Glucose Homeostasis and Pancreatic Islet Size Are Regulated by the Transcription Factors Elk-1 and Egr-1 and the Protein Phosphatase Calcineurin
Abstract
:1. Introduction
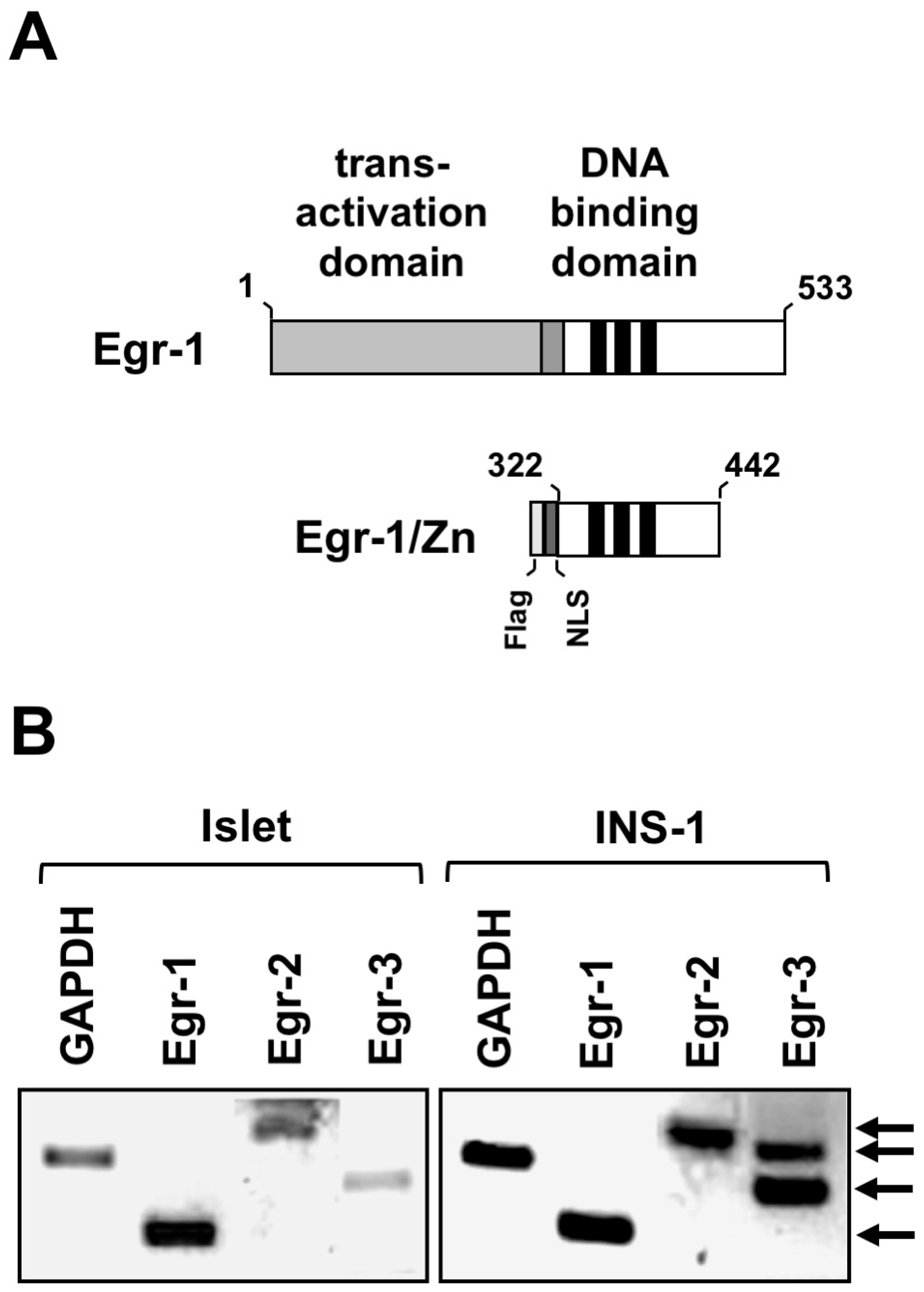
2. Stimulation of Pancreatic β-Cells Induces the Expression of Egr-1
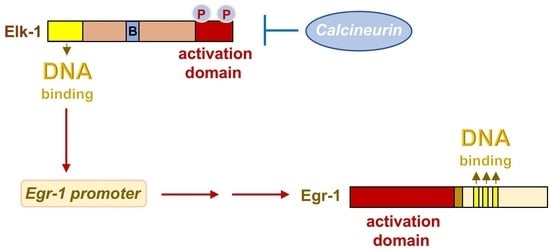
3. Stimulation of Pancreatic β-Cells Induces Phosphorylation and Activation of the Ternary Complex Factor Elk-1
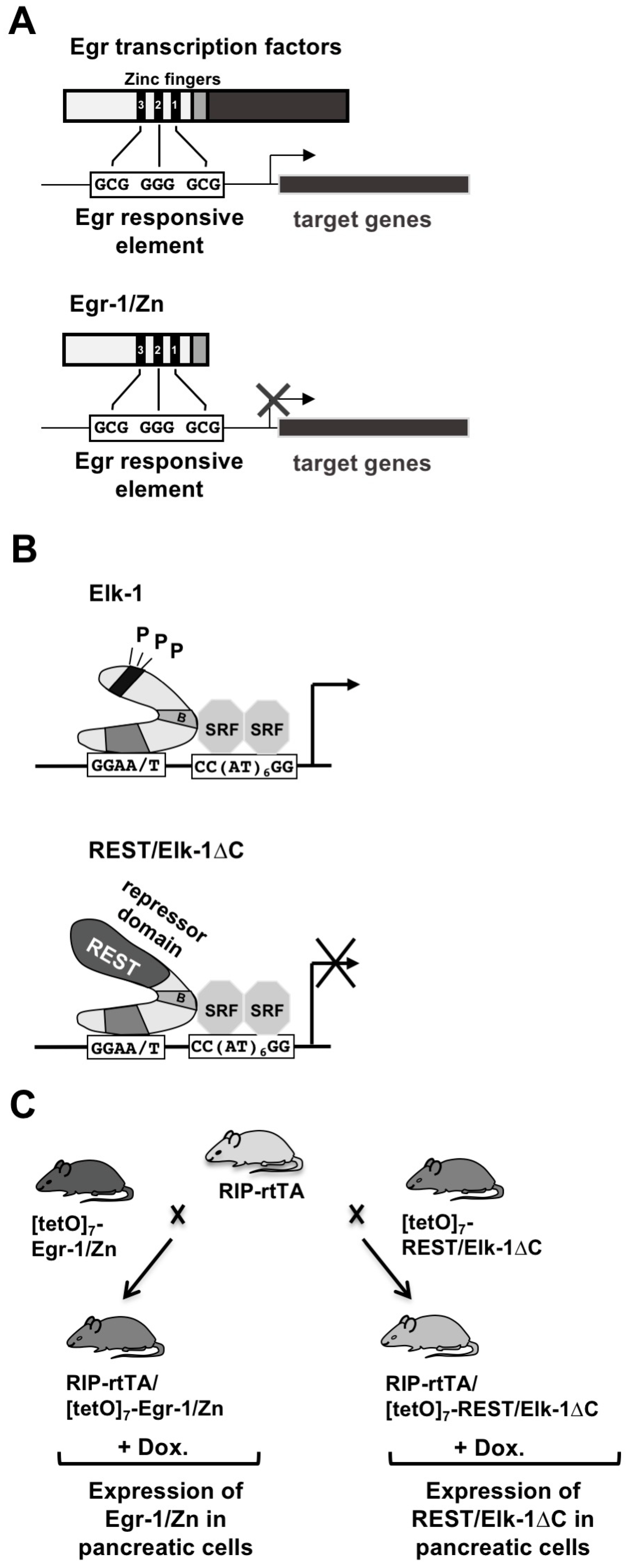
4. Experimentally Overcoming Functional Redundancy: A Transgenic Strategy
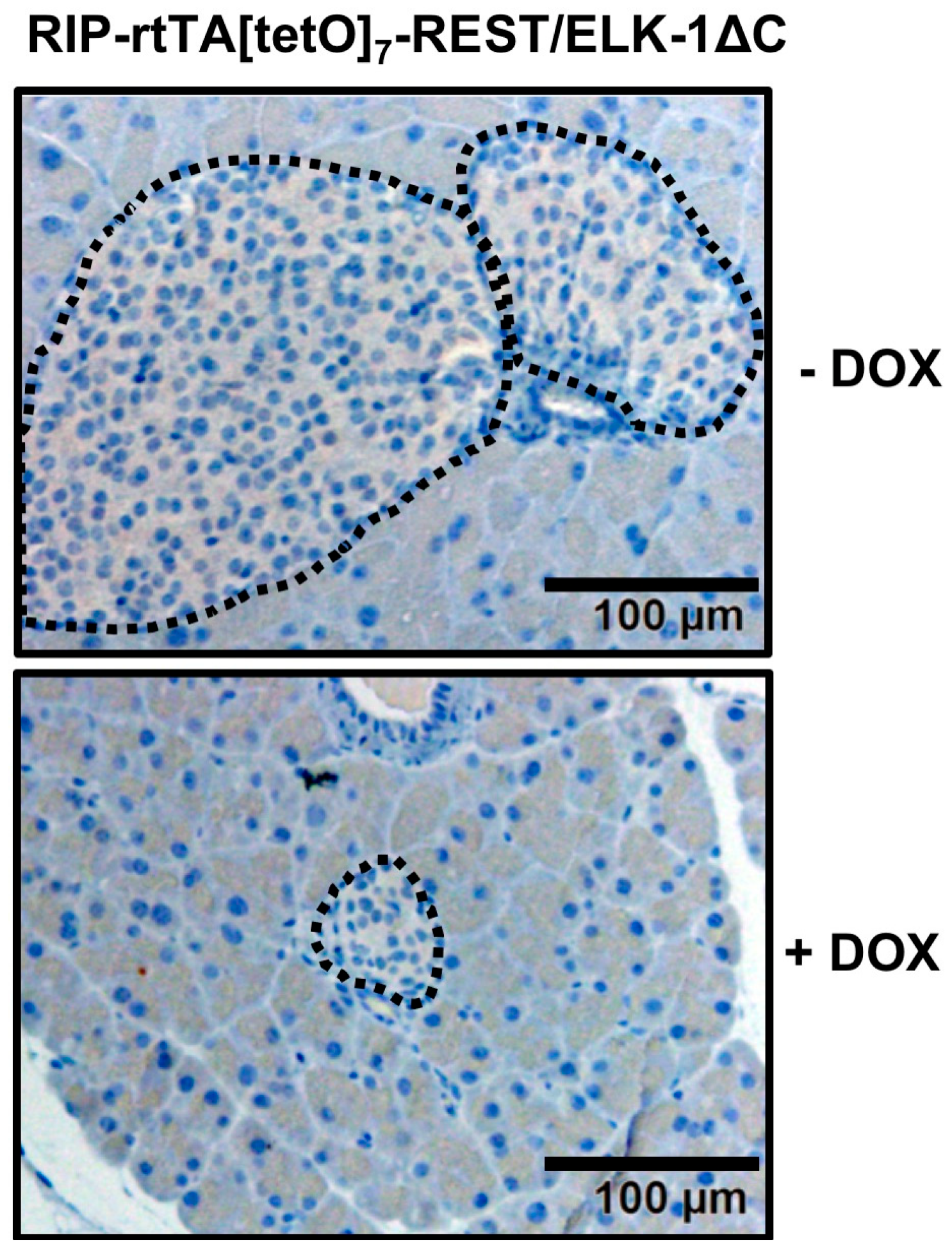
5. Expression of Egr-1/Zn or REST/Elk-1∆C Leads to a Reduction in the Size of Pancreatic Islets
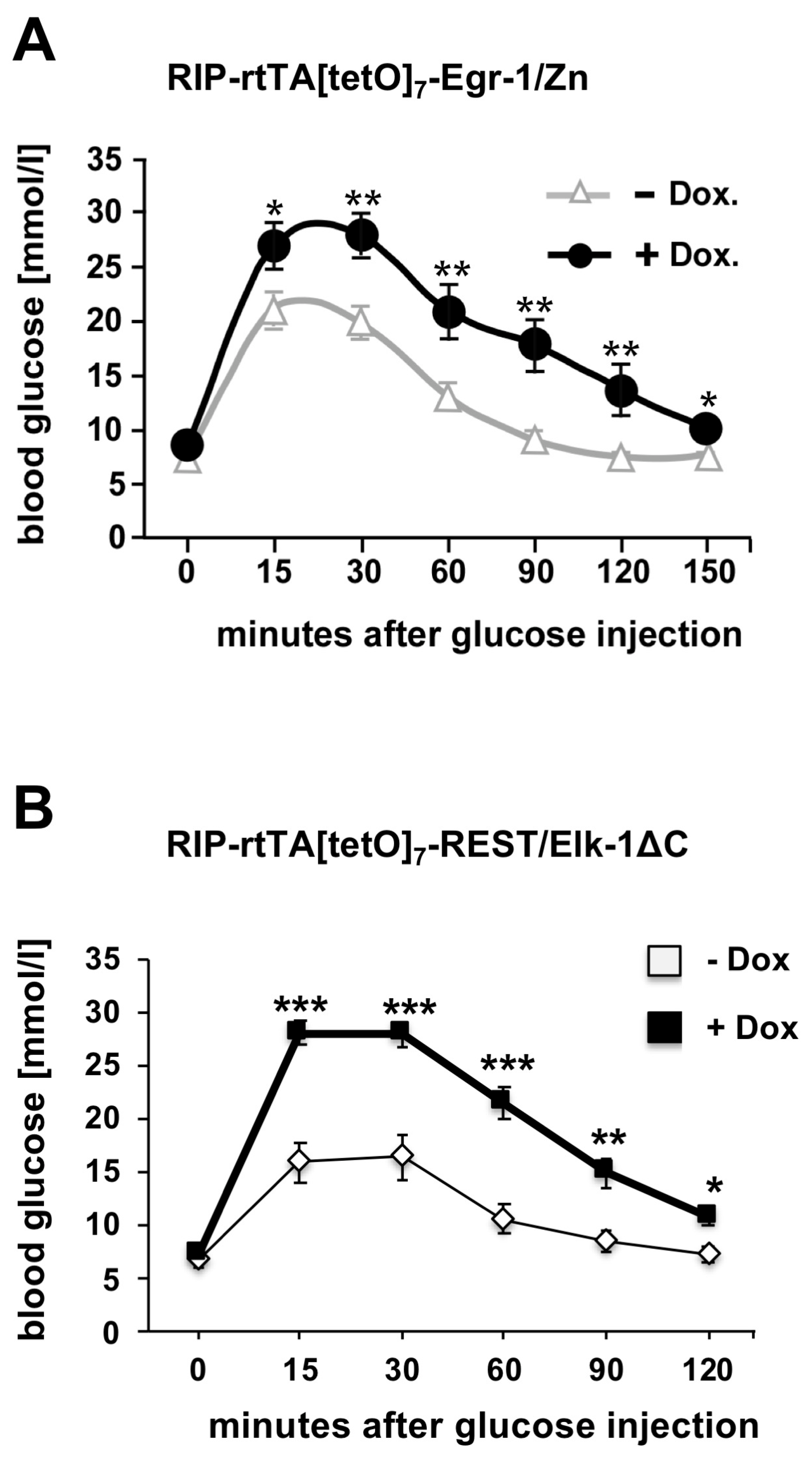
6. Impaired Glucose Tolerance in Transgenic Mice Expressing Either Egr-1/Zn or REST/Elk-1ΔC in Pancreatic β-Cells
7. Expression of Activated Calcineurin A Leads to the Formation of Smaller Islets and Impaired Glucose Homeostasis
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nitert, M.D.; Nagorny, C.L.F.; Wendt, A.; Eliasson, L.; Mulder, H. Cav1.2 rather than Cav1.3 is coupled to glucose-stimulated insulin secretion in INS-1 832/13 cells. J. Mol. Endocrinol. 2008, 41, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiel, G.; Müller, I.; Rössler, O.G. Signal transduction via TRPM3 channels in pancreatic β-cells. J. Mol. Endocrinol. 2013, 50, R75–R83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautam, D.; Han, S.