Overexpression of EgrIAA20 from Eucalyptus grandis, a Non-Canonical Aux/IAA Gene, Specifically Decouples Lignification of the Different Cell-Types in Arabidopsis Secondary Xylem
Abstract
:1. Introduction
2. Results
2.1. EgrIAA20 Transcripts Accumulate Preferentially in Cambium in Eucalyptus
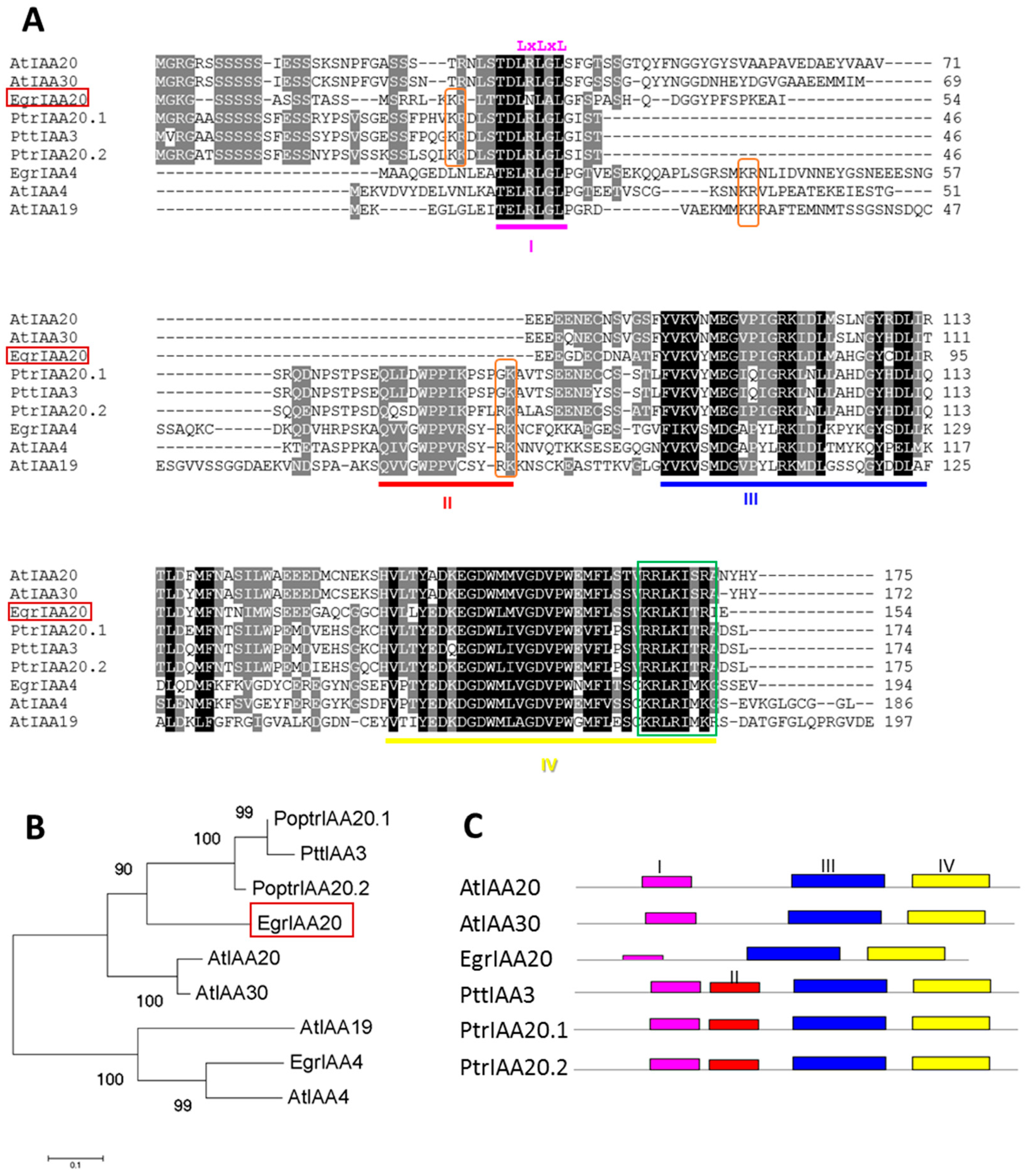
2.2. EgrIAA20 Defines a Non-Canonical Aux/IAA Protein Which Lacks Highly Conserved Aux/IAA Characteristic DOMAIN II
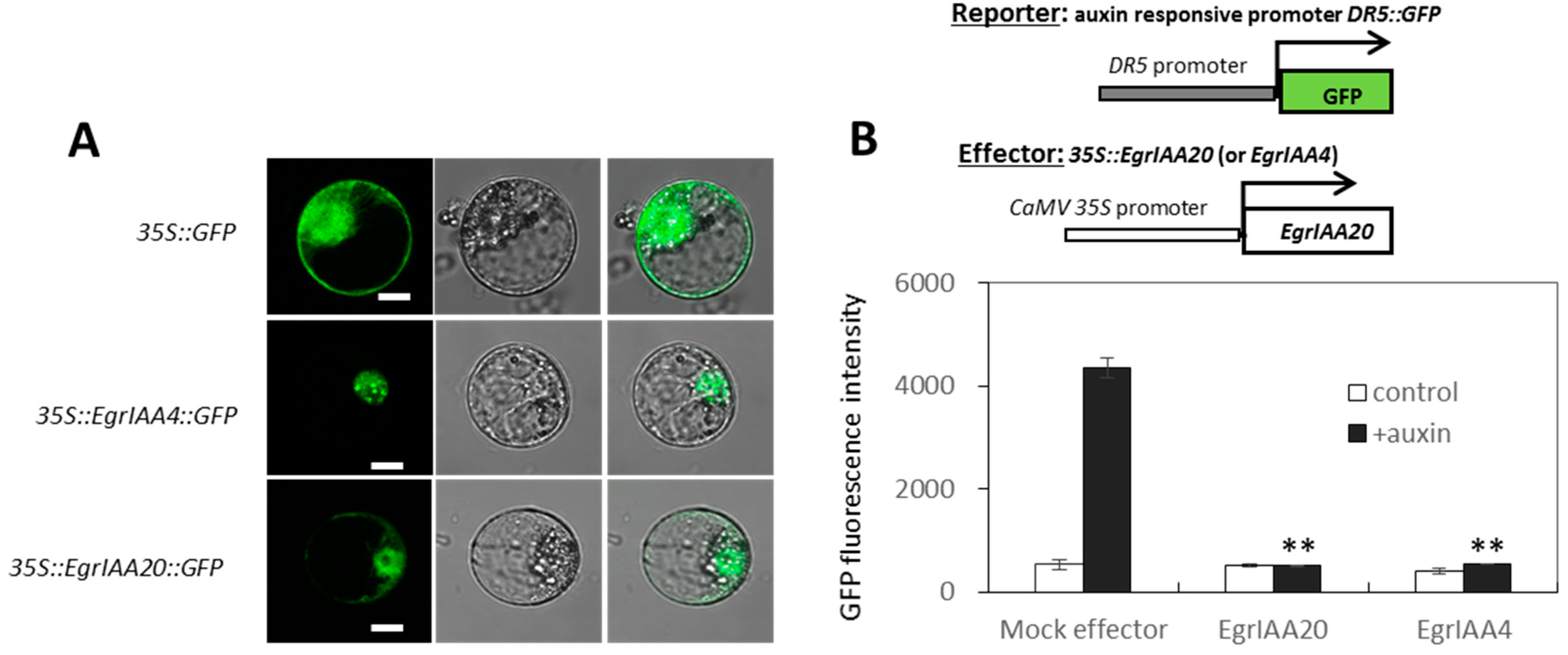
2.3. EgrIAA20 Is Mainly but Not Exclusively Nuclear Localized
2.4. EgrIAA20 Acts as a Potent Transcriptional Repressor of Auxin Response In Vivo
2.5. Over-Expression of EgrIAA20 in Transgenic Arabidopsis Results in Smaller and Bushier Plants with Floppy Stems
2.6. EgrIAA20 Over-Expression Reduces Fiber-Specific Lignin in Arabidopsis Hypocotyls
2.7. EgrIAA20 Overexpression Alters Primary Xylem Development in Cotyledons
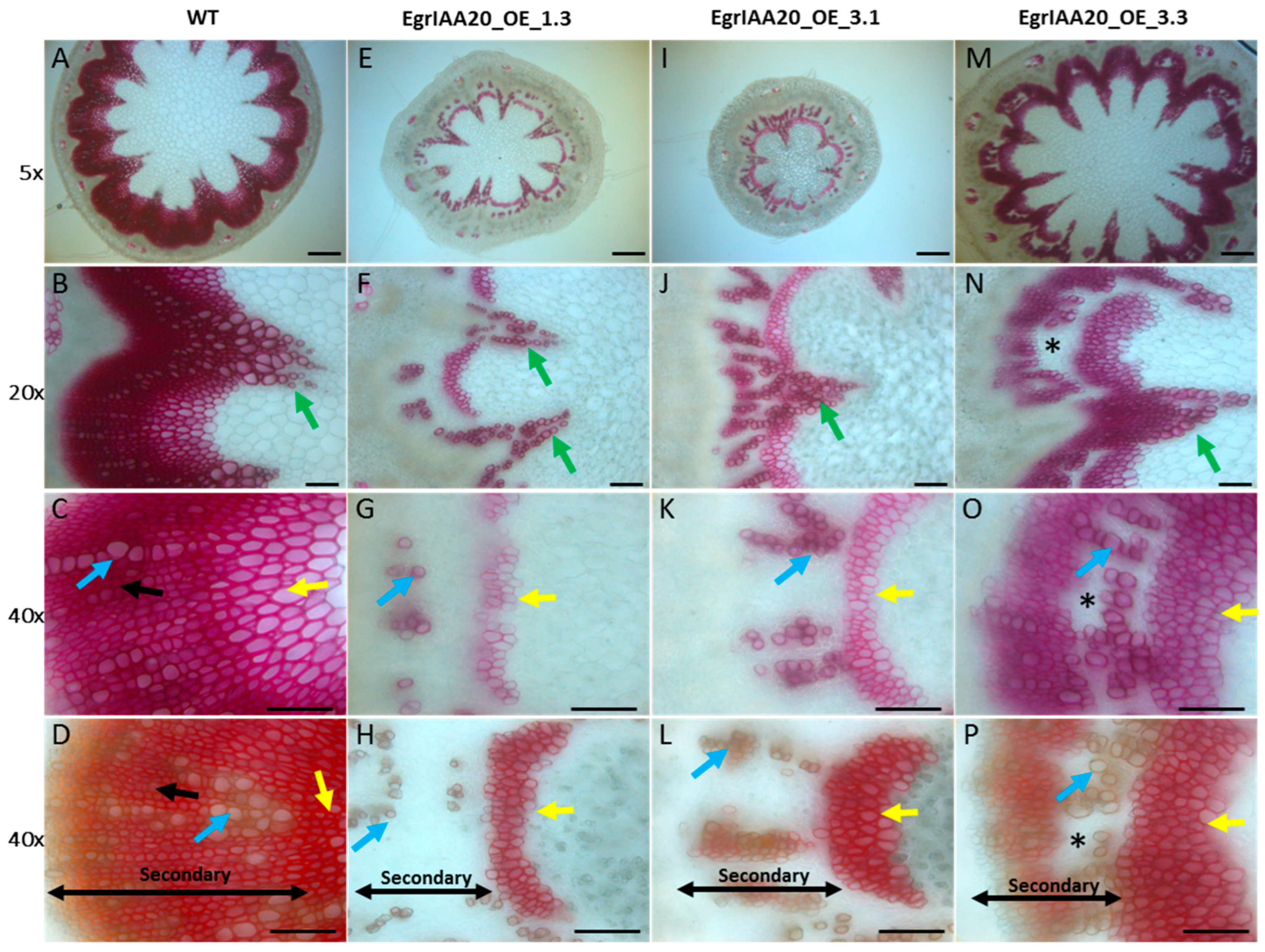
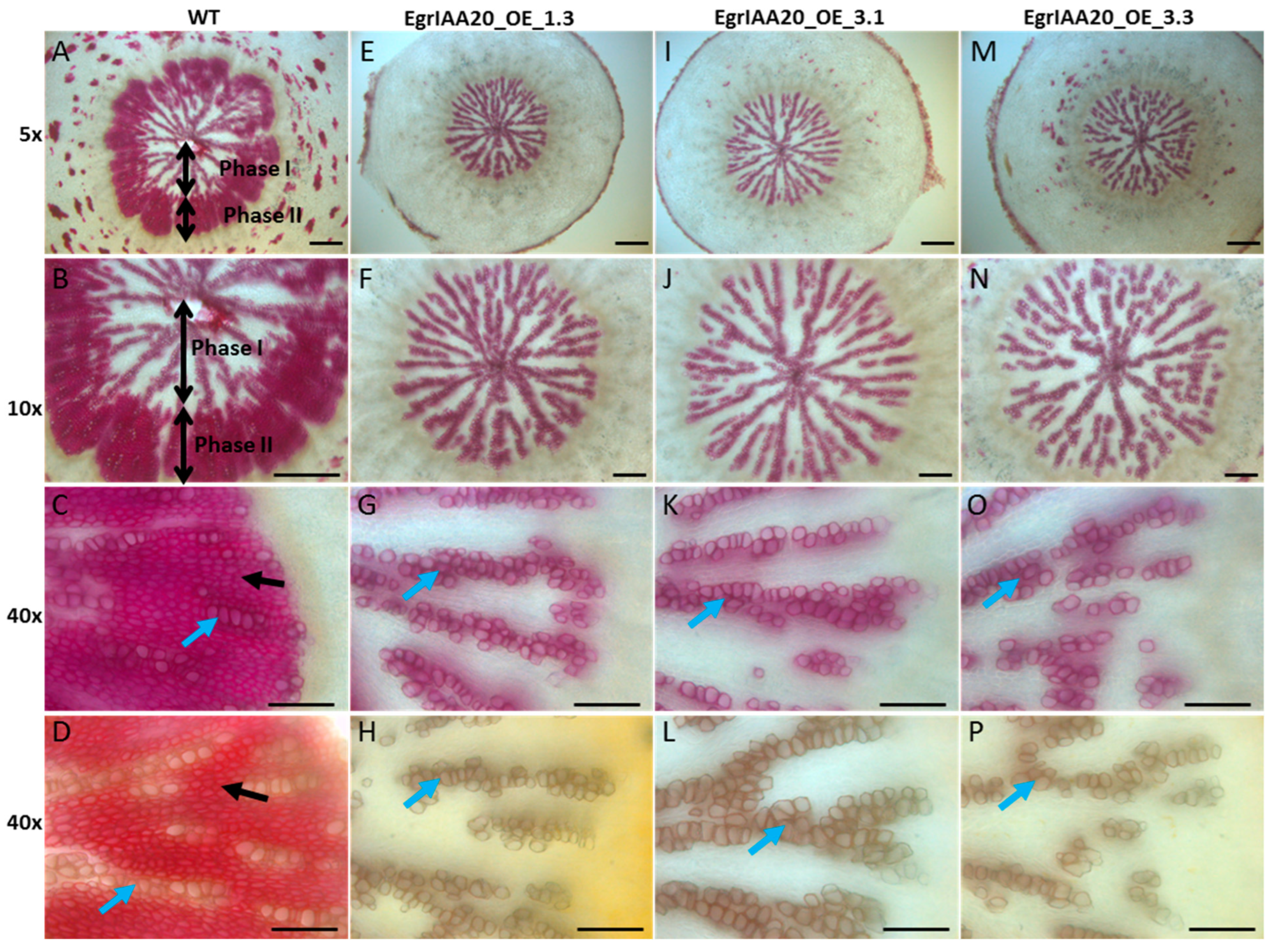
2.8. EgrIAA20 Overexpression Specifically Represses Xylary Fibers of Secondary Xylem in Arabidopsis Stem and Hypocotyl
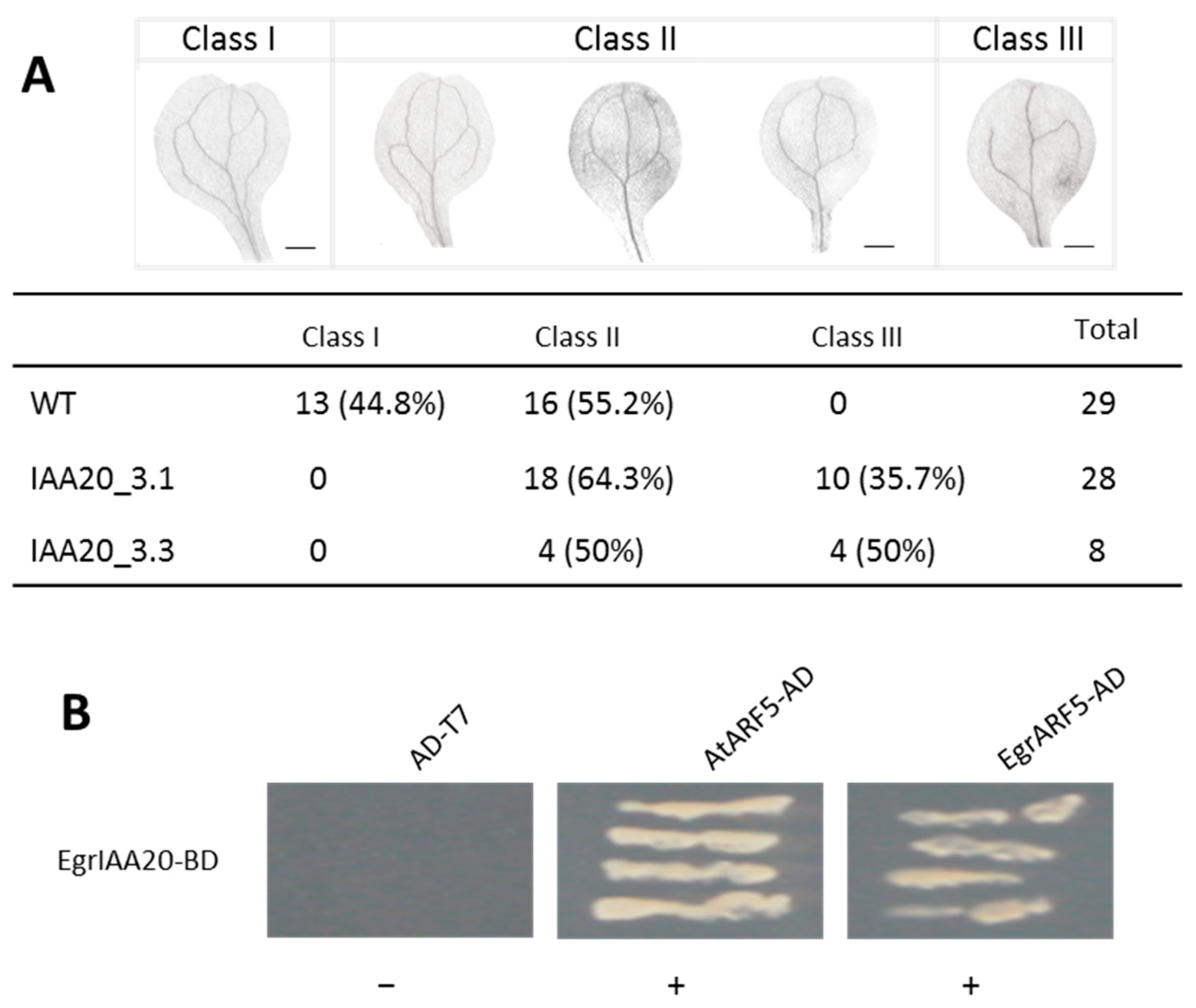
2.9. Identification of EgrIAA9A as the Main Interacting Protein of EgrIAA20
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Microfluidic Quantitative PCR Expression Analysis
4.3. Gene Cloning, Vector Construction, and Genetic Transformation
4.4. Transient Expression in Protoplasts for Subcellular Localization and Transactivation Assay
4.5. Microscopy Analysis
4.6. Pyrolysis Analysis
4.7. Yeast Two-Hybrid (Y2H) Screening
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boudet, A.M.; Kajita, S.; Grima-Pettenati, J.; Goffner, D. Lignins and lignocellulosics: A better control of synthesis for new and improved uses. Trends Plant Sci. 2003, 8, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.H.; Zhong, R. Molecular control of wood formation in trees. J. Exp. Bot. 2015, 66, 4119–4131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barros, J.; Serk, H.; Granlund, I.; Pesquet, E. The