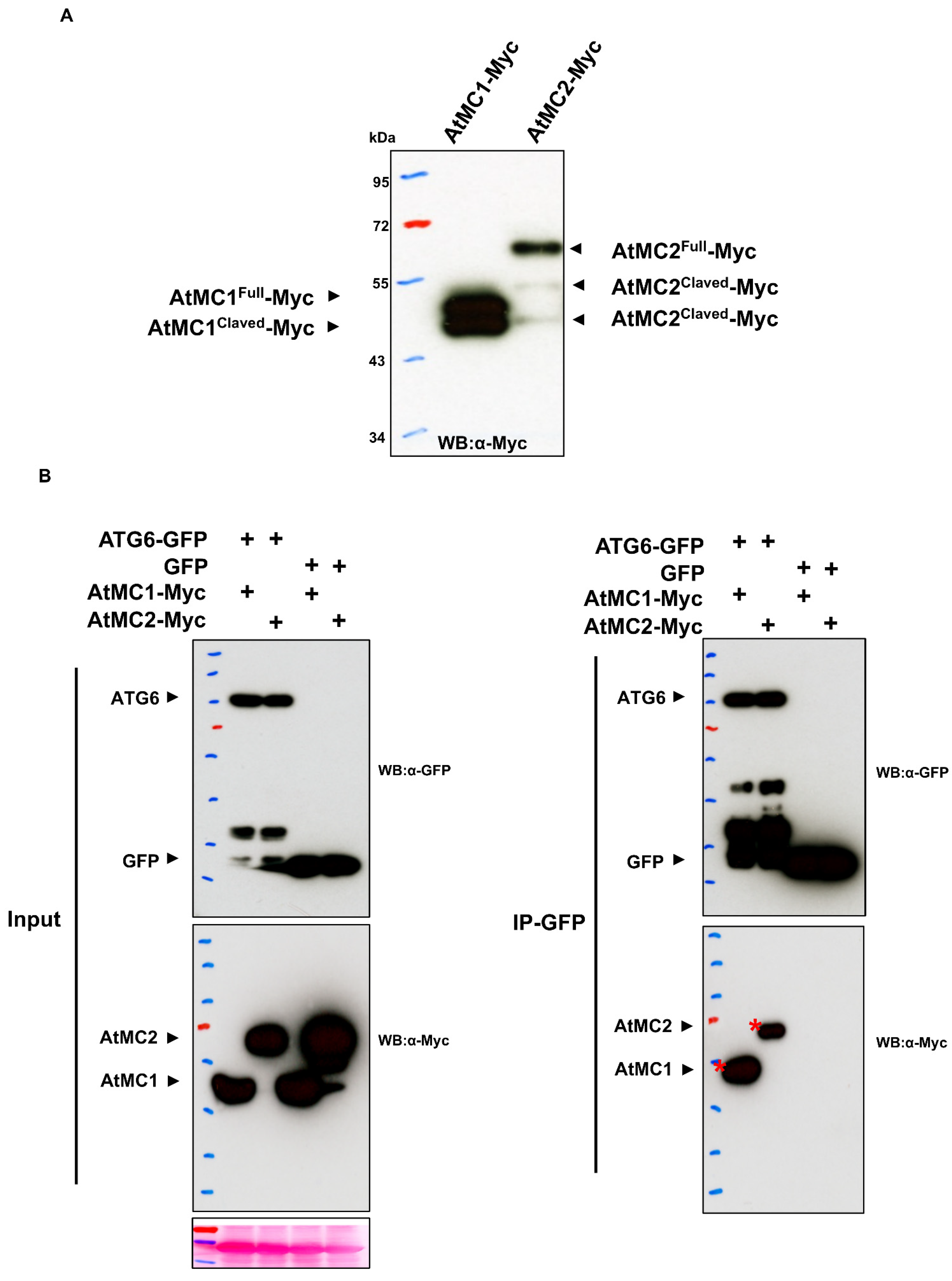
5. Emerging Physical Interactions between Metacaspase and Autophagy
6. Conclusions
Funding
Conflicts of Interest
References
- Galluzzi, L.; Vitale, I.; Abrams, J.M.; Alnemri, E.S.; Baehrecke, E.H.; Blagosklonny, M.V.; Dawson, T.M.; Dawson, V.L.; El-Deiry, W.S.; Fulda, S.; et al. Molecular definitions of cell death subroutines: Recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012, 19, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Alnemri, E.S.; Livingston, D.J.; Nicholson, D.W.; Salvesen, G.; Thornberry, N.A.; Wong, W.W.; Yuan, J. Human ICE/CED-3 protease nomenclature. Cell 1996, 87, 171. [Google Scholar] [CrossRef] [Green Version]
- Man, S.M.; Kanneganti, T.D. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat. Rev. Immunol. 2016, 16, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665. [Google Scholar] [CrossRef]
- Kayagaki, N.; Stowe, I.B.; Lee, B.L.; O’Rourke, K.; Anderson, K.; Warming, S.; Cuellar, T.; Haley, B.; Roose-Girma, M.; Phung, Q.T.; et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015, 526, 666–671. [Google Scholar] [CrossRef]
- Ball, D.P.; Taabazuing, C.Y.; Griswold, A.R.; Orth, E.L.; Rao, S.D.; Kotliar, I.B.; Vostal, L.E.; Johnson, D.C.; Bachovchin, D.A. Caspase-1 interdomain linker cleavage is required for pyroptosis. Life Sci. Alliance 2020, 3. [Google Scholar] [CrossRef] [Green Version]
- Wong, A.H.; Yan, C.; Shi, Y. Crystal structure of the yeast metacaspase Yca1. J. Biol. Chem. 2012, 287, 29251–29259. [Google Scholar] [CrossRef] [Green Version]
- Longo, V.; Zdralevic, M.; Guaragnella, N.; Giannattasio, S.; Zolla, L.; Timperio, A.M. Proteome and metabolome profiling of wild-type and YCA1-knock-out yeast cells during acetic acid-induced programmed cell death. J. Proteom. 2015, 128, 173–188. [Google Scholar] [CrossRef]
- Zdralevic, M.; Longo, V.; Guaragnella, N.; Giannattasio, S.; Timperio, A.M.; Zolla, L. Differential proteome-metabolome profiling of YCA1-knock-out and wild type cells reveals novel metabolic pathways and cellular processes dependent on the yeast metacaspase. Mol. Biosyst. 2015, 11, 1573–1583. [Google Scholar] [CrossRef]
- Chaves, S.R.; Rego, A.; Martins, V.M.; Santos-Pereira, C.; Sousa, M.J.; Corte-Real, M. Regulation of Cell Death Induced by Acetic Acid in Yeasts. Front. Cell Dev. Biol. 2021, 9, 642375. [Google Scholar] [CrossRef]
- Shrestha, A.; Puente, L.G.; Brunette, S.; Megeney, L.A. The role of Yca1 in proteostasis. Yca1 regulates the composition of the insoluble proteome. J. Proteom. 2013, 81, 24–30. [Google Scholar] [CrossRef]
- Fernandez, J.; Lopez, V.; Kinch, L.; Pfeifer, M.A.; Gray, H.; Garcia, N.; Grishin, N.V.; Khang, C.H.; Orth, K. Role of Two Metacaspases in Development and Pathogenicity of the Rice Blast Fungus Magnaporthe oryzae. mBio 2021, 12, e03471-20. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- McLuskey, K.; Mottram, J.C. Comparative structural analysis of the caspase family with other clan CD cysteine peptidases. Biochem. J. 2015, 466, 219–232. [Google Scholar] [CrossRef] [Green Version]
