Isolation of Potato Endophytes and Screening of Chaetomium globosum Antimicrobial Genes
Abstract
:1. Introduction
2. Results
2.1. Isolation, Purification, and Screening of Potato Endophytes
2.2. Identification of Endophytic Fungus F5
2.3. Screening of Antibacterial Genes from C. globosum cDNA Library by B. subtilis System
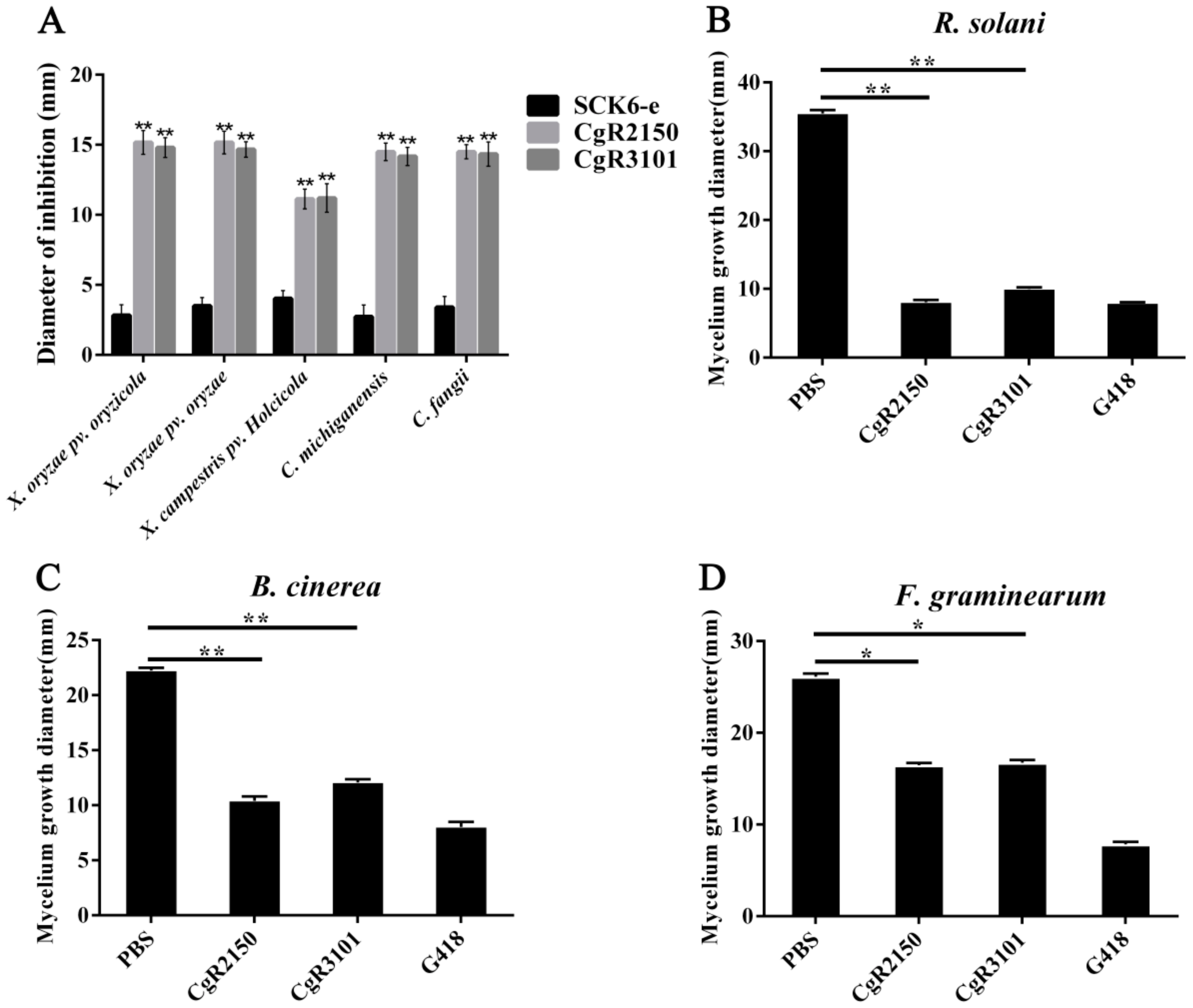
2.4. Antibacterial Activities of Antimicrobial Genes against Pathogenic Bacteria and Fungi
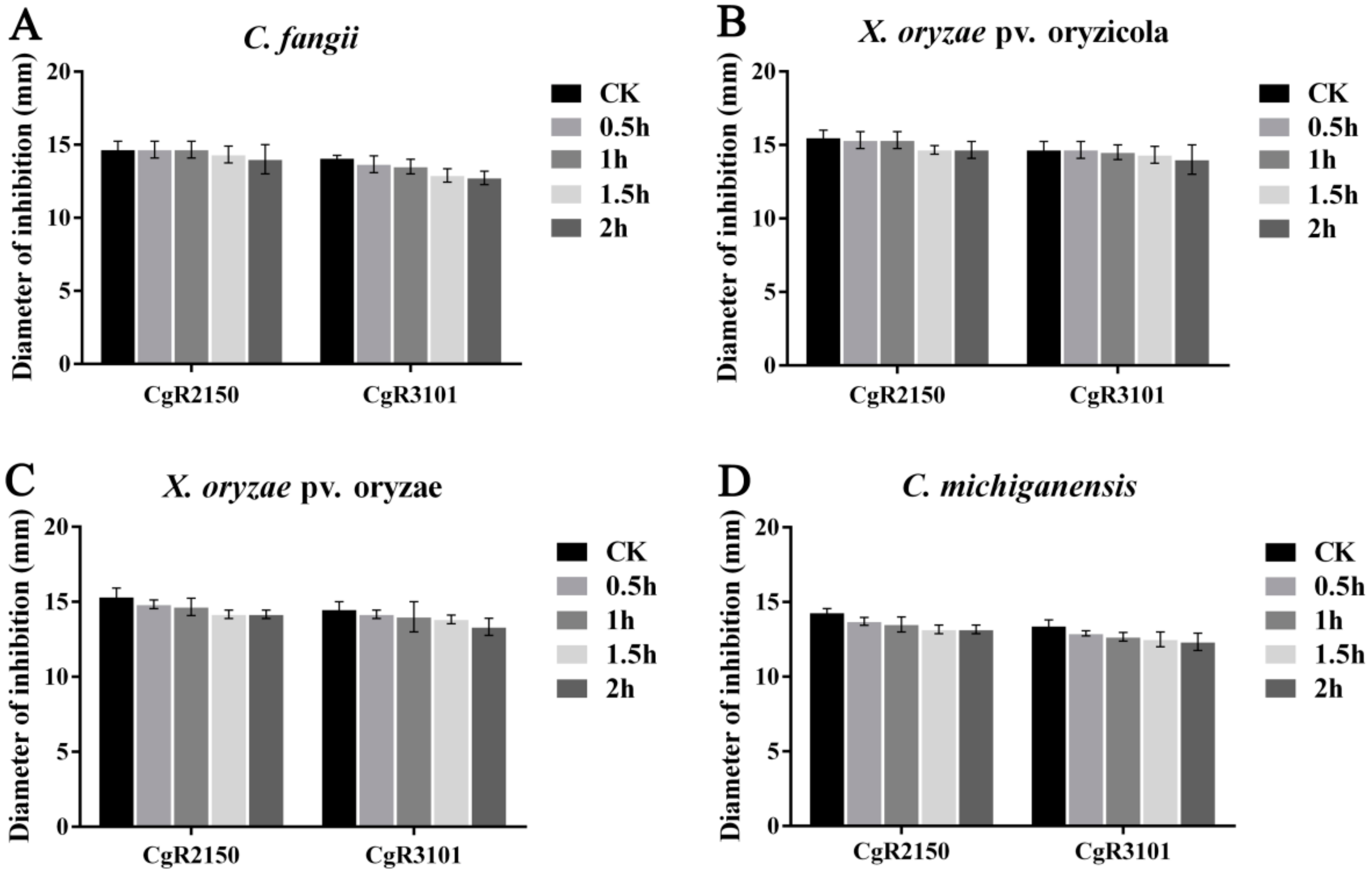
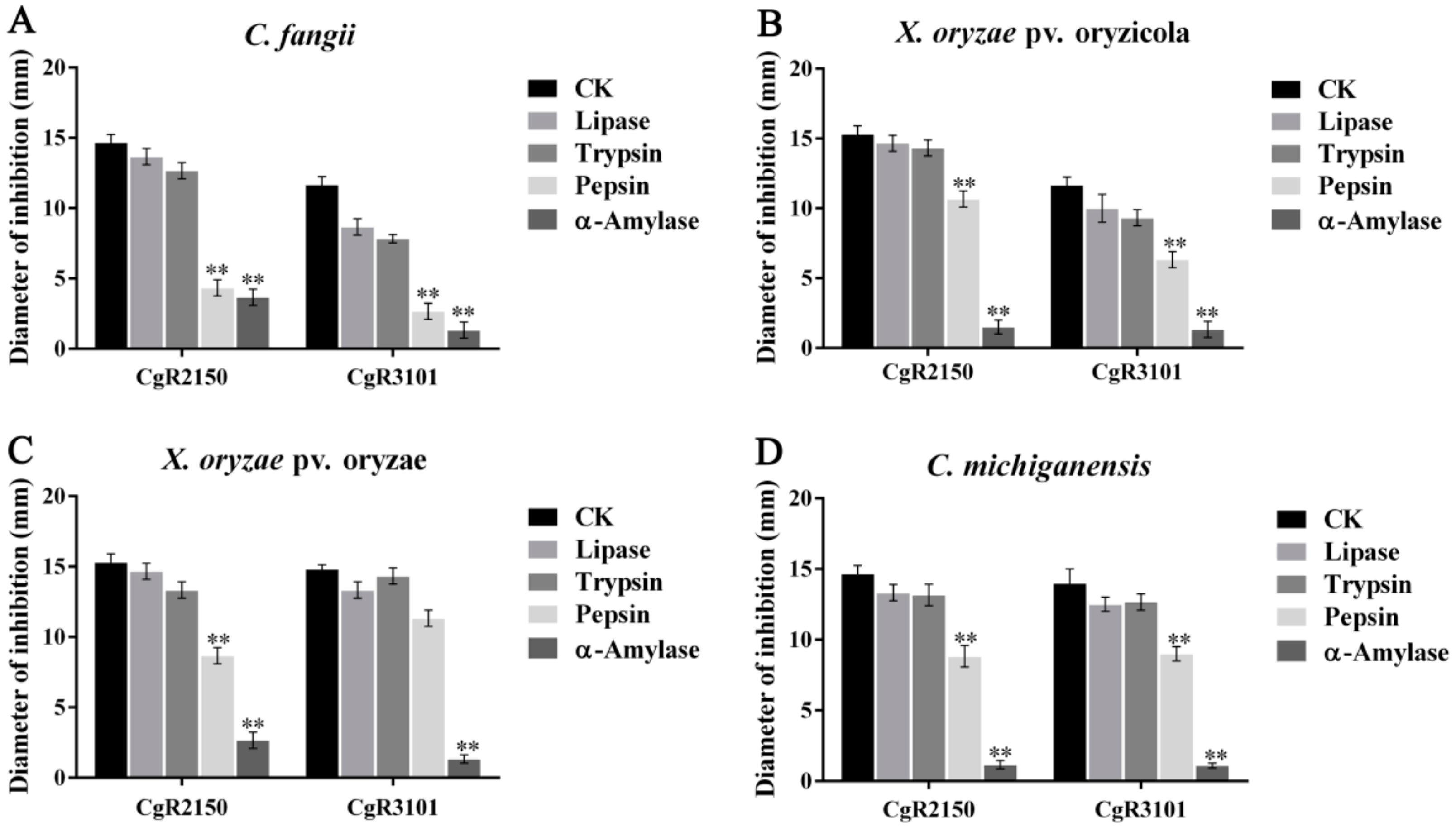
2.5. Effects of UV, Temperature, and Enzymes on Antibacterial Activities of Antimicrobial Peptides
2.6. SDS-PAGE Analysis and Western Blot Reveal the Expression of Antimicrobial Peptides
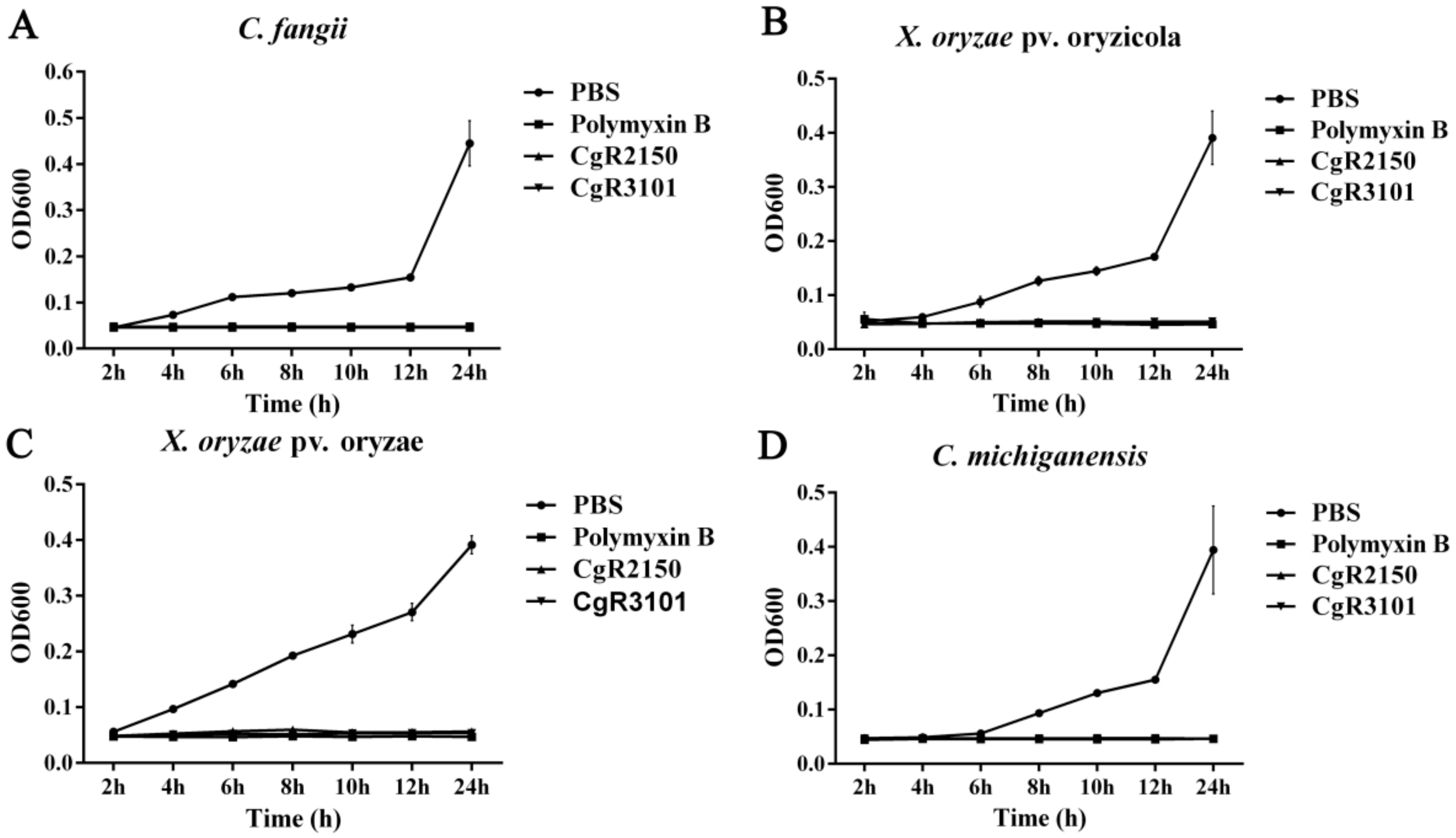
2.7. Minimum Inhibitory Concentration (MIC) Assay for Peptides CgR2150 and CgR3101
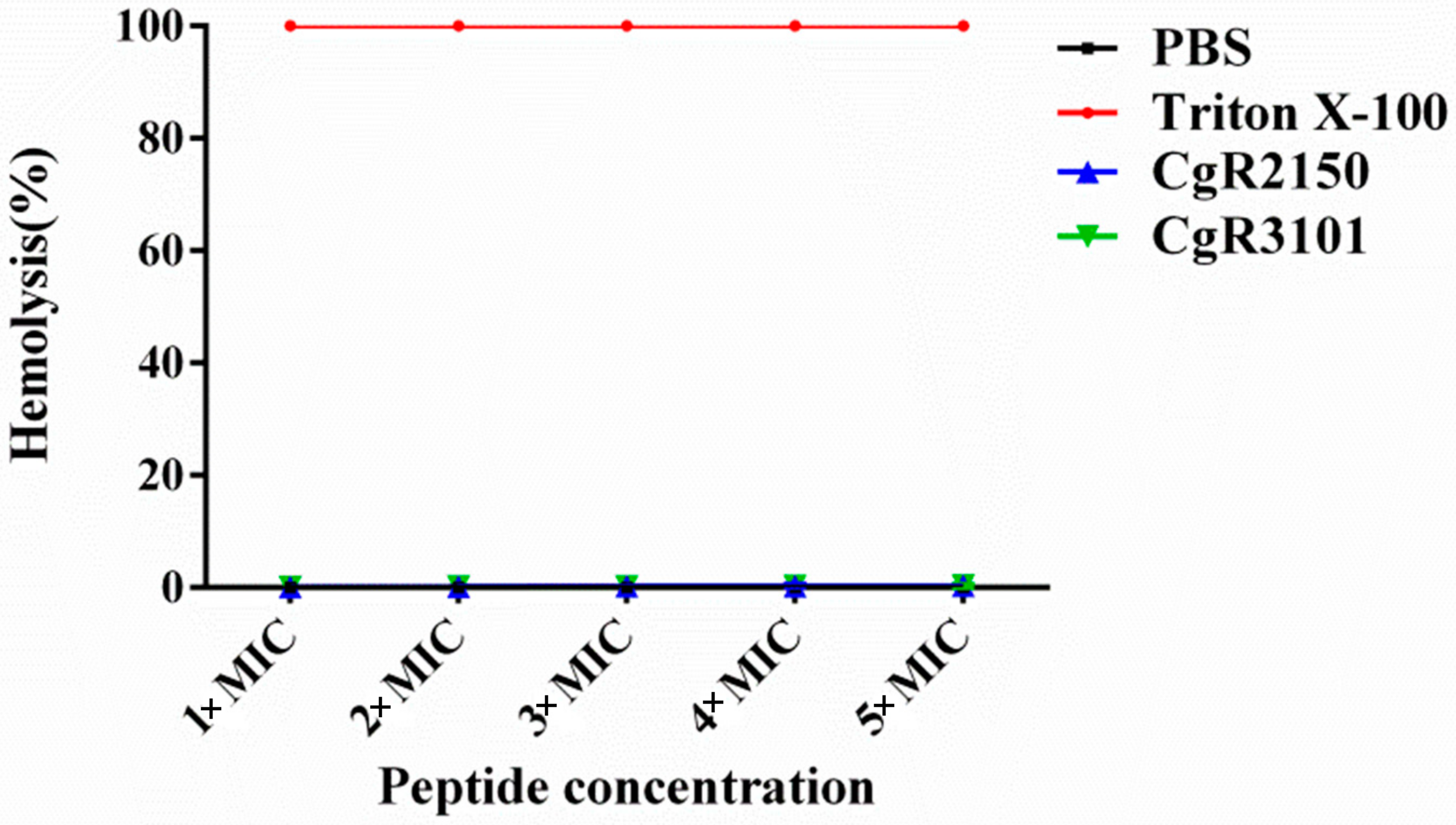
2.8. Hemolytic Activity Assay of CgR2150 and CgR3101 on Mammalian Cells
2.9. Destructive Effects of CgR2150 and CgR3101 on X. oryzae pv. oryzae and C. michiganensis
3. Discussion
4. Materials and Methods
4.1. Isolation, Purification, and Screening of Endophytic Fungi
4.2. Identification of Endophytic Fungi
4.3. Activation of C. globosum
4.4. Construction of C. globosum cDNA Library
4.5. Screening of Candidate Resistance Genes
4.6. Expression of Extracellular Peptides
4.7. Determination of the Inhibitory Effect of Antimicrobial Peptides on Pathogenic Bacteria and Fungi
4.8. Stability Determination of Antimicrobial Peptides
4.9. Purification of His-Tag Fusion Peptides
4.10. Tris-Tricine SDS-PAGE and Western Blotting
4.11. Determination of the Minimum Inhibitory Concentration (MIC)
4.12. Hemolytic Assay
4.13. Scanning Electron Microscope Analysis
4.14. Pathogenicity Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dzhioev, Y.P.; Zlobin, V.I.; Salovarova, V.P.; Stepanenko, L.A.; Bukinet, Y.S. Analysis of the “superbacteria” issue and contemporary approaches to its solution. Proc. Univ. Appl. Chem. Biotechnol. 2019, 9, 665–678. [Google Scholar] [CrossRef]
