A Simple Protocol for Sample Preparation for Scanning Electron Microscopic Imaging Allows Quick Screening of Nanomaterials Adhering to Cell Surface
Abstract
:1. Introduction
2. Results
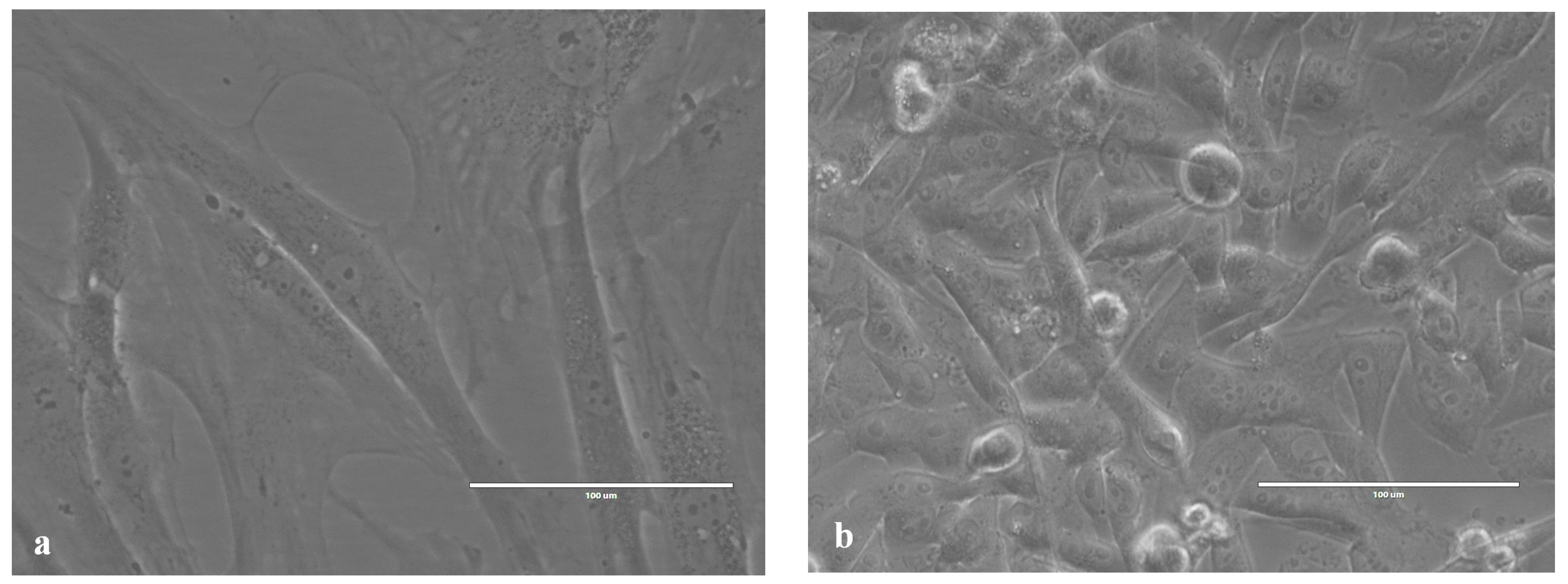
2.1. Images of Cells Obtained Using an Inverted Microscope
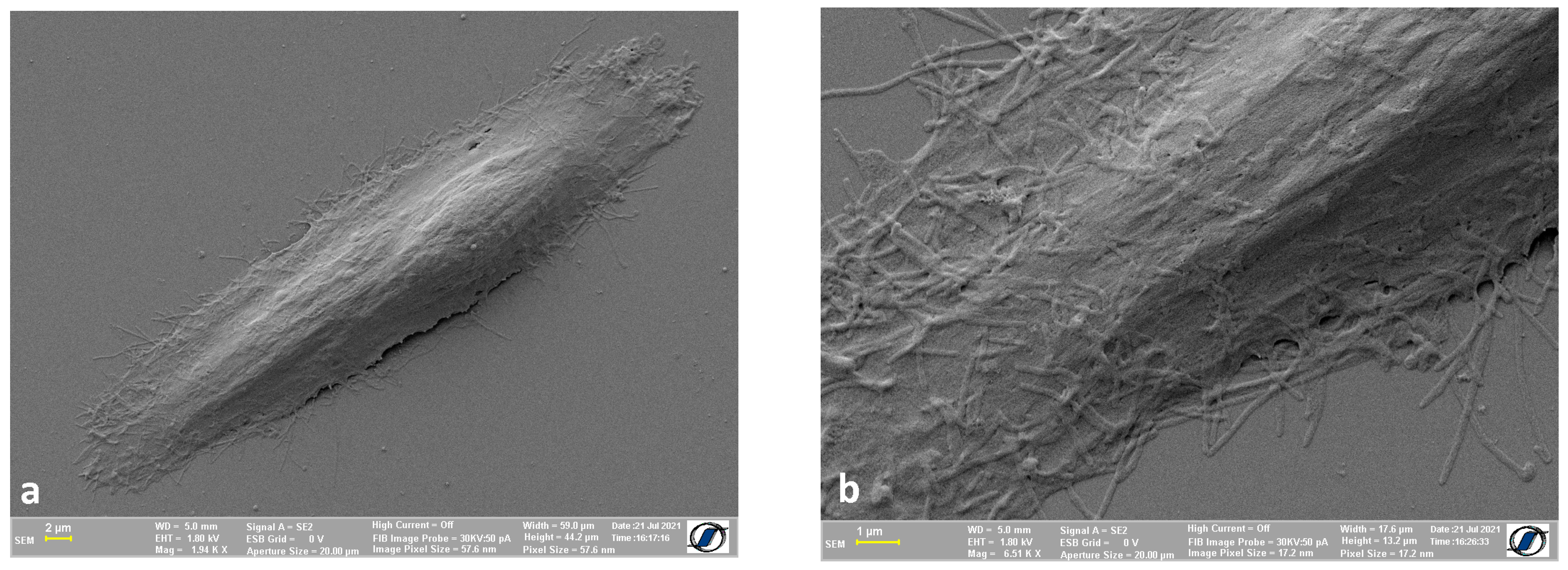
2.2. Images of Cells without Nanomaterials Obtained Using the Proposed Protocol
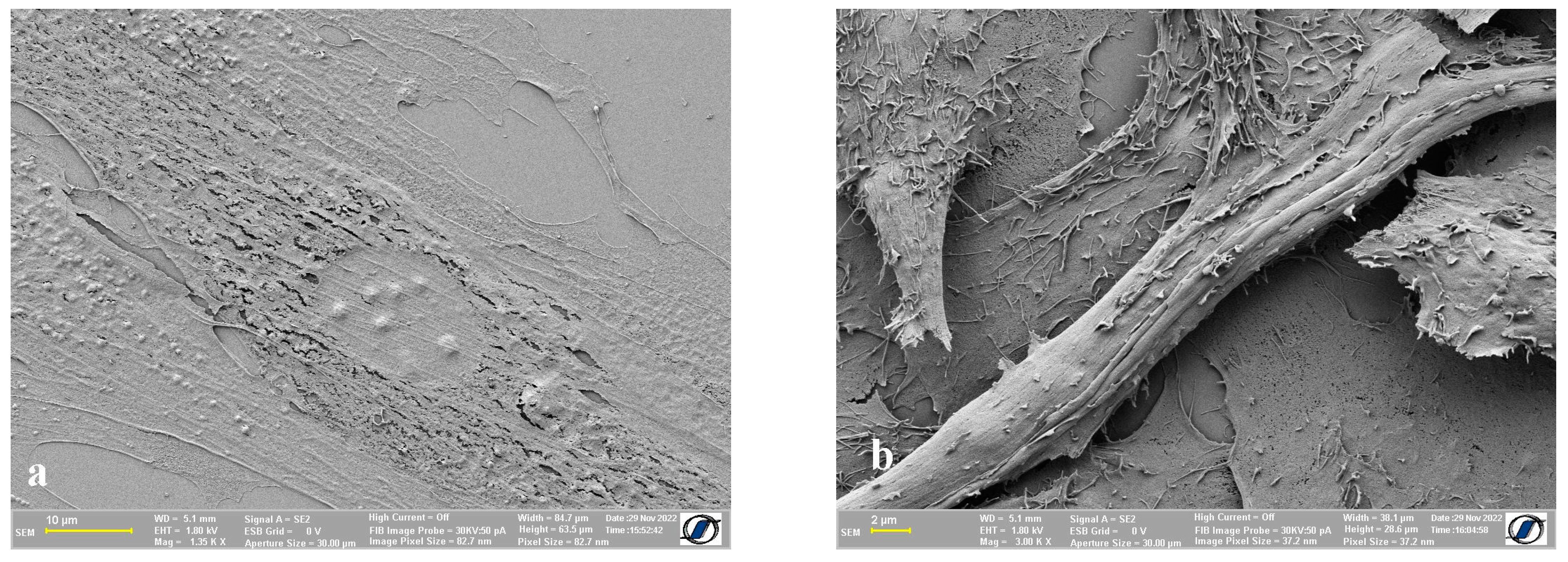
2.3. Images of Control Cells (without Nanomaterials) Obtained Using CPD
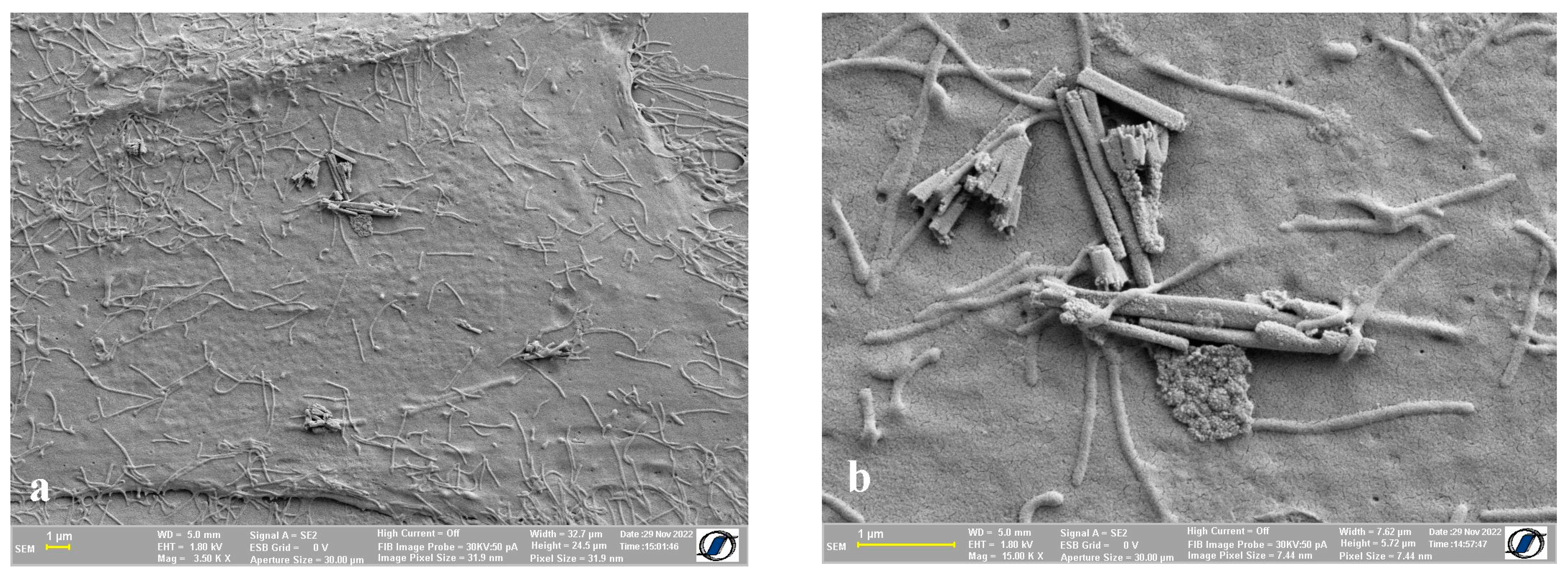
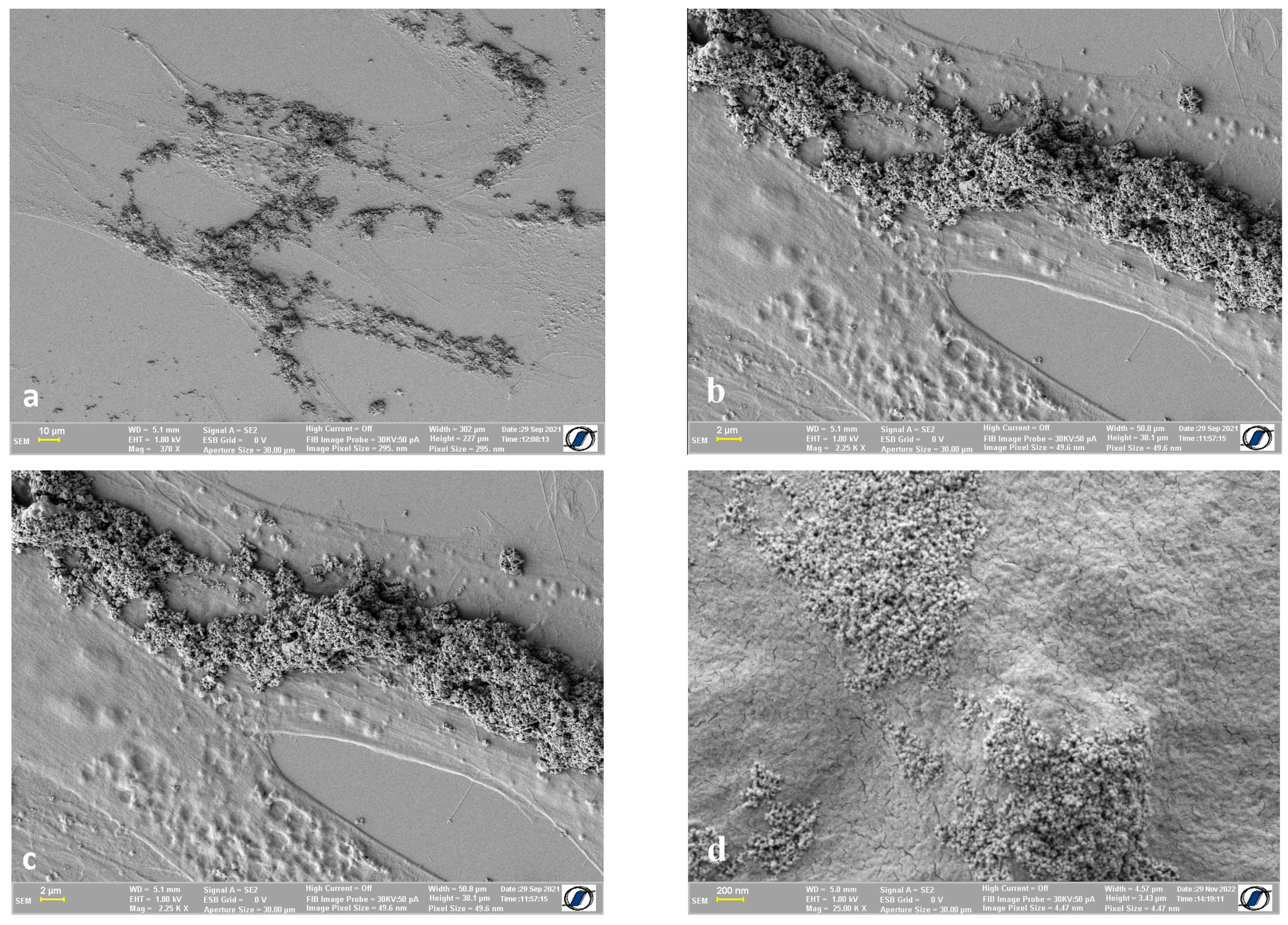
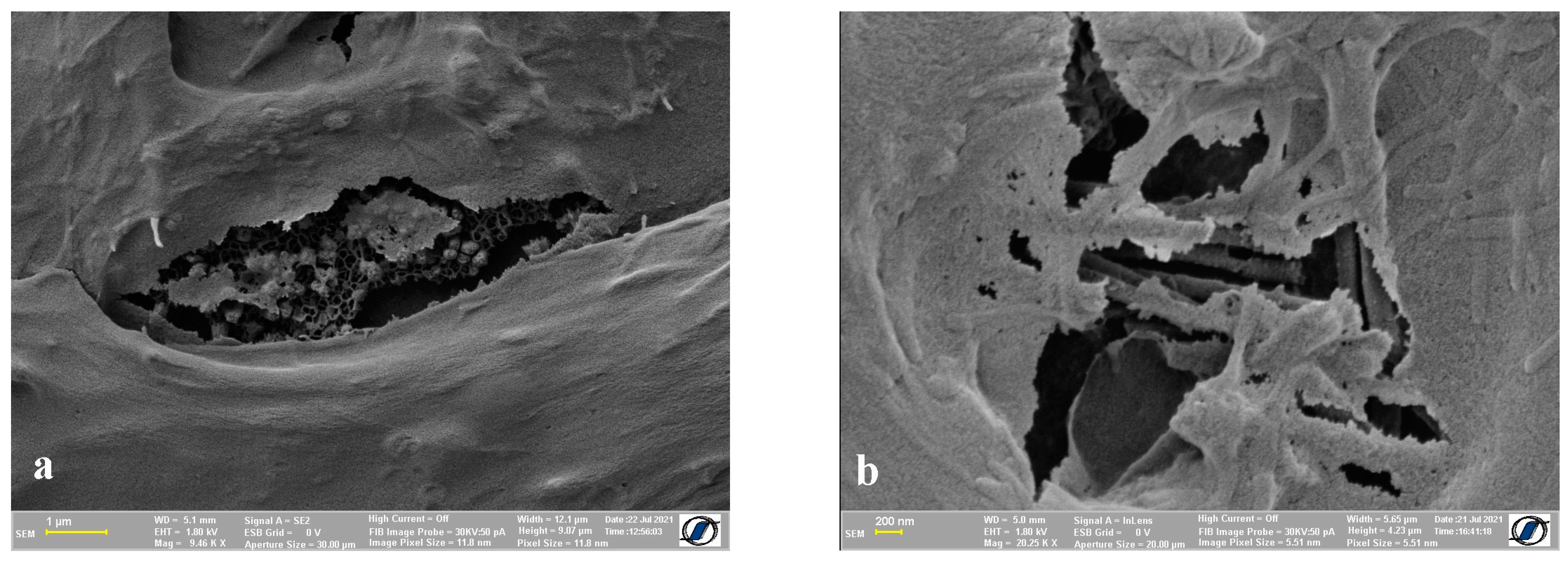
2.4. Images of Cells with Nanomaterials on the Cell Surface
3. Discussion
4. Materials and Methods
4.1. Cell Growth and Nanomaterial Adherence on the Cell Surface
4.2. Cell Fixation and Preparation for SEM Imaging
