Characterization of the Skeletal Muscle Proteome in Undernourished Old Rats
Abstract
:1. Introduction
2. Results
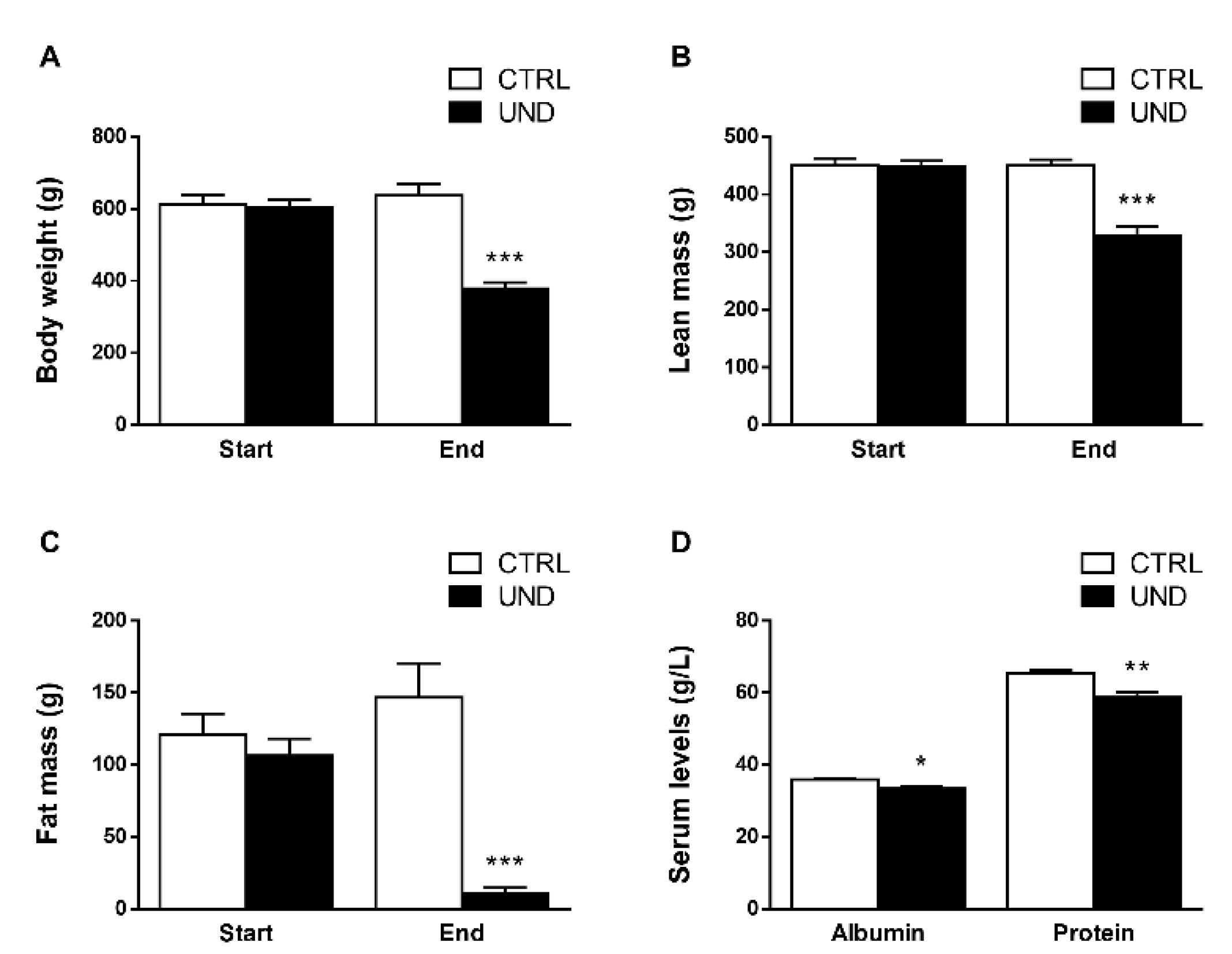
2.1. Undernutrition Affects Muscle Mass and Function in Old Rats
2.2. Undernutrition Induces Profound Changes in Muscle Proteome Independently of Muscle Mass
2.3. Regulation of Metabolism-Related Proteins in Muscle from Undernourished Old Rats
2.4. Regulation of Proteostasis-Related Proteins in Muscle from Undernourished Old Rats
2.5. Proteomic Signature of Undernourished Aged Muscle
3. Discussion
4. Materials and Methods
4.1. Animals and Undernutrition Protocol
4.2. Whole Body Composition and Grip Strength Analysis
4.3. Biological Sample Collection
4.4. Nutritional Status Analyses
4.5. Histological Analyses
4.6. Muscle Protein Extraction
4.7. Nano-LC-MS/MS Analysis
4.8. Gene Ontology Analysis
4.9. Western Blots
4.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cederholm, T.; Bosaeus, I.; Barazzoni, R.; Bauer, J.; Van Gossum, A.; Klek, S.; Muscaritoli, M.; Nyulasi, I.; Ockenga, J.; Schneider, S.M.; et al. Diagnostic criteria for malnutrition—An espen consensus statement. Clin. Nutr. 2015, 34, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Elia, M. Defining, recognizing, and reporting malnutrition. Int. J. Low Extrem. Wounds 2017, 16, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Morley, J.E. Sarcopenia is recognized as an independent condition by an international classification of disease, tenth revision, clinical modification (icd-10-cm) code. J. Am. Med. Dir. Assoc. 2016, 17, 675–677. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Jentoft, A.J.; Bahat, G.; Bauer, J.; Boirie, Y.; Bruyère, O.; Cederholm, T.; Cooper, C.; Landi, F.; Rolland, Y.; Sayer, A.A.; et al. Sarcopenia: Revised european consensus on definition and diagnosis. Age Ageing 2019, 48, 16–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argilés, J.M.; Campos, N.; Lopez-Pedrosa, J.M.; Rueda, R.; Rodriguez-Mañas, L. Skeletal muscle regulates metabolism via interorgan crosstalk: Roles in health and disease. J. Am. Med. Dir. Assoc. 2016, 17, 789–796. [Google Scholar] [CrossRef] [Green Version]
- Boirie, Y.; Gachon, P.; Beaufrère, B. Splanchnic and whole-body leucine kinetics in young and elderly men. Am. J. Clin. Nutr. 1997, 65, 489–495. [Google Scholar] [CrossRef]
- Jourdan, M.; Cynober, L.; Moinard, C.; Blanc, M.C.; Neveux, N.; De Bandt, J.P.; Aussel, C. Splanchnic sequestration of amino acids in aged rats: In vivo and ex vivo experiments using a model of isolated perfused liver. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 294, R748–R755. [Google Scholar] [CrossRef] [Green Version]