-J.; Hamdan, F.F.; Jeon, J.; Li, B.; Li, J.H.; Cui, Y.; Mears, D.; Lu, H.; Deng, C.; et al. A critical role for β cell M3 muscarinic acetylcholine receptors in regulating insulin release and blood glucose homeostasis in vivo. Cell Metabol. 2006, 3, 449–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frödin, M.; Sekine, N.; Roche, E.; Filloux, C.; Prentki, M.; Wollheim, C.B.; Van Obberghen, E.V. Glucose, other secretagogues, and nerve growth factor stimulate mitogen-activated protein kinase in the insulin-secreting beta cell line, INS-1. J. Biol. Chem. 1995, 270, 7882–7889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Josefsen, K.; Sorensen, L.R.; Buschard, K.; Birkenbach, M. Glucose induces early growth response gene (Egr-1) expression in pancreatic beta cells. Diabetologia 1999, 42, 195–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, S.I.; Thiel, G. Calcium influx into MIN6 insulinoma cells induces expression of Egr-1 involving extracellular signal-regulated protein kinase and the transcription factors Elk-1 and CREB. Eur. J. Cell Biol. 2009, 88, 19–33. [Google Scholar] [CrossRef]
- Mayer, S.I.; Müller, I.; Mannebach, S.; Endo, T.; Thiel, G. Signal transduction of pregnenolone sulfate in insulinoma cells. Activation of Egr-1 expression involving TRPM3, voltage-gated calcium channels, ERK, and ternary complex factors. J. Biol. Chem. 2011, 286, 10084–10096. [Google Scholar] [CrossRef] [Green Version]