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Perez, J.E.; Ravasi, T.; Kosel, J. Mesenchymal stem cells cultured on magnetic nanowire substrates. Nanotechnology 2017, 28, 055703. [Google Scholar] [CrossRef] [PubMed]
- Anderson, T.F. Techniques for the preservation of three-dimensionalstructure in preparing specimens for the electron microscope. Trans. N. Y. Acad. Sci. 1951, 13, 130–133. [Google Scholar] [CrossRef]
- Bray, D.F.; Bagu, J.; Koegler, P. Comparison of hexamethyldisilazane (HMDS), Peldri II, and critical point drying methods for scanning electronmicroscopy of biological specimens. Microsc. Res. Tech. 1993, 26, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Chiriac, H.; Petreus, T.; Carasevici, E.; Labusca, L.; Herea, D.-D.; Danceanu, C.; Lupu, N. In vitro cytotoxicity of Fe–Cr–Nb–B magnetic nanoparticles under high frequency electromagnetic field. J. Magn. Magn. Mater. 2015, 380, 13–19. [Google Scholar] [CrossRef]
- Mendes, R.G.; Mandarino, A.; Koch, B.; Meyer, A.K.; Bachmatiuk, A.; Hirsch, C.; Gemming, T.; Schmidt, O.G.; Liu, A.; Rümmeli, M.H. Size and time dependent internalization of label-free nano-graphene oxide in human macrophages. Nano Res. 2017, 10, 1980–1995. [Google Scholar] [CrossRef]
- Avasthi, A.; Caro, C.; Pozo-Torres, E.; Leal, M.P.; García-Martín, M.L. Magnetic Nanoparticles as MRI Contrast Agents. Top. Curr. Chem. 2020, 378, 40. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.F.; Jang, B.; Issadore, D.; Tsourkas, A. Use of magnetic fields and nanoparticles to trigger drug release and improve tumor targeting. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2019, 11, e1571. [Google Scholar] [CrossRef] [PubMed]
- Labusca, L.; Herea, D.D.; Mashayekhi, K. Stem cells as delivery vehicles for regenerative medicine-challenges and perspectives. World J. Stem Cells 2018, 10, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Ortega, L.; Rojas, J.M.; Portilla, Y.; Pérez-Yagüe, S.; Barber, D.F. Magnetic Nanoparticles Attached to the NK Cell Surface for Tumor Targeting in Adoptive Transfer Therapies Does Not Affect Cellular Effector Functions. Front. Immunol. 