- Dardevet, D.; Rémond, D.; Peyron, M.A.; Papet, I.; Savary-Auzeloux, I.; Mosoni, L. Muscle wasting and resistance of muscle anabolism: The “anabolic threshold concept” for adapted nutritional strategies during sarcopenia. Sci. World J. 2012, 2012, 269531. [Google Scholar] [CrossRef]
- Guyonnet, S.; Rolland, Y. Screening for malnutrition in older people. Clin. Geriatr. Med. 2015, 31, 429–437. [Google Scholar] [CrossRef]
- Roberts, H.C.; Lim, S.E.R.; Cox, N.J.; Ibrahim, K. The challenge of managing undernutrition in older people with frailty. Nutrients 2019, 11, 808. [Google Scholar] [CrossRef] [Green Version]
- Barrows, C.H., Jr.; Roeder, L.M. The effect of reduced dietary intake on enzymatic activities and life span of rats. J. Gerontol. 1965, 20, 69–71. [Google Scholar] [CrossRef] [PubMed]
- Chambon-Savanovitch, C.; Felgines, C.; Farges, M.C.; Pernet, P.; Cézard, J.; Raul, F.; Cynober, L.; Vasson, M.P. Severe dietary restriction initiated in aged rats: Evidence for poor adaptation in terms of protein metabolism and intestinal functions. Eur. J. Clin. Investig. 1999, 29, 504–511. [Google Scholar] [CrossRef]
- Felgines, C.; Savanovitch, C.; Farges, M.C.; Cynober, L.; Vasson, M.P. Protein metabolism in rats during long-term dietary restriction: Influence of aging. JPEN J. Parenter Enter. Nutr. 1999, 23, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Walrand, S.; Chambon-Savanovitch, C.; Felgines, C.; Chassagne, J.; Raul, F.; Normand, B.; Farges, M.C.; Beaufrere, B.; Vasson, M.P.; Cynober, L. Aging: A barrier to renutrition? Nutritional and immunologic evidence in rats. Am. J. Clin. Nutr. 2000, 72, 816–824. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, D.H.; Walls, R.C.; Bopp, M.M. Protein-energy undernutrition and the risk of mortality within one year of hospital discharge: A follow-up study. J. Am. Geriatr. Soc. 1995, 43, 507–512. [Google Scholar] [CrossRef]
- Nations Unies. World population prospects 2019: Highlights; Department of Economic and Social Affairs: New York, NY, USA, 2019. [Google Scholar]
- Salles, J.; Cardinault, N.; Patrac, V.; Berry, A.; Giraudet, C.; Collin, M.L.; Chanet, A.; Tagliaferri, C.; Denis, P.; Pouyet, C.; et al. Bee pollen improves muscle protein and energy metabolism in malnourished old rats through interfering with the mtor signaling pathway and mitochondrial activity. Nutrients 2014, 6, 5500–5516. [Google Scholar] [CrossRef] [Green Version]
- Salles, J.; Chanet, A.; Berry, A.; Giraudet, C.; Patrac, V.; Domingues-Faria, C.; Rocher, C.; Guillet, C.; Denis, P.; Pouyet, C.; et al. Fast digestive, leucine-rich, soluble milk proteins improve muscle protein anabolism, and mitochondrial function in undernourished old rats. Mol. Nutr. Food Res. 2017, 61, 1700287. [Google Scholar] [CrossRef] [Green Version]