- Uren, A.G.; O’Rourke, K.; Aravind, L.A.; Pisabarro, M.T.; Seshagiri, S.; Koonin, E.V.; Dixit, V.M. Identification of paracaspases and metacaspases: Two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell 2000, 6, 961–967. [Google Scholar] [CrossRef]
- Minina, E.A.; Coll, N.S.; Tuominen, H.; Bozhkov, P.V. Metacaspases versus caspases in development and cell fate regulation. Cell Death Differ. 2017, 24, 1314–1325. [Google Scholar] [CrossRef]
- Zhu, P.; Yu, X.H.; Wang, C.; Zhang, Q.; Liu, W.; McSweeney, S.; Shanklin, J.; Lam, E.; Liu, Q. Structural basis for Ca(2+)-dependent activation of a plant metacaspase. Nat. Commun. 2020, 11, 2249. [Google Scholar] [CrossRef]
- Klemencic, M.; Funk, C. Type III metacaspases: Calcium-dependent activity proposes new function for the p10 domain. New Phytol. 2018, 218, 1179–1191. [Google Scholar] [CrossRef]
- Amcheslavsky, A.; Wang, S.; Fogarty, C.E.; Lindblad, J.L.; Fan, Y.; Bergmann, A. Plasma Membrane Localization of Apoptotic Caspases for Non-apoptotic Functions. Dev. Cell 2018, 45, 450–464.e3. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Zhang, H.; Hong, Y.; Liu, S.; Li, D.; Song, F. Stress-Responsive Expression, Subcellular Localization and Protein-Protein Interactions of the Rice Metacaspase Family. Int. J. Mol. Sci. 2015, 16, 16216–16241. [Google Scholar] [CrossRef] [Green Version]
- Bryan, N.B.; Dorfleutner, A.; Rojanasakul, Y.; Stehlik, C. Activation of inflammasomes requires intracellular redistribution of the apoptotic speck-like protein containing a caspase recruitment domain. J. Immunol. 2009, 182, 3173–3182. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.L.; Zhao, G.; Zhu, W.; Dong, X.M.; Liu, T.; Li, Y.Y.; Song, W.G.; Wang, Y.Q. Herpes simplex virus-1 infection or Simian virus 40-mediated immortalization of corneal cells causes permanent translocation of NLRP3 to the nuclei. Int. J. Ophthalmol. 2015, 8, 46–51. [Google Scholar]
- Mao, P.L.; Jiang, Y.; Wee, B.Y.; Porter, A.G. Activation of caspase-1 in the nucleus requires nuclear translocation of pro-caspase-1 mediated by its prodomain. J. Biol. Chem. 1998, 273, 23621–23624. [Google Scholar] [CrossRef] [Green Version]
- Aries, A.; Whitcomb, J.; Shao, W.; Komati, H.; Saleh, M.; Nemer, M. Caspase-1 cleavage of transcription factor GATA4 and regulation of cardiac cell fate. Cell Death Dis. 2014, 5, e1566. [Google Scholar] [CrossRef] [Green Version]
- He, C.; Cheng, H.; Zhou, R. GATA family of transcription factors of vertebrates: Phylogenetics and chromosomal synteny. J. Biosci. 2007, 32, 1273–1280. [Google Scholar] [CrossRef]