- Kaufmann, K.; Thiel, G. Epidermal growth factor and thrombin induced proliferation of immortalized human keratinocytes is coupled to the synthesis of Egr-1, a zinc finger transcriptional regulator. J. Cell. Biochem. 2002, 85, 381–391. [Google Scholar] [CrossRef]
- Kim, M.-J.; Kang, J.-H.; Chang, S.-Y.; Jang, H.-J.; Ryu, G.R.; Ko, S.H.; Jeong, I.-K.; Kim, M.-S.; Jo, Y.-H. Exendin-4 induction of Egr-1 in INS-1 b-cells: Interaction of SRF, not YY1, with SRE site of rat Egr-1 promoter. J. Cell. Biochem. 2008, 104, 2261–2271. [Google Scholar] [CrossRef]
- Müller, I.; Rössler, O.G.; Wittig, C.; Menger, M.D.; Thiel, G. Critical role of Egr transcription factors in regulating insulin biosynthesis, blood glucose homeostasis and islet size. Endocrinology 2012, 153, 3040–3053. [Google Scholar] [CrossRef]
- Wang, W.; Walker, J.R.; Wang, X.; Tremblay, M.S.; Lee, J.W.; Wu, X.; Schultz, P.G. Identification of small-molecule inducers of pancreatic β-cell expansion. Proc. Natl. Acad. Sci. USA 2009, 106, 1427–1432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Sadovsky, Y.; Swirnoff, A.H.; Polish, J.A.; Goda, P.; Gavrilina, G.; Milbrandt, J. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (egr-1). Science 1996, 273, 1219–1221. [Google Scholar] [CrossRef] [PubMed]
- Topilko, P.; Schneider-Maunoury, S.; Levi, G.; Trembleau, A.; Gourji, D.; Driancourt, M.-A.; Rao, C.V.; Charnay, P. Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. Mol. Endocrinol. 1997, 12, 107–122. [Google Scholar] [CrossRef]