- Subramaniam, G.; Girish, M. Antibiotic Resistance—A Cause for Reemergence of Infections. Indian J. Pediatr. 2020, 87, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Carter, V.; Underhill, A.; Baber, I.; Sylla, L.; Baby, M.; Larget-Thiery, I.; Zettor, A.; Bourgouin, C.; Langel, U.; Faye, I.; et al. Killer bee molecules: Antimicrobial peptides as effector molecules to target sporogonic stages of Plasmodium. PLoS Pathog. 2013, 9, e1003790. [Google Scholar] [CrossRef] [Green Version]
- Marcocci, M.E.; Amatore, D.; Villa, S.; Casciaro, B.; Aimola, P.; Franci, G.; Grieco, P.; Galdiero, M.; Palamara, A.T.; Mangoni, M.L.; et al. The Amphibian Antimicrobial Peptide Temporin B Inhibits In Vitro Herpes Simplex Virus 1 Infection. Antimicrob. Agents Chemother. 2018, 62, e02367-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, M.S.; Mahmud, S.; Sultana, R.; Dong, W. Identification and in silico molecular model-ling study of newly isolated Bacillus subtilis SI-18 strain against S9 protein of Rhizoctonia solani. Arab. J. Chem. 2020, 13, 8600–8612. [Google Scholar] [CrossRef]
- Martin, E.; Ganz, T.; Lehrer, R.I. Defensins and other endogenous peptide antibiotics of vertebrates. J. Leukoc. Biol. 1995, 58, 128–136. [Google Scholar] [CrossRef]
- Franco, O.L.; Murad, A.M.; Leite, J.R.; Mendes, P.A.; Prates, M.V.; Bloch, C. Identification of a cowpea gamma-thionin with bactericidal activity. FEBS J. 2006, 273, 489–497. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Boparai, J.K.; Sharma, P.K. Mini Review on Antimicrobial Peptides, Sources, Mechanism and Recent Applications. Protein Pept. Lett. 2020, 27, 4–16. [Google Scholar] [CrossRef]
- Diamond, G.; Beckloff, N.; Weinberg, A.; Kisich, K.O. The roles of antimicrobial peptides in innate host defense. Curr. Pharm. Des. 2009, 15, 2377–2392. [Google Scholar] [CrossRef] [Green Version]
- Sarika; Iquebal, M.A.; Rai, A. Biotic stress resistance in agriculture through antimicrobial peptides. Peptides 2012, 36, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Bruni, N.; Capucchio, M.T.; Biasibetti, E.; Pessione, E.; Cirrincione, S.; Giraudo, L.; Corona, A.; Dosio, F. Antimicrobial Activity of Lactoferrin-Related Peptides and Applications in Human and Veterinary Medicine. Molecules 2016, 21, 752. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zeng, X.; Yang, Q.; Qiao, S. Antimicrobial Peptides as Potential Alternatives to Antibiotics in Food Animal Industry. Int. J. Mol. Sci. 2016, 17, 603. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Cheng, L.; Zhang, S.; Wang, L.; Hu, J. The role of natural antimicrobial peptides during infection and chronic inflammation. Antonie Van Leeuwenhoek 2018, 111, 5–26. [Google Scholar] [CrossRef]