- Reyes, J.C.; Muro-Pastor, M.I.; Florencio, F.J. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004, 134, 1718–1732. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Xi, H.; Park, J. Genome-wide comparative analyses of GATA transcription factors among 19 Arabidopsis ecotype genomes: Intraspecific characteristics of GATA transcription factors. PLoS ONE 2021, 16, e0252181. [Google Scholar] [CrossRef]
- Kim, M.; Xi, H.; Park, S.; Yun, Y.; Park, J. Genome-wide comparative analyses of GATA transcription factors among seven Populus genomes. Sci. Rep. 2021, 11, 16578. [Google Scholar] [CrossRef]
- Manzoor, M.A.; Sabir, I.A.; Shah, I.H.; Wang, H.; Yu, Z.; Rasool, F.; Mazhar, M.Z.; Younas, S.; Abdullah, M.; Cai, Y. Comprehensive Comparative Analysis of the GATA Transcription Factors in Four Rosaceae Species and Phytohormonal Response in Chinese Pear (Pyrus bretschneideri) Fruit. Int. J. Mol. Sci. 2021, 22, 12492. [Google Scholar] [CrossRef]
- Yu, M.; Yu, J.; Cao, H.; Yong, M.; Liu, Y. Genome-wide identification and analysis of the GATA transcription factor gene family in Ustilaginoidea virens. Genome 2019, 62, 807–816. [Google Scholar] [CrossRef]
- Tanaka, H.; Takizawa, Y.; Takaku, M.; Kato, D.; Kumagawa, Y.; Grimm, S.A.; Wade, P.A.; Kurumizaka, H. Interaction of the pioneer transcription factor GATA3 with nucleosomes. Nat. Commun. 2020, 11, 4136. [Google Scholar] [CrossRef]
- Lopez-Cruzan, M.; Sharma, R.; Tiwari, M.; Karbach, S.; Holstein, D.; Martin, C.R.; Lechleiter, J.D.; Herman, B. Caspase-2 resides in the mitochondria and mediates apoptosis directly from the mitochondrial compartment. Cell Death Dis. 2016, 2, 16005. [Google Scholar] [CrossRef] [Green Version]
- Brown-Suedel, A.N.; Bouchier-Hayes, L. Caspase-2 Substrates: To Apoptosis, Cell Cycle Control, and Beyond. Front. Cell Dev. Biol. 2020, 8, 610022. [Google Scholar] [CrossRef]
- McKenzie, B.A.; Fernandes, J.P.; Doan, M.A.L.; Schmitt, L.M.; Branton, W.G.; Power, C. Activation of the executioner caspases-3 and -7 promotes microglial pyroptosis in models of multiple sclerosis. J. Neuroinflam. 2020, 17, 253. [Google Scholar] [CrossRef]
- Lee, R.E.; Brunette, S.; Puente, L.G.; Megeney, L.A. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc. Natl. Acad. Sci. USA 2010, 107, 13348–13353. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, A.; Brunette, S.; Stanford, W.L.; Megeney, L.A. The metacaspase Yca1 maintains proteostasis through multiple interactions with the ubiquitin system. Cell Discov. 2019, 5, 6. [Google Scholar] [CrossRef]
- Coll, N.S.; Smidler, A.; Puigvert, M.; Popa, C.; Valls, M.; Dangl, J.L. The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: Functional linkage with autophagy. Cell Death Differ. 2014, 21, 1399–1408. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Lv, Y.; Zhou, Z.; Mei, F.; Wang, L. Type II metacaspase protein localization and gene transcription during programmed cell semi-death of sieve elements in developing caryopsis of Tritium aestivum. Biologia 2017, 72, 398–406. [Google Scholar] [CrossRef]