- Zangarelli, A.; Chanseaume, E.; Morio, B.; Brugère, C.; Mosoni, L.; Rousset, P.; Giraudet, C.; Patrac, V.; Gachon, P.; Boirie, Y.; et al. Synergistic effects of caloric restriction with maintained protein intake on skeletal muscle performance in 21-month-old rats: A mitochondria-mediated pathway. FASEB J. 2006, 20, 2439–2450. [Google Scholar] [CrossRef]
- Sartori, R.; Romanello, V.; Sandri, M. Mechanisms of muscle atrophy and hypertrophy: Implications in health and disease. Nat. Commun. 2021, 12, 330. [Google Scholar] [CrossRef]
- Patterson, S.D.; Aebersold, R.H. Proteomics: The first decade and beyond. Nat. Genet. 2003, 33, 311–323. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, P.; Liu, H.; Fan, M.; Chen, X. Proteomic analysis of mouse soleus muscles affected by hindlimb unloading and reloading. Muscle Nerve 2015, 52, 803–811. [Google Scholar] [CrossRef]
- Massart, I.S.; Paulissen, G.; Loumaye, A.; Lause, P.; Pötgens, S.A.; Thibaut, M.M.; Balan, E.; Deldicque, L.; Atfi, A.; Louis, E.; et al. Marked increased production of acute phase reactants by skeletal muscle during cancer cachexia. Cancers 2020, 12, 3221. [Google Scholar] [CrossRef] [PubMed]
- Ubaida-Mohien, C.; Lyashkov, A.; Gonzalez-Freire, M.; Tharakan, R.; Shardell, M.; Moaddel, R.; Semba, R.D.; Chia, C.W.; Gorospe, M.; Sen, R.; et al. Discovery proteomics in aging human skeletal muscle finds change in spliceosome, immunity, proteostasis and mitochondria. Elife 2019, 8, e49874. [Google Scholar] [CrossRef] [PubMed]
- Walrand, S.; Zangarelli, A.; Guillet, C.; Salles, J.; Soulier, K.; Giraudet, C.; Patrac, V.; Boirie, Y. Effect of fast dietary proteins on muscle protein synthesis rate and muscle strength in ad libitum-fed and energy-restricted old rats. Br. J. Nutr. 2011, 106, 1683–1690. [Google Scholar] [CrossRef] [Green Version]
- Vainshtein, A.; Sandri, M. Signaling pathways that control muscle mass. Int. J. Mol. Sci. 2020, 21, 4759. [Google Scholar] [CrossRef] [PubMed]
- Isumi, Y.; Hirata, T.; Saitoh, H.; Miyakawa, T.; Murakami, K.; Kudoh, G.; Doi, H.; Ishibashi, K.; Nakajima, H. Transgenic overexpression of usp15 in the heart induces cardiac remodeling in mice. Biochem. Biophys. Res. Commun. 2011, 405, 216–221. [Google Scholar] [CrossRef]
- Liu, C.C.; Chen, R.H. Klhl20 links the ubiquitin-proteasome system to autophagy termination. Autophagy 2016, 12, 890–891. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Luo, L.; Eash, J.; Ibebunjo, C.; Glass, D.J. The scf-fbxo40 complex induces irs1 ubiquitination in skeletal muscle, limiting igf1 signaling. Dev. Cell 2011, 21, 835–847. [Google Scholar] [CrossRef] [Green Version]
- Damas, J.; Bourdon, V.; Lefebvre, P.J. Insulin sensitivity, clearance and release in kininogen-deficient rats. Exp. Physiol. 1999, 84, 549–557. [Google Scholar] [CrossRef]