- Cho, J.; Hwang, I.S.; Choi, H.; Hwang, J.H.; Hwang, J.S.; Lee, D.G. The novel biological action of antimicrobial peptides via apoptosis induction. J. Microbiol. Biotechnol. 2012, 22, 1457–1466. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.H.; Hall, K.N.; Aguilar, M.I. Antimicrobial Peptide Structure and Mechanism of Action: A Focus on the Role of Membrane Structure. Curr. Top. Med. Chem. 2016, 16, 25–39. [Google Scholar] [CrossRef]
- Porowińska, D.; Wujak, M.; Roszek, K.; Komoszyński, M. Prokariotyczne systemy ekspresyjne [Prokaryotic expression systems]. Postepy Hig. Med. Dosw. 2013, 67, 119–129. [Google Scholar] [CrossRef]
- Rhayat, L.; Maresca, M.; Nicoletti, C.; Perrier, J.; Brinch, K.S.; Christian, S.; Devillard, E.; Eckhardt, E. Effect of Bacillus subtilis Strains on Intestinal Barrier Function and Inflammatory Response. Front. Immunol. 2019, 10, 564. [Google Scholar] [CrossRef]
- Dijl, J.M.; Hecker, M. Bacillus subtilis: From soil bacterium to super-secreting cell factory. Microb. Cell Factoris 2013, 12, 3. [Google Scholar] [CrossRef] [Green Version]
- Fu, T.; Islam, M.S.; Ali, M.; Wu, J.; Dong, W. Two antimicrobial genes from Aegilops tauschii Cosson identified by the Bacillus subtilis expression system. Sci. Rep. 2020, 10, 13346. [Google Scholar] [CrossRef]
- Islam, M.S.; Mohamed, G.; Polash, S.A.; Hasan, M.A.; Sultana, R.; Saiara, N.; Dong, W. Antimicrobial Peptides from Plants: A cDNA-Library Based Isolation, Purification, Characterization Approach and Elucidating Their Modes of Action. Int. J. Mol. Sci. 2021, 22, 8712. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Abbas, H.M.; Li, J.; Yuan, Y.; Liu, Y.; Wang, G.; Dong, W. Cell Membrane-Interrupting Antimicrobial Peptides from Isatis indigotica Fortune Isolated by a Bacillus subtilis Expression System. Biomolecules 2019, 10, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Islam, S.; Guo, P.; Hu, X.; Dong, W. Isolation of Antimicrobial Genes from Oryza rufipogon Griff by Using a Bacillus subtilis Expression System with Potential Antimicrobial Activities. Int. J. Mol. Sci. 2020, 21, 8722. [Google Scholar] [CrossRef]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef]