- Gong, P.; Riemann, M.; Dong, D.; Stoeffler, N.; Gross, B.; Markel, A.; Nick, P. Two grapevine metacaspase genes mediate ETI-like cell death in grapevine defence against infection of Plasmopara viticola. Protoplasma 2019, 256, 951–969. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Lam, E. Calcium-dependent activation and autolysis of Arabidopsis metacaspase 2d. J. Biol. Chem. 2011, 286, 10027–10040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, N.; Lam, E. Arabidopsis metacaspase 2d is a positive mediator of cell death induced during biotic and abiotic stresses. Plant J. 2011, 66, 969–982. [Google Scholar] [CrossRef]
- Luan, Q.L.; Zhu, Y.X.; Ma, S.; Sun, Y.; Liu, X.Y.; Liu, M.; Balint-Kurti, P.J.; Wang, G.F. Maize metacaspases modulate the defense response mediated by the NLR protein Rp1-D21 likely by affecting its subcellular localization. Plant J. 2021, 105, 151–166. [Google Scholar] [CrossRef]
- Liu, H.; Liu, J.; Wei, Y. Identification and analysis of the metacaspase gene family in tomato. Biochem. Biophys. Res. Commun. 2016, 479, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Dubey, N.; Trivedi, M.; Varsani, S.; Vyas, V.; Farsodia, M.; Singh, S.K. Genome-wide characterization, molecular evolution and expression profiling of the metacaspases in potato (Solanum tuberosum L.). Heliyon 2019, 5, e01162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapin, L.J.; Moon, Y.; Jones, M.L. Downregulating a Type I Metacaspase in Petunia Accelerates Flower Senescence. J. Am. Soc. Hortic. Sci. 2017, 142, 405–414. [Google Scholar] [CrossRef] [Green Version]
- Choi, C.J.; Berges, J.A. New types of metacaspases in phytoplankton reveal diverse origins of cell death proteases. Cell Death Dis. 2013, 4, e490. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Fan, S.; Liu, A.; Zhang, Z.; Zou, X.; Jiang, X.; Huang, J.; Fan, L.; Zhang, Z.; Deng, X.; Ge, Q.; et al. Genome-Wide Identification and Expression Analysis of the Metacaspase Gene Family in Gossypium Species. Genes 2019, 10, 527. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Gong, P.; Wei, R.; Li, S.; Zhang, X.; Yu, Y.; Wang, Y. The metacaspase gene family of Vitis vinifera L.: Characterization and differential expression during ovule abortion in stenospermocarpic seedless grapes. Gene 2013, 528, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, H. Genomewide survey and characterization of metacaspase gene family in rice (Oryza sativa). J. Genet. 2014, 93, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.H.; Ok, S.H.; Yoo, K.S.; Jung, K.W.; Yoo, S.D.; Shin, J.S. An Arabidopsis cell growth defect factor-related protein, CRS, promotes plant senescence by increasing the production of hydrogen peroxide. Plant Cell Physiol. 