- Dietze, G.J.; Henriksen, E.J. Angiotensin-converting enzyme in skeletal muscle: Sentinel of blood pressure control and glucose homeostasis. J. Renin Angiotensin Aldosterone Syst. 2008, 9, 75–88. [Google Scholar] [CrossRef] [Green Version]
- Henriksen, E.J.; Jacob, S.; Fogt, D.L.; Dietze, G.J. Effect of chronic bradykinin administration on insulin action in an animal model of insulin resistance. Am. J. Physiol. 1998, 275, R40–R45. [Google Scholar] [CrossRef] [PubMed]
- Miyata, T.; Taguchi, T.; Uehara, M.; Isami, S.; Kishikawa, H.; Kaneko, K.; Araki, E.; Shichiri, M. Bradykinin potentiates insulin-stimulated glucose uptake and enhances insulin signal through the bradykinin b2 receptor in dog skeletal muscle and rat l6 myoblasts. Eur. J. Endocrinol. 1998, 138, 344–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carnagarin, R.; Dharmarajan, A.M.; Dass, C.R. Pedf attenuates insulin-dependent molecular pathways of glucose homeostasis in skeletal myocytes. Mol. Cell. Endocrinol. 2016, 422, 115–124. [Google Scholar] [CrossRef]
- Crowe, S.; Wu, L.E.; Economou, C.; Turpin, S.M.; Matzaris, M.; Hoehn, K.L.; Hevener, A.L.; James, D.E.; Duh, E.J.; Watt, M.J. Pigment epithelium-derived factor contributes to insulin resistance in obesity. Cell Metab. 2009, 10, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Famulla, S.; Lamers, D.; Hartwig, S.; Passlack, W.; Horrighs, A.; Cramer, A.; Lehr, S.; Sell, H.; Eckel, J. Pigment epithelium-derived factor (pedf) is one of the most abundant proteins secreted by human adipocytes and induces insulin resistance and inflammatory signaling in muscle and fat cells. Int. J. Obes. 2011, 35, 762–772. [Google Scholar] [CrossRef] [Green Version]
- McAinch, A.J.; Cornall, L.M.; Watts, R.; Hryciw, D.H.; O’Brien, P.E.; Cameron-Smith, D. Increased pyruvate dehydrogenase kinase expression in cultured myotubes from obese and diabetic individuals. Eur. J. Nutr. 2015, 54, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Hwang, B.; Jeoung, N.H.; Harris, R.A. Pyruvate dehydrogenase kinase isoenzyme 4 (pdhk4) deficiency attenuates the long-term negative effects of a high-saturated fat diet. Biochem. J. 2009, 423, 243–252. [Google Scholar] [CrossRef] [Green Version]
- Thoudam, T.; Ha, C.M.; Leem, J.; Chanda, D.; Park, J.S.; Kim, H.J.; Jeon, J.H.; Choi, Y.K.; Liangpunsakul, S.; Huh, Y.H.; et al. Pdk4 augments er-mitochondria contact to dampen skeletal muscle insulin signaling during obesity. Diabetes 2019, 68, 571–586. [Google Scholar] [CrossRef]
- Pin, F.; Novinger, L.J.; Huot, J.R.; Harris, R.A.; Couch, M.E.; O’Connell, T.M.; Bonetto, A. Pdk4 drives metabolic alterations and muscle atrophy in cancer cachexia. FASEB J. 2019, 33, 7778–7790. [Google Scholar] [CrossRef] [Green Version]