- Aravind, R.; Kumar, A.; Eapen, S.J.; Ramana, K.V. Endophytic bacterial flora in root and stem tissues of black pepper (Piper nigrum L.) genotype: Isolation, identification and evaluation against Phytophthora capsici. Lett. Appl. Microbiol. 2009, 48, 58–64. [Google Scholar] [CrossRef]
- Sheoran, N.; Valiya, N.A.; Munjal, V.; Kundu, A.; Subaharan, K.; Venugopal, V.; Rajamma, S.; Eapen, S.J.; Kumar, A. Genetic analysis of plant endophytic Pseudomonas putida BP25 and chemo-profiling of its antimicrobial volatile organic compounds. Microbiol. Res. 2015, 173, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Luginbühl, M.; Müller, E. Endophytische Pilze in den oberirdischen organen von vier gemeinsam an gleichen Standorten wachsenden Pflanzen (Buxus, Hedera, Ilex, Ruscus). Sydowia. 1980, 33, 185–209. [Google Scholar]
- Kanokmedhakul, S.; Kanokmedhakul, K.; Phonkerd, N.; Soytong, K.; Kongsaeree, P.; Suksamrarn, A. Antimycobacterial anthraquinone-chromanone compound and diketopiperazine alkaloid from the fungus Chaetomium globosum KMITL-N0802. Planta Med. 2002, 68, 834–836. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.S.; Zhang, Y.Y.; Yan, W.; Cao, L.L.; Xiao, Y.; Ye, Y.H. Chaetomium globosum CDW7, a potential biological control strain and its antifungal metabolites. FEMS Microbiol. Lett. 2017, 364, fnw287. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Xu, Y.; Gao, Y.; Huang, Q.; Luo, X.; An, H.; Dong, J. Chemical and bioactive diversities of the genera Stachybotrys and Memnoniella secondary metabolites. Phytochem. Rev. 2014, 14, 623–655. [Google Scholar] [CrossRef]
- Khan, B.; Yan, W.; Wei, S.; Wang, Z.; Zhao, S.; Cao, L.; Rajput, N.A.; Ye, Y. Nematicidal metabolites from endophytic fungus Chaetomium globosum YSC5. FEMS Microbiol. Lett. 2019, 366, fnz169. [Google Scholar] [CrossRef]
- Qi, J.; Jiang, L.; Zhao, P.; Chen, H.; Jia, X.; Zhao, L.; Dai, H.; Hu, J.; Liu, C.; Shim, S.H.; et al. Chaetoglobosins and azaphilones from Chaetomium globosum associated with Apostichopus japonicus. Appl. Microbiol. Biotechnol. 2020, 104, 1545–1553. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Yang, M.; Abbas, H.M.; Wu, J.; Li, M.; Dong, W. Antimicrobial genes from Allium sativum and Pinellia ternata revealed by a Bacillus subtilis expression system. Sci. Rep. 2018, 8, 14514. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.M.; Xiang, J.; Ahmad, Z.; Wang, L.; Dong, W. Enhanced Nicotiana benthamiana immune responses caused by heterologous plant genes from Pinellia ternata. BMC Plant Biol. 2018, 18, 57. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.F.; Qutb, A.M.; Dong, W. Prediction and Activity of a Cationic α-Helix Antimicrobial Peptide ZM-804 from Maize. Int. J. Mol. Sci. 2021, 22, 2643. [Google Scholar] [CrossRef]
- Hallmann, J.; Quadt-Hallmann, A.; Mahaffee, W.; Kloepper, J. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997, 43, 895–914. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Hurek, T. Living inside plants: Bacterial endophytes. Curr. Opin. Plant Biol. 2011, 14, 435–443. [Google Scholar] [CrossRef]
- Singh, L.P.; Gill, S.S.; Tuteja, N. Unraveling the role of fungal symbionts in plant abiotic stress tolerance. Plant Signal. Behav. 2011, 6, 175–191. [Google Scholar] [CrossRef] [Green Version]
- Rosenblueth, M.; Martínez-Romero, E. Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 2006, 19, 827–837. [Google Scholar] [CrossRef] [Green Version]
- Senthilkumar, M.; Anandham, R.; Madhaiyan, M.; Venkateswaran, V.; Sa, T. Endophytic Bacteria: Perspectives and Applications in Agricultural Crop Production; Springer: Berlin/Heidelberg, Germany, 2011; pp. 61–96. [Google Scholar] [CrossRef]
- Santoyo, G.; Moreno-Hagelsieb, G.; Orozco-Mosqueda, M.C.; Glick, B.R. Plant growth-promoting bacterial endophytes. Res. Microbiol. 2016, 183, 92–99. [Google Scholar] [CrossRef]
- Khare, E.; Mishra, J.; Arora, N.K. Multifaceted Interactions Between Endophytes and Plant: Developments and Prospects. Front. Microbiol. 2018, 9, 2732. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Ahmad, Z.; Ashraf, M.F.; Dong, W. Maize endophytic microbial-communities revealed by removing PCR and 16S rRNA sequencing and their synthetic applications to suppress maize banded leaf and sheath blight. Res. Microbiol. 2021, 242, 126639. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, Z.; Wu, J.; Chen, L.; Dong, W. Isolated Bacillus subtilis strain 330-2 and its antagonistic genes identified by the removing PCR. Sci. Rep. 2017, 7, 1777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Res. Microbiol. 2019, 221, 36–49. [Google Scholar] [CrossRef]