cell biology of lignification in higher plants. Ann. Bot. London 2015, 115, 1053–1074. [Google Scholar] [CrossRef] [Green Version]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Pesquet, E.; Wagner, A.; Grabber, J.H. Cell culture systems: Invaluable tools to investigate lignin formation and cell wall properties. Curr. Opin. Biotech. 2019, 56, 215–222. [Google Scholar] [CrossRef]
- Brackmann, K.; Qi, J.; Gebert, M.; Jouannet, V.; Schlamp, T.; Grunwald, K.; Wallner, E.S.; Novikova, D.D.; Levitsky, V.G.; Agusti, J.; et al. Spatial specificity of auxin responses coordinates wood formation. Nat. Commun. 2018, 9, 875. [Google Scholar] [CrossRef] [Green Version]
- De Zio, E.; Montagnoli, A.; Karady, M.; Terzaghi, M.; Sferra, G.; Antoniadi, I.; Scippa, G.S.; Ljung, K.; Chiatante, D.; Trupiano, D. Reaction Wood Anatomical Traits and Hormonal Profiles in Poplar Bent Stem and Root. Front. Plant Sci. 2020, 11, 590985. [Google Scholar] [CrossRef]
- Fischer, U.; Kucukoglu, M.; Helariutta, Y.; Bhalerao, R.P. The Dynamics of Cambial Stem Cell Activity. Annu. Rev. Plant Biol. 2019, 70, 293–319. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, H.; Komamine, A. Establishment of an Experimental System for the Study of Tracheary Element Differentiation from Single Cells Isolated from the Mesophyll of Zinnia elegans. Plant Physiol. 1980, 65, 57–60. [Google Scholar] [CrossRef] [Green Version]
- Pesquet, E.; Ranocha, P.; Legay, S.; Digonnet, C.; Barbier, O.; Pichon, M.; Goffner, D. Novel markers of xylogenesis in zinnia are differentially regulated by auxin and cytokinin. Plant Physiol. 2005, 139, 1821–1839. [Google Scholar] [CrossRef] [Green Version]
- Johnsson, C.; Jin, X.; Xue, W.; Dubreuil, C.; Lezhneva, L.; Fischer, U. The plant hormone auxin directs timing of xylem development by inhibition of secondary cell wall deposition through repression of secondary wall NAC-domain transcription factors. Physiol. Plant. 2018, 165, 673–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morey, P.R.; Dahl, B.E. Histological and Morphological Effects of Auxin Transport Inhibitors on Honey Mesquite. Bot. Gaz. 1975, 136, 274–280. [Google Scholar] [CrossRef]