- Kanazawa, Y.; Ikegami, K.; Sujino, M.; Koinuma, S.; Nagano, M.; Oi, Y.; Onishi, T.; Sugiyo, S.; Takeda, I.; Kaji, H.; et al. Effects of aging on basement membrane of the soleus muscle during recovery following disuse atrophy in rats. Exp. Gerontol. 2017, 98, 153–161. [Google Scholar] [CrossRef]
- Oguro, A.; Sakurai, T.; Fujita, Y.; Lee, S.; Kubota, H.; Nagata, K.; Atomi, Y. The molecular chaperone hsp47 rapidly senses gravitational changes in myoblasts. Genes Cells 2006, 11, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- Nagata, K. Hsp47: A collagen-specific molecular chaperone. Trends Biochem. Sci. 1996, 21, 22–26. [Google Scholar] [CrossRef]
- Allen, D.L.; Bandstra, E.R.; Harrison, B.C.; Thorng, S.; Stodieck, L.S.; Kostenuik, P.J.; Morony, S.; Lacey, D.L.; Hammond, T.G.; Leinwand, L.L.; et al. Effects of spaceflight on murine skeletal muscle gene expression. J. Appl. Physiol. (1985) 2009, 106, 582–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slack, C. Ras signaling in aging and metabolic regulation. Nutr. Healthy Aging 2017, 4, 195–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbé, C.; Bray, F.; Gueugneau, M.; Devassine, S.; Lause, P.; Tokarski, C.; Rolando, C.; Thissen, J.P. Comparative proteomic and transcriptomic analysis of follistatin-induced skeletal muscle hypertrophy. J. Proteome Res. 2017, 16, 3477–3490. [Google Scholar] [CrossRef]
- Steelman, C.A.; Recknor, J.C.; Nettleton, D.; Reecy, J.M. Transcriptional profiling of myostatin-knockout mice implicates wnt signaling in postnatal skeletal muscle growth and hypertrophy. FASEB J. 2006, 20, 580–582. [Google Scholar] [CrossRef]
- Sun, T.; Yi, H.; Yang, C.; Kishnani, P.S.; Sun, B. Starch binding domain-containing protein 1 plays a dominant role in glycogen transport to lysosomes in liver. J. Biol. Chem. 2016, 291, 16479–16484. [Google Scholar] [CrossRef] [Green Version]
- Mubaid, S.; Ma, J.F.; Omer, A.; Ashour, K.; Lian, X.J.; Sanchez, B.J.; Robinson, S.; Cammas, A.; Dormoy-Raclet, V.; Di Marco, S.; et al. Hur counteracts mir-330 to promote stat3 translation during inflammation-induced muscle wasting. Proc. Natl. Acad. Sci. USA 2019, 116, 17261–17270. [Google Scholar] [CrossRef] [Green Version]
- Bonetto, A.; Aydogdu, T.; Jin, X.; Zhang, Z.; Zhan, R.; Puzis, L.; Koniaris, L.G.; Zimmers, T.A. Jak/stat3 pathway inhibition blocks skeletal muscle wasting downstream of il-6 and in experimental cancer cachexia. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E410–E421. [Google Scholar] [CrossRef] [Green Version]
- Sieber, C.C. Malnutrition and sarcopenia. Aging Clin. Exp. Res. 2019, 31, 793–798. [Google Scholar] [CrossRef]