2019, 10, 2073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharya, R.; Saha, S.; Kostina, O.; Muravnik, L.; Mitra, A. Replacing critical point drying with a low-cost chemical drying provides comparable surface image quality of glandular trichomes from leaves of Millingtonia hortensis L. f. in scanning electron micrograph. Appl. Microsc. 2020, 50, 15. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.Y.; Hwang, K.R.; Park, J.Y.; Lee, J.P.; Kim, J.S.; Lee, J.S. Critical Point Drying: An Effective Drying Method for Direct Measurement of the Surface Area of a Pretreated Cellulosic Biomass. Polymers 2018, 10, 676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albrecht, R.M.; Rasmussen, D.H.; Keller, C.S.; Hinsdill, R.D. Preparation of cultured cells for SEM: Air drying from organic solvents. J. Microsc. 1976, 108, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Osahor, A.; Deekonda, K.; Lee, C.W.; Sim, E.U.; Radu, A.; Narayanan, K. Rapid preparation of adherent mammalian cells for basic scanning electron microscopy (SEM) analysis. Anal. Biochem. 2017, 534, 46–48. [Google Scholar] [CrossRef] [PubMed]
- Ali, R.; El-Boubbou, K.; Boudjelal, M. An easy, fast and inexpensive method of preparing a biological specimen for scanning electron microscopy (SEM). MethodsX 2021, 8, 101521. [Google Scholar] [CrossRef] [PubMed]
- Minuti, A.E.; Stoian, G.; Herea, D.-D.; Radu, E.; Lupu, N.; Chiriac, H. Fe-Cr-Nb-B Ferrofluid for Biomedical Applications. Nanomaterials 2022, 12, 1488. [Google Scholar] [CrossRef] [PubMed]
- Chiriac, H.; Minuti, A.E.; Lupu, N. Cancer Cell Destruction by Magneto-Mechanical Actuation of Nanowires Compared with Nano/Micromagnetic Particles. J. Biomed. Res. Environ. Sci. 2022, 3, 905–907. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minuti, A.E.; Labusca, L.; Herea, D.-D.; Stoian, G.; Chiriac, H.; Lupu, N. A Simple Protocol for Sample Preparation for Scanning Electron Microscopic Imaging Allows Quick Screening of Nanomaterials Adhering to Cell Surface. Int. J. Mol. Sci. 2023, 24, 430. https://doi.org/10.3390/ijms24010430
Minuti AE, Labusca L, Herea D-D, Stoian G, Chiriac H, Lupu N. A Simple Protocol for Sample Preparation for Scanning Electron Microscopic Imaging Allows Quick Screening of Nanomaterials Adhering to Cell Surface. International Journal of Molecular Sciences. 2023; 24(1):430. https://doi.org/10.3390/ijms24010430
Chicago/Turabian StyleMinuti, Anca Emanuela, Luminita Labusca, Dumitru-Daniel Herea, George Stoian, Horia Chiriac, and Nicoleta Lupu. 2023. "A Simple Protocol for Sample Preparation for Scanning Electron Microscopic Imaging Allows Quick Screening of Nanomaterials Adhering to Cell Surface" International Journal of Molecular Sciences 24, no. 1: 430. https://doi.org/10.3390/ijms24010430