- Mitter, B.; Brader, G.; Pfaffenbichler, N.; Sessitsch, A. Next generation microbiome applications for crop production-limitations and the need of knowledge-based solutions. Curr. Opin. Microbiol. 2019, 49, 59–65. [Google Scholar] [CrossRef]
- Yan, L.; Zhu, J.; Zhao, X.; Shi, J.; Jiang, C.; Shao, D. Beneficial effects of endophytic fungi colonization on plants. Appl. Microbiol. Biotechnol. 2019, 103, 3327–3340. [Google Scholar] [CrossRef]
- Kushwaha, P.; Kashyap, P.L.; Bhardwaj, A.K.; Kuppusamy, P.; Srivastava, A.K.; Tiwari, R.K. Bacterial endophyte mediated plant tolerance to salinity: Growth responses and mechanisms of action. World J. Microbiol. Biotechnol. 2020, 36, 26. [Google Scholar] [CrossRef]
- Awad, N.E.; Kassem, H.A.; Hamed, M.A.; El-Naggar, M.A.; El-Feky, A.M. Bioassays guided isolation of compounds from Chaetomium globosum. J. Mycol. Med. 2014, 24, e35–e42. [Google Scholar] [CrossRef]
- Aminov, R.I. The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 2009, 11, 2970–2988. [Google Scholar] [CrossRef]
- Alexander, J.L.; Thompson, Z.; Cowan, J.A. Antimicrobial Metallopeptides. ACS Chem. Biol. 2018, 13, 844–853. [Google Scholar] [CrossRef]
- Fry, D.E. Antimicrobial Peptides. Surg. Infect. 2018, 19, 804–811. [Google Scholar] [CrossRef] [PubMed]
- Kampshoff, F.; Willcox, M.D.; Dutta, D. A Pilot Study of the Synergy between Two Antimicrobial Peptides and Two Common Antibiotics. Antibiotics 2019, 8, 60. [Google Scholar] [CrossRef] [Green Version]
- Lewies, A.; Plessis, L.H.; Wentzel, J.F. Antimicrobial Peptides: The Achilles’ Heel of Antibiotic Resistance? Probiotics Antimicrob. Proteins 2019, 11, 370–381. [Google Scholar] [CrossRef]
- Ho, K.M.; Lim, B.L. Co-expression of a prophage system and a plasmid system in Bacillus subtilis. Protein Expr. Purif. 2003, 32, 293–301. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.; Hsueh, Y.H.; Cheng, Y.; Wu, C.; Liu, S. Water surface tension modulates the swarming mechanics of Bacillus subtilis. Front. Microbiol. 2015, 12, 1017–1029. [Google Scholar] [CrossRef] [Green Version]