- Hirabayashi, T.; Nakanishi, R.; Tanaka, M.; Nisa, B.U.; Maeshige, N.; Kondo, H.; Fujino, H. Reduced metabolic capacity in fast and slow skeletal muscle via oxidative stress and the energy-sensing of ampk/sirt1 in malnutrition. Physiol. Rep. 2021, 9, e14763. [Google Scholar] [CrossRef] [PubMed]
- Maurissen, J.P.; Marable, B.R.; Andrus, A.K.; Stebbins, K.E. Factors affecting grip strength testing. Neurotoxicol. Teratol. 2003, 25, 543–553. [Google Scholar] [CrossRef]
- Reid, K.F.; Pasha, E.; Doros, G.; Clark, D.J.; Patten, C.; Phillips, E.M.; Frontera, W.R.; Fielding, R.A. Longitudinal decline of lower extremity muscle power in healthy and mobility-limited older adults: Influence of muscle mass, strength, composition, neuromuscular activation and single fiber contractile properties. Eur. J. Appl. Physiol. 2014, 114, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Madapallimattam, A.G.; Law, L.; Jeejeebhoy, K.N. Effect of hypoenergetic feeding on muscle oxidative phosphorylation and mitochondrial complex i-iv activities in rats. Am. J. Clin. Nutr. 2002, 76, 1031–1039. [Google Scholar] [CrossRef] [Green Version]
- Rolfe, D.F.; Brown, G.C. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol. Rev. 1997, 77, 731–758. [Google Scholar] [CrossRef] [Green Version]
- Schiaffino, S.; Dyar, K.A.; Ciciliot, S.; Blaauw, B.; Sandri, M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013, 280, 4294–4314. [Google Scholar] [CrossRef]
- Piec, I.; Listrat, A.; Alliot, J.; Chambon, C.; Taylor, R.G.; Bechet, D. Differential proteome analysis of aging in rat skeletal muscle. FASEB J. 2005, 19, 1143–1145. [Google Scholar] [CrossRef]
- Gelfi, C.; Vigano, A.; Ripamonti, M.; Pontoglio, A.; Begum, S.; Pellegrino, M.A.; Grassi, B.; Bottinelli, R.; Wait, R.; Cerretelli, P. The human muscle proteome in aging. J. Proteome Res. 2006, 5, 1344–1353. [Google Scholar] [CrossRef]
- Kim, J.A.; Vetrivel, P.; Kim, S.M.; Ha, S.E.; Kim, H.H.; Bhosale, P.B.; Heo, J.D.; Lee, W.S.; Senthil, K.; Kim, G.S. Quantitative proteomics analysis for the identification of differential protein expression in calf muscles between young and old sd rats using mass spectrometry. ACS Omega 2021, 6, 7422–7433. [Google Scholar] [CrossRef]
- Theron, L.; Gueugneau, M.; Coudy, C.; Viala, D.; Bijlsma, A.; Butler-Browne, G.; Maier, A.; Bechet, D.; Chambon, C. Label-free quantitative protein profiling of vastus lateralis muscle during human aging. Mol. Cell. Proteom. 2014, 13, 283–294. [Google Scholar] [CrossRef] [Green Version]