- Perrot-Rechenmann, C. Cellular responses to auxin: Division versus expansion. Cold Spring Harb. Perspect. Biol. 2010, 2, a001446. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, S.; Friml, J. Auxin: A Trigger for Change in Plant Development. Cell 2009, 136, 1005–1016. [Google Scholar] [CrossRef]
- Bhalerao, R.P.; Fischer, U. Auxin gradients across wood-instructive or incidental? Physiol. Plant. 2014, 151, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Leyser, O. Auxin Signaling. Plant Physiol. 2018, 176, 465–479. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.H.; Estelle, M. Diversity and specificity: Auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 2014, 21, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Berleth, T.; Jurgens, G. The Role of the Monopteros Gene in Organizing the Basal Body Region of the Arabidopsis Embryo. Development 1993, 118, 575–587. [Google Scholar] [CrossRef]
- Hamann, T.; Benkova, E.; Baurle, I.; Kientz, M.; Jurgens, G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002, 16, 1610–1615. [Google Scholar] [CrossRef] [Green Version]
- Hamann, T.; Mayer, U.; Jurgens, G. The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 1999, 126, 1387–1395. [Google Scholar] [CrossRef]
- Muller, C.J.; Valdes, A.E.; Wang, G.; Ramachandran, P.; Beste, L.; Uddenberg, D.; Carlsbecker, A. PHABULOSA Mediates an Auxin Signaling Loop to Regulate Vascular Patterning in Arabidopsis. Plant Physiol. 2016, 170, 956–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenzel, C.L.; Schuetz, M.; Yu, Q.; Mattsson, J. Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in Arabidopsis thaliana. Plant J. 2007, 49, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Ruonala, R.; Ko, D.; Helariutta, Y. Genetic Networks in Plant Vascular Development. Annu. Rev. Genet. 2017, 51, 335–359. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Karlberg, A.; Antti, H.; Lopez-Vernaza, M.; Mellerowicz, E.; Perrot-Rechenmann, C.; Sandberg, G.; Bhalerao, R.P. Dissecting the molecular basis of the regulation of wood formation by auxin in hybrid aspen. Plant Cell 2008, 20, 843–855. [Google Scholar] [CrossRef] [Green Version]
- Karannagoda, N.; Spokevicius, A.; Hussey, S.; Cassan-Wang, H.; Grima-Pettenati, J.; Bossinger, G. Eucalyptus grandis AUX/INDOLE-3-ACETIC ACID 13 (EgrIAA13) is a novel transcriptional regulator of xylogenesis. Plant Mol. Biol. 2022. [Google Scholar] [CrossRef]
- Yu, H.; Soler, M.; San Clemente, H.; Mila, I.; Paiva, J.A.; Myburg, A.A.; Bouzayen, M.; Grima-Pettenati, J.; Cassan-Wang, H. Comprehensive genome-wide analysis of the Aux/IAA gene family in Eucalyptus: Evidence for the role of EgrIAA4 in wood formation. Plant Cell Physiol. 2015, 56, 700–714. [Google Scholar] [CrossRef] [Green Version]
- Barra-Jimenez, A.; Ragni, L. Secondary development in the stem: When Arabidopsis and trees are closer than it seems. Curr. Opin. Plant Biol. 2017, 35, 145–151. [Google Scholar] [CrossRef]
- Chaffey, N.; Cholewa, E.; Regan, S.; Sundberg, B. Secondary xylem development in Arabidopsis: A model for wood formation. Physiol. Plant. 2002, 114, 594–600. [Google Scholar] [CrossRef]
- Legay, S.; Sivadon, P.; Blervacq, A.S.; Pavy, N.; Baghdady, A.; Tremblay, L.; Levasseur, C.; Ladouce, N.; Lapierre, C.; Seguin, A.; et al. EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. New Phytol. 2010, 188, 774–786. [Google Scholar] [CrossRef]
- Soler, M.; Plasencia, A.; Larbat, R.; Pouzet, C.; Jauneau, A.; Rivas, S.; Pesquet, E.; Lapierre, C.; Truchet, I.; Grima-Pettenati, J. The Eucalyptus linker histone variant EgH1.3 cooperates with the transcription factor EgMYB1 to control lignin biosynthesis during wood formation. New Phytol. 2017, 213, 287–299. [Google Scholar] [CrossRef]