2013, 54, 155–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawai-Yamada, M.; Saito, Y.; Jin, L.; Ogawa, T.; Kim, K.M.; Yu, L.H.; Tone, Y.; Hirata, A.; Umeda, M.; Uchimiya, H. A novel Arabidopsis gene causes Bax-like lethality in Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 39468–39473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yusof, N.Y.M.; Saparin, N.F.; Ahmad Seman, Z.; Ab Rahman, Z.; Sew, Y.S.; Shaharuddin, N.A.; Roslan, M.A.M.; Abdul Rahman, N.A.; Gallois, P.; Sobri, Z.M. Overexpression of Rice Metacaspase, OsMC4, Increases Endoplasmic Reticulum Stress Tolerance in Transgenic Rice Calli. Preprints 2021, 2021, 2021020361. [Google Scholar] [CrossRef]
- Wang, S.; Xue, M.; He, C.; Shen, D.; Jiang, C.; Zhao, H.; Niu, D. AtMC1 Associates With LSM4 to Regulate Plant Immunity Through Modulating Pre-mRNA Splicing. Mol. Plant Microbe Interact. 2021, 34, 1423–1432. [Google Scholar] [CrossRef]
- Coll, N.S.; Vercammen, D.; Smidler, A.; Clover, C.; Van Breusegem, F.; Dangl, J.L.; Epple, P. Arabidopsis type I metacaspases control cell death. Science 2010, 330, 1393–1397. [Google Scholar] [CrossRef]
- Patel, S.; Wall, D.M.; Castillo, A.; McCormick, B.A. Caspase-3 cleavage of Salmonella type III secreted effector protein SifA is required for localization of functional domains and bacterial dissemination. Gut Microbes 2019, 10, 172–187. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Liu, W.; Fu, J.; Cheng, S.; Xu, Y.; Wang, Z.; Liu, X.; Shi, X.; Liu, Y.; Qi, X.; et al. Shigella evades pyroptosis by arginine ADP-riboxanation of caspase-11. Nature 2021, 599, 290–295. [Google Scholar] [CrossRef]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef]
- Wu, H.; Che, X.; Zheng, Q.; Wu, A.; Pan, K.; Shao, A.; Wu, Q.; Zhang, J.; Hong, Y. Caspases: A molecular switch node in the crosstalk between autophagy and apoptosis. Int. J. Biol. Sci. 2014, 10, 1072–1083. [Google Scholar] [CrossRef]
- Tsapras, P.; Nezis, I.P. Caspase involvement in autophagy. Cell Death Differ. 2017, 24, 1369–1379. [Google Scholar] [CrossRef] [Green Version]
- Gu, Q.; Jiao, S.; Duan, K.; Wang, Y.X.; Petralia, R.S.; Li, Z. The BAD-BAX-Caspase-3 Cascade Modulates Synaptic Vesicle Pools via Autophagy. J. Neurosci. 2021, 41, 1174–1190. [Google Scholar] [CrossRef]
- Norman, J.M.; Cohen, G.M.; Bampton, E.T. The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy 2010, 6, 1042–1056. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.; Xu, Y.; Wan, W.; Shou, X.; Qian, J.; You, Z.; Liu, B.; Chang, C.; Zhou, T.; Lippincott-Schwartz, J.; et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 2015, 57, 456–466. [Google Scholar] [CrossRef] [Green Version]
- Sancho, A.; Duran, J.; Garcia-Espana, A.; Mauvezin, C.; Alemu, E.A.; Lamark, T.; Macias, M.J.; DeSalle, R.; Royo, M.; Sala, D.; et al. DOR/Tp53inp2 and Tp53inp1 constitute a metazoan gene family encoding dual regulators of autophagy and transcription. PLoS ONE 2012, 7, e34034. [Google Scholar] [CrossRef]