- Gueugneau, M.; d’Hose, D.; Barbe, C.; de Barsy, M.; Lause, P.; Maiter, D.; Bindels, L.B.; Delzenne, N.M.; Schaeffer, L.; Gangloff, Y.G.; et al. Increased serpina3n release into circulation during glucocorticoid-mediated muscle atrophy. J. Cachexia Sarcopenia Muscle 2018, 9, 929–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The pride database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Kaspric, N.; Picard, B.; Reichstadt, M.; Tournayre, J.; Bonnet, M. Proteinside to easily investigate proteomics data from ruminants: Application to mine proteome of adipose and muscle tissues in bovine foetuses. PLoS ONE 2015, 10, e0128086. [Google Scholar] [CrossRef] [PubMed] [Green Version]







Proteins (  ) ) | PL | SOL | |||
|---|---|---|---|---|---|
| Symbol | Name | FC | P | FC | P |
| PDK4 | Pyruvate dehydrogenase kinase isozyme 4 | −2.2 | 0.002 | −3.8 | 0.000 |
| SERPINF1 | Alpha-2 antiplasmin | −2.1 | 0.012 | −2.1 | 0.000 |
| NDUFA8 | NADH dehydrogenase 1 alpha subcomplex subunit 8 | −1.6 | 0.048 | −2.1 | 0.015 |
| HSPD1 | 60 kDa heat shock protein | −1.6 | 0.003 | −1.4 | 0.012 |
| FH | Fumarate hydratase | −1.5 | 0.014 | −2.0 | 0.029 |
| HADH | Hydroxyacyl-coenzyme A dehydrogenase | −1.5 | 0.028 | −1.7 | 0.014 |
| BCAT2 | Branched-chain amino acid aminotransferase | −1.4 | 0.037 | −1.4 | 0.023 |
| CYCS | Cytochrome c, somatic | −1.4 | 0.047 | −1.6 | 0.011 |
| ACADM | Acetyl-coenzyme A dehydrogenase medium-chain | −1.4 | 0.040 | −1.8 | 0.001 |
| UQCRC1 | Cytochrome b-c1 complex subunit 1 | −1.4 | 0.040 | −1.5 | 0.039 |
| IMPDH2 | Inosine-5′-monophosphate dehydrogenase 2 | +1.7 | 0.045 | +1.7 | 0.011 |
| KNG2L | T-kininogen 2 | +2.2 | 0.041 | +3.1 | 0.044 |
Proteins (  & &  ) ) | PL | SOL | |||
|---|---|---|---|---|---|
| Symbol | Name | FC | P | FC | P |
| SERPINH1 | Serpin H1 | −2.0 | 0.031 | ND | ND |
| CD163 | Scavenger receptor cysteine-rich type 1 protein M130 | +2.2 | 0.005 | ND | ND |
| PSME4 | Proteasome activator subunit 4 | +2.2 | 0.014 | ND | ND |
| MPC2 | Mitochondrial pyruvate carrier 2 | −5.7 | 0.033 | +1.1 | 0.914 |
| LASP1 | LIM and SH3 domain protein 1 | −2.3 | 0.037 | +1.0 | 0.858 |
| MTCO3 | Cytochrome c oxidase subunit 3 | −2.3 | 0.023 | +1.1 | 0.602 |
| NT5C2 | 5′-nucleotidase, cytosolic II | +2.0 | 0.040 | −1.2 | 0.252 |
| AK2 | Adenylate kinase 2, mitochondrial | +2.1 | 0.045 | +1.3 | 0.154 |
| GSTM2 | Glutathione S-transferase Mu 2 | +2.1 | 0.003 | +1.3 | 0.609 |
| MRC1 | Mannose receptor C-type 1 | +2.2 | 0.028 | +1.2 | 0.943 |
| CUTC | CutC copper transporter | +2.2 | 0.004 | +1.3 | 0.711 |
| INMT | Indolethylamine N-methyltransferase | +2.2 | 0.025 | −1.1 | 0.707 |
| MYOT | Myotilin | +2.5 | 0.035 | −1.1 | 0.538 |
| PHYKPL | 5-phosphohydroxy-L-lysine phospho-lyase | +7.7 | 0.017 | −1.2 | 0.705 |
Proteins (  & &  ) ) | PL | SOL | |||
|---|---|---|---|---|---|
| Symbol | Name | FC | P | FC | P |
| L2HGDH | L-2-hydroxyglutarate dehydrogenase | ND | ND | −2.1 | 0.015 |
| AKAP1 | A-kinase anchor protein 1 | ND | ND | +2.0 | 0.036 |