- Myburg, A.A.; Grattapaglia, D.; Tuskan, G.A.; Hellsten, U.; Hayes, R.D.; Grimwood, J.; Jenkins, J.; Lindquist, E.; Tice, H.; Bauer, D.; et al. The genome of Eucalyptus grandis. Nature 2014, 510, 356–362. [Google Scholar] [CrossRef] [Green Version]
- Worley, C.K.; Zenser, N.; Ramos, J.; Rouse, D.; Leyser, O.; Theologis, A.; Callis, J. Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. 2000, 21, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Dreher, K.A.; Brown, J.; Saw, R.E.; Callis, J. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 2006, 18, 699–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, A.; Yamamoto, K.T. What’s the physiological role of domain II-less Aux/IAA proteins? Plant Signal. Behav. 2008, 3, 496–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szemenyei, H.; Hannon, M.; Long, J.A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 2008, 319, 1384–1386. [Google Scholar] [CrossRef]
- Audran-Delalande, C.; Bassa, C.; Mila, I.; Regad, F.; Zouine, M.; Bouzayen, M. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol. 2012, 53, 659–672. [Google Scholar] [CrossRef] [Green Version]
- Ulmasov, T.; Murfett, J.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997, 9, 1963–1971. [Google Scholar] [CrossRef] [Green Version]
- Reed, J.W. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 2001, 6, 420–425. [Google Scholar] [CrossRef]
- Zhong, R.; Taylor, J.J.; Ye, Z.H. Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 1997, 9, 2159–2170. [Google Scholar] [CrossRef] [Green Version]
- Schilmiller, A.L.; Stout, J.; Weng, J.K.; Humphreys, J.; Ruegger, M.O.; Chapple, C. Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 2009, 60, 771–782. [Google Scholar] [CrossRef]
- Roschzttardtz, H.; Paez-Valencia, J.; Dittakavi, T.; Jali, S.; Reyes, F.C.; Baisa, G.; Anne, P.; Gissot, L.; Palauqui, J.C.; Masson, P.H.; et al. The VASCULATURE COMPLEXITY AND CONNECTIVITY gene encodes a plant-specific protein required for embryo provasculature development. Plant Physiol. 2014, 166, 889–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piya, S.; Shrestha, S.K.; Binder, B.; Stewart, C.N., Jr.; Hewezi, T. Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front. Plant Sci. 2014, 5, 744. [Google Scholar] [CrossRef] [Green Version]