- Wei, F.; Xu, Z.C.; Qu, Z.; Milbrandt, J.; Zhuo, M. Role of EGR1 in hippocampal synaptic enhancement induced by tetanic stimulation and amputation. J. Cell Biol. 2000, 149, 1325–1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, S.-F.; Fujita, T.; Lu, J.; Okada, K.; Zou, Y.S.; Mackman, N.; Pinsky, D.J.; Stern, D.M. Egr-1, a master switch coordinating upregulation of divergent gene families underlying ischemic stress. Nat. Med. 2000, 6, 1355–1361. [Google Scholar] [CrossRef]
- Mayer, S.I.; Rössler, O.G.; Endo, T.; Charnay, P.; Thiel, G. Epidermal growth factor-induced proliferation of astrocytes requires Egr transcription factors. J. Cell Sci. 2009, 122, 3340–3350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, B.; Wang, C.; Zheng, Z.; Song, B.; Ma, C.; Thiel, G.; Li, M. Egr-1 transactivates Bim gene expression to promote neuronal apoptosis. J. Neurosci. 2011, 31, 5032–5044. [Google Scholar] [CrossRef] [Green Version]
- Eto, K.; Kaur, V.; Thomas, M.K. Regulation of insulin gene transcription by the immediate early response gene Egr-1. Endocrinology 2006, 147, 2923–2935. [Google Scholar] [CrossRef]
- Eto, K.; Kaur, V.; Thomas, M.K. Regulation of pancreatic duodenum homeobox-1 expression by early growth response-1. J. Biol. Chem. 2007, 282, 5973–5983. [Google Scholar] [CrossRef] [Green Version]
- Thiel, G.; Backes, T.M.; Guethlein, L.A.; Rössler, O.G. Critical protein—Protein interactions determine the biological activity of Elk-1, a master regulator of stimulus-induced gene transcription. Molecules 2021, 26, 6125. [Google Scholar] [CrossRef]