- Ivanova, S.; Polajnar, M.; Narbona-Perez, A.J.; Hernandez-Alvarez, M.I.; Frager, P.; Slobodnyuk, K.; Plana, N.; Nebreda, A.R.; Palacin, M.; Gomis, R.R.; et al. Regulation of death receptor signaling by the autophagy protein TP53INP2. EMBO J. 2019, 38, e99300. [Google Scholar] [CrossRef]
- Mnich, K.; Koryga, I.; Pakos-Zebrucka, K.; Thomas, M.; Logue, S.E.; Eriksson, L.A.; Gorman, A.M.; Samali, A. The stressosome, a caspase-8-activating signalling complex assembled in response to cell stress in an ATG5-mediated manner. J. Cell. Mol. Med. 2021, 25, 8809–8820. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, S.; Zhang, Y.; Lin, X.; Song, Y.; Xue, Z.; Qian, H.; Wang, S.; Wan, G.; Zheng, X.; et al. Induction of autophagy by spermidine is neuroprotective via inhibition of caspase 3-mediated Beclin 1 cleavage. Cell Death Dis. 2017, 8, e2738. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhao, L.; Liu, L.; Gao, P.; Tian, W.; Wang, X.; Jin, H.; Xu, H.; Chen, Q. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell 2010, 1, 468–477. [Google Scholar] [CrossRef] [Green Version]
- Berenguer, E.; Minina, E.A.; Carneros, E.; Bárány, I.; Bozhkov, P.V.; Testillano, P.S. Suppression of Metacaspase- and Autophagy-Dependent Cell Death Improves Stress-Induced Microspore Embryogenesis in Brassica napus. Plant Cell Physiol. 2021, 61, 2097–2110. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Wang, S.; Han, S.; Xie, K.; Wang, Y.; Li, J.; Liu, Y. Plant Bax Inhibitor-1 interacts with ATG6 to regulate autophagy and programmed cell death. Autophagy 2017, 13, 1161–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.; Jones, M.L. Silencing ATG6 and PI3K accelerates petal senescence and reduces flower number and shoot biomass in petunia. Plant Sci. 2021, 302, 110713. [Google Scholar] [CrossRef] [PubMed]
- Vesela, B.; Svandova, E.; Ramesova, A.; Kratochvilova, A.; Tucker, A.S.; Matalova, E. Caspase Inhibition Affects the Expression of Autophagy-Related Molecules in Chondrocytes. Cartilage 2021, 13 (Suppl. S2), 956S–968S. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huh, S.U. Evolutionary Diversity and Function of Metacaspases in Plants: Similar to but Not Caspases. Int. J. Mol. Sci. 2022, 23, 4588. https://doi.org/10.3390/ijms23094588
Huh SU. Evolutionary Diversity and Function of Metacaspases in Plants: Similar to but Not Caspases. International Journal of Molecular Sciences. 2022; 23(9):4588. https://doi.org/10.3390/ijms23094588
Chicago/Turabian StyleHuh, Sung Un. 2022. "Evolutionary Diversity and Function of Metacaspases in Plants: Similar to but Not Caspases" International Journal of Molecular Sciences 23, no. 9: 4588. https://doi.org/10.3390/ijms23094588