- Vernoux, T.; Brunoud, G.; Farcot, E.; Morin, V.; Van den Daele, H.; Legrand, J.; Oliva, M.; Das, P.; Larrieu, A.; Wells, D.; et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 2011, 7, 508. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Soler, M.; Mila, I.; San Clemente, H.; Savelli, B.; Dunand, C.; Paiva, J.A.; Myburg, A.A.; Bouzayen, M.; Grima-Pettenati, J.; et al. Genome-wide characterization and expression profiling of the AUXIN RESPONSE FACTOR (ARF) gene family in Eucalyptus grandis. PLoS ONE 2014, 9, e108906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaschek, L.; Champagne, A.; Dimotakis, C.; Nuoendagula; Decou, R.; Hishiyama, S.; Kratzer, S.; Kajita, S.; Pesquet, E. Cellular and Genetic Regulation of Coniferaldehyde Incorporation in Lignin of Herbaceous and Woody Plants by Quantitative Wiesner Staining. Front. Plant Sci. 2020, 11, 109. [Google Scholar] [CrossRef]
- Mitra, P.P.; Loque, D. Histochemical Staining of Arabidopsis thaliana Secondary Cell Wall Elements. Jove-J. Vis. Exp. 2014, 87, e51381. [Google Scholar] [CrossRef] [Green Version]
- Sehr, E.M.; Agusti, J.; Lehner, R.; Farmer, E.E.; Schwarz, M.; Greb, T. Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J. 2010, 63, 811–822. [Google Scholar] [CrossRef]
- Azevedo, C.; Betsuyaku, S.; Peart, J.; Takahashi, A.; Noel, L.; Sadanandom, A.; Casais, C.; Parker, J.; Shirasu, K. Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 2006, 25, 2007–2016. [Google Scholar] [CrossRef] [Green Version]
- Sato, A.; Yamamoto, K.T. Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol. Plant. 2008, 133, 397–405. [Google Scholar] [CrossRef]
- Lv, B.; Yu, Q.; Liu, J.; Wen, X.; Yan, Z.; Hu, K.; Li, H.; Kong, X.; Li, C.; Tian, H.; et al. Non-canonical AUX/IAA protein IAA33 competes with canonical AUX/IAA repressor IAA5 to negatively regulate auxin signaling. EMBO J. 2020, 39, e101515. [Google Scholar] [CrossRef]