- Lipsky, B.A. Bone of contention: Diagnosing diabetic foot osteomyelitis. Clin. Infect. Dis. 2008, 47, 528–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velden, W.J.; Iersel, T.M.; Blijlevens, N.M.; Donnelly, J.P. Safety and tolerability of the antimicrobial peptide human lactoferrin 1-11 (hLF1-11). BMC Med. 2009, 7, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2011, 11, 37–51. [Google Scholar] [CrossRef]
- Hancock, R.E.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef]
- Villarreal-Ramirez, E.; Eliezer, D.; Garduo-Juarez, R.; Gericke, A.; Perez-Aguilar, J.M.; Boskey, A. Phosphorylation regulates the secondary structure and function of dentin phosphoprotein peptides. Bone 2017, 95, 65–75. [Google Scholar] [CrossRef] [Green Version]
- Lata, S.; Sharma, B.K.; Raghava, G.P. Analysis and prediction of antibacterial peptides. BMC Bioinform. 2007, 8, 263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, P.E. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 1989, 8, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Olsufyeva, E.N.; Tevyashova, A.N. Synthesis, Properties, and Mechanism of Action of New Generation of Polycyclic Glycopeptide Antibiotics. Curr. Top. Med. Chem. 2017, 17, 2166–2198. [Google Scholar] [CrossRef] [PubMed]
- Moravej, H.; Moravej, Z.; Yazdanparast, M.; Heiat, M.; Mirhosseini, A.; Moosazadeh, M.; Mirnejad, R. Antimicrobial Peptides: Features, Action, and Their Resistance Mechanisms in Bacteria. Microb. Drug Resist. 2018, 25, 747–767. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Jenssen, H.; Hamill, P.; Hancock, R.E. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [Green Version]
- Gautam, A.; Chaudhary, K.; Singh, S.; Joshi, A.; Anand, P.; Tuknait, A.; Mathur, D.; Varshney, G.C.; Raghava, G.P. Hemolytik: A database of experimentally determined hemolytic and non-hemolytic peptides. Nucleic Acids Res. 2014, 42, D444–D449. [Google Scholar] [CrossRef] [Green Version]
- Kamaruzzaman, M.; Islam, M.S.; Mahmud, S.; Polash, S.A.; Sultana, R.; Hasan, M.A.; Wang, C.; Jiang, C. In vitro and in silico approach of fungal growth inhibition by Trichoderma asperellum HbGT6-07 derived volatile organic compounds. Arab. J. Chem. 2021, 14, 103290. [Google Scholar] [CrossRef]
- Islam, M.; Sultana, R.; Hasan, M.; Alam, M.; Sikdar, B.; Kamaruzzaman, M. Characterization and biocontrol measures of Pseudomonas syringae pv. syringae associated with citrus blast disease. Vegetos 2020, 33, 555–569. [Google Scholar] [CrossRef]