| ENPP3 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 3 | ND | ND | +2.2 | 0.023 |
| NOMO1 | Nodal modulator 1 | ND | ND | +2.2 | 0.012 |
| UGDH | UDP-glucose 6-dehydrogenase | ND | ND | +2.3 | 0.040 |
| CYP4B1 | Cytochrome P450 4B1 | ND | ND | +2.4 | 0.013 |
| CHMP3 | Charged multivesicular body protein 3 | ND | ND | +2.6 | 0.017 |
| RRAS2 | RAS-related protein 2 | ND | ND | +2.6 | 0.017 |
| MXRA7 | Matrix-remodeling-associated protein 7 | ND | ND | +2.8 | 0.017 |
| PDLIM4 | PDZ and LIM domain protein 4 | ND | ND | +2.9 | 0.048 |
| TTF1 | Transcription termination factor 1 | ND | ND | +2.9 | 0.017 |
| TNNT2 | Troponin T, cardiac muscle | ND | ND | +3.1 | 0.027 |
| PFKFB1 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | −1.1 | 0.719 | −3.2 | 0.004 |
| PGAM2 | Phosphoglycerate mutase 2 | −1.1 | 0.663 | −2.7 | 0.006 |
| CKM | Creatine kinase M-type | +1.1 | 0.868 | −2.6 | 0.201 |
| CDC37 | Hsp90 co-chaperone Cdc37 | −1.2 | 0.427 | −2.6 | 0.034 |
| MRPS36 | Mitochondrial ribosomal protein S36 | −1.0 | 0.760 | −2.4 | 0.028 |
| STAT3 | Signal transducer and activator of transcription 3 | −1.1 | 0.862 | −2.3 | 0.050 |
| ACADS | Acetyl-coenzyme A dehydrogenase, short chain | −1.2 | 0.113 | −2.2 | 0.015 |
| GNPDA1 | Glucosamine-6-phosphate isomerase | +1.2 | 0.484 | −2.2 | 0.016 |
| PHKG1 | Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform | −1.1 | 0.978 | −2.1 | 0.048 |
| DDX5 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | −1.1 | 0.755 | +2.0 | 0.008 |
| HNRNPH2 | Heterogeneous nuclear ribonucleoprotein H2 | +1.2 | 0.837 | +2.1 | 0.037 |
| STBD1 | Starch-binding domain-containing protein 1 | +1.1 | 0.402 | +2.1 | 0.049 |
| FASN | Fatty acid synthase | +1.0 | 0.914 | +4.3 | 0.002 |
| ELAVL1 | ELAV-like protein 1 | −1.3 | 0.260 | +7.2 | 0.047 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbé, C.; Salles, J.; Chambon, C.; Giraudet, C.; Sanchez, P.; Patrac, V.; Denis, P.; Boirie, Y.; Walrand, S.; Gueugneau, M. Characterization of the Skeletal Muscle Proteome in Undernourished Old Rats. Int. J. Mol. Sci. 2022, 23, 4762. https://doi.org/10.3390/ijms23094762
Barbé C, Salles J, Chambon C, Giraudet C, Sanchez P, Patrac V, Denis P, Boirie Y, Walrand S, Gueugneau M. Characterization of the Skeletal Muscle Proteome in Undernourished Old Rats. International Journal of Molecular Sciences. 2022; 23(9):4762. https://doi.org/10.3390/ijms23094762
Chicago/Turabian StyleBarbé, Caroline, Jérôme Salles, Christophe Chambon, Christophe Giraudet, Phelipe Sanchez, Véronique Patrac, Philippe Denis, Yves Boirie, Stéphane Walrand, and Marine Gueugneau. 2022. "Characterization of the Skeletal Muscle Proteome in Undernourished Old Rats" International Journal of Molecular Sciences 23, no. 9: 4762. https://doi.org/10.3390/ijms23094762