- Yang, F.; Mitra, P.; Zhang, L.; Prak, L.; Verhertbruggen, Y.; Kim, J.S.; Sun, L.; Zheng, K.; Tang, K.; Auer, M.; et al. Engineering secondary cell wall deposition in plants. Plant Biotechnol. J. 2013, 11, 325–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Meester, B.; de Vries, L.; Ozparpucu, M.; Gierlinger, N.; Corneillie, S.; Pallidis, A.; Goeminne, G.; Morreel, K.; De Bruyne, M.; De Rycke, R.; et al. Vessel-Specific Reintroduction of CINNAMOYL-COA REDUCTASE1 (CCR1) in Dwarfed ccr1 Mutants Restores Vessel and Xylary Fiber Integrity and Increases Biomass. Plant Physiol. 2018, 176, 611–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Meester, B.; Vanholme, R.; de Vries, L.; Wouters, M.; Van Doorsselaere, J.; Boerjan, W. Vessel- and ray-specific monolignol biosynthesis as an approach to engineer fiber-hypolignification and enhanced saccharification in poplar. Plant J. 2021, 108, 752–765. [Google Scholar] [CrossRef] [PubMed]
- Cassan-Wang, H.; Soler, M.; Yu, H.; Camargo, E.L.O.; Carocha, V.; Ladouce, N.; Savelli, B.; Paiva, J.A.P.; Leple, J.C.; Grima-Pettenati, J. Reference Genes for High-Throughput Quantitative Reverse Transcription-PCR Analysis of Gene Expression in Organs and Tissues of Eucalyptus Grown in Various Environmental Conditions. Plant Cell Physiol. 2012, 53, 2101–2116. [Google Scholar] [CrossRef] [Green Version]
- Paux, E.; Tamasloukht, M.; Ladouce, N.; Sivadon, P.; Grima-Pettenati, J. Identification of genes preferentially expressed during wood formation in Eucalyptus. Plant Mol. Biol. 2004, 55, 263–280. [Google Scholar] [CrossRef]
- Arvidsson, S.; Kwasniewski, M.; Riano-Pachon, D.M.; Mueller-Roeber, B. QuantPrime-a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinform. 2008, 9, 465. [Google Scholar] [CrossRef] [Green Version]
- Shimada, T.L.; Shimada, T.; Hara-Nishimura, I. A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 2010, 61, 519–528. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Abel, S.; Theologis, A. Transient transformation of Arabidopsis leaf protoplasts: A versatile experimental system to study gene expression. Plant J. 1994, 5, 421–427. [Google Scholar] [CrossRef]