- Davis, E.G.; Sang, Y.; Rush, B.; Zhang, G.; Blecha, F. Molecular cloning and characterization of equine NK-lysin. Vet. Immunol. Immunopathol. 2005, 105, 163–169. [Google Scholar] [CrossRef]
- Yin, M.; Liu, D.; Xu, F.; Xiao, L.; Wang, Q.; Wang, B.; Chang, Y.; Zheng, J.; Tao, X.; Liu, G.; et al. A specific antimicrobial protein CAP-1 from Pseudomonas sp. isolated from the jellyfish Cyanea capillata. Int. J. Biol. Macromol. 2016, 82, 488–496. [Google Scholar] [CrossRef] [PubMed]







| Treatment Group | Disease Index | Incidence (%) | Control Efficacy (%) |
|---|---|---|---|
| PBS Buffer | 78.33 a | 86.67 a | - |
| Polymyxin B | 3.33 c | 10.00 d | 95.75 a |
| CgR2150 | 5.83 bc | 13.33 c | 92.56 b |
| CgR3101 | 7.50 b | 16.67 b | 90.43 c |
| Peptides | Amino Acid Sequence | Accession Number |
|---|---|---|
| CgR2150 | LAVHHLHSIRGRHHSCTIAQTQANHRHNSYQPNNSSCLAKETDLPTTRSTPATTSSTAPARKSPPASPAPTRQRLCPSPPRGPVSSRHPRRRGQARGRTRAAGVRRTKGYGGKCV | XP_001222255.1 |
| CgR3101 | LAVPNFEPPTRTTCPAPTSPGNPCGPPPPASQTRRPPFHPTSPLRR | XP_001225253.1 |
| Antimicrobial Peptides | Amino Acid Number | Second Structure | PI | MV |
|---|---|---|---|---|
| CgR2150 | 115 | α-helix Random coil | 11.96 | 12,469.01 |
| CgR3101 | 46 | Random coil | 11.40 | 4899.59 |
| Strain | MIC (µg/mL) | |
|---|---|---|
| CgR2150 | CgR3101 | |
| X. oryzae pv. oryzae | 10 | 12 |
| X. oryzae pv. oryzicola | 10 | 12 |
| C. fangii | 15 | 15 |
| X. campestris pv. Holcicola | 25 | 24 |
| C. michiganensis | 18 | 18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Islam, M.S.; Wang, J.; Zhao, Y.; Dong, W. Isolation of Potato Endophytes and Screening of Chaetomium globosum Antimicrobial Genes. Int. J. Mol. Sci. 2022, 23, 4611. https://doi.org/10.3390/ijms23094611
Zhang J, Islam MS, Wang J, Zhao Y, Dong W. Isolation of Potato Endophytes and Screening of Chaetomium globosum Antimicrobial Genes. International Journal of Molecular Sciences. 2022; 23(9):4611. https://doi.org/10.3390/ijms23094611
Chicago/Turabian StyleZhang, Jiaxin, Md. Samiul Islam, Jieyu Wang, Yang Zhao, and Wubei Dong. 2022. "Isolation of Potato Endophytes and Screening of Chaetomium globosum Antimicrobial Genes" International Journal of Molecular Sciences 23, no. 9: 4611. https://doi.org/10.3390/ijms23094611