- Vickers, E.R.; Kasza, A.; Kurnaz, I.A.; Seifert, A.; Zeef, L.A.H.; O’Donnell, A.; Hayes, A.; Sharrocks, A.D. Ternary complex factor-serum response factor complex-regulated gene activity is required for cellular proliferation and inhibition of apoptotic cell death. Mol. Cell. Biol. 2004, 24, 10340–10351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costello, P.; Nicolas, R.; Willoughby, J.; Wasylyk, B.; Nordheim, A.; Treisman, R. Ternary complex factors SAP-1 and Elk-1, but not Net, are functionally equivalent in thymocyte development. J. Immunol. 2010, 185, 1082–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patki, M.; Chari, V.; Sivakumaran, S.; Gonit, M.; Trumbly, R.; Ratnam, M. The ETS domain transcription factor ELK1 directs a critical component of growth signaling by the androgen receptor in prostate cancer cells. J. Biol. Chem. 2013, 288, 11047–11065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gualdrini, F.; Esnault, C.; Horswell, S.; Stewart, A.; Matthews, N.; Treisman, R. SRF Co-factors control the balance between cell proliferation and contractility. Mol. Cell 2016, 64, 1048–1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernal-Mizrachi, E.; Wen, W.; Srinivasan, S.; Klenk, A.; Cohen, D.; Permutt, M.A. Activation of Elk-1, an Ets transcription factor, by glucose and EGF treatment of insulinoma cells. Am. J. Physiol. Endocrinol. Metabol. 2001, 281, E1286–E1299. [Google Scholar] [CrossRef]