- Gerber, L.; Eliasson, M.; Trygg, J.; Moritz, T.; Sundberg, B. Multivariate curve resolution provides a high-throughput data processing pipeline for pyrolysis-gas chromatography/mass spectrometry. J. Anal. Appl. Pyrol. 2012, 95, 95–100. [Google Scholar] [CrossRef]





| Genotype | Carbohydrate | Guaiacyl | Syringyl | p-Hydroxy-Phenol | Phenolic | Lignin | S/G | C/L |
|---|---|---|---|---|---|---|---|---|
| WT | 77.5 ± 2.0 | 8.5 ± 1.2 | 2.9 ± 0.7 | 1.7 ± 0.2 | 1.1 ± 0.1 | 14.2 ± 2.0 | 0.34 ± 0.04 | 5.55 ± 0.82 |
| EgrIAA201 | 76.9 ± 3.0 | 8.3 ± 0.8 | 2.0 ± 0.4 * | 1.9 ± 0.2 * | 1.0 ± 0.1 * | 13.2 ± 2.4 | 0.25 ± 0.03 ** | 6.03 ± 1.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.; Liu, M.; Zhu, Z.; Wu, A.; Mounet, F.; Pesquet, E.; Grima-Pettenati, J.; Cassan-Wang, H. Overexpression of EgrIAA20 from Eucalyptus grandis, a Non-Canonical Aux/IAA Gene, Specifically Decouples Lignification of the Different Cell-Types in Arabidopsis Secondary Xylem. Int. J. Mol. Sci. 2022, 23, 5068. https://doi.org/10.3390/ijms23095068
Yu H, Liu M, Zhu Z, Wu A, Mounet F, Pesquet E, Grima-Pettenati J, Cassan-Wang H. Overexpression of EgrIAA20 from Eucalyptus grandis, a Non-Canonical Aux/IAA Gene, Specifically Decouples Lignification of the Different Cell-Types in Arabidopsis Secondary Xylem. International Journal of Molecular Sciences. 2022; 23(9):5068. https://doi.org/10.3390/ijms23095068
Chicago/Turabian StyleYu, Hong, Mingjun Liu, Zhangsheng Zhu, Aiming Wu, Fabien Mounet, Edouard Pesquet, Jacqueline Grima-Pettenati, and Hua Cassan-Wang. 2022. "Overexpression of EgrIAA20 from Eucalyptus grandis, a Non-Canonical Aux/IAA Gene, Specifically Decouples Lignification of the Different Cell-Types in Arabidopsis Secondary Xylem" International Journal of Molecular Sciences 23, no. 9: 5068. https://doi.org/10.3390/ijms23095068