- Lesch, A.; Backes, T.M.; Langfermann, D.S.; Rössler, O.G.; Laschke, M.W.; Thiel, G. Ternary complex factor regulates pancreatic islet size and blood glucose homeostasis in transgenic mice. Pharmacol. Res. 2020, 159, 104983. [Google Scholar] [CrossRef] [PubMed]
- Tourtellotte, W.G.; Nagarajan, R.; Bartke, A.; Milbrandt, J. 2000 Functional compensation by Egr4 in Egr1-dependent luteinizing hormone regulation and Leydig cell steroidogenesis. Mol. Cell. Biol. 2000, 20, 5261–5268. [Google Scholar] [CrossRef] [Green Version]
- Cesari, F.; Brecht, S.; Vintersten, K.; Vuong, L.G.; Hofmann, M.; Klingel, K.; Schnorr, J.J.; Arsenian, S.; Schild, H.; Herdegen, T.; et al. Mice deficient for the Ets transcription factor Elk-1 show normal immune responses and mildly impaired neuronal gene activation. Mol. Cell. Biol. 2004, 24, 294–305. [Google Scholar] [CrossRef] [Green Version]
- Tourtellotte, W.G.; Milbrandt, J. Sensory ataxia and muscle spindle agenesis in mice lacking the transcription factor Egr3. Nat. Genet. 1998, 20, 87–91. [Google Scholar] [CrossRef]
- Butler, A.E.; Janson, J.; Bonner-Weir, S.; Ritzel, R.; Rizza, R.A.; Butler, F.C. β-cell deficit and increased β-cell apoptosis in humans with type 2 diabetes. Diabetes 2003, 52, 102–110. [Google Scholar] [CrossRef]
- Marchetti, P.; Del Guerra, S.; Marselli, L.; Lupi, R.; Masini, M.; Pollera, M.; Bugliani, M.; Boggi, U.; Vistoli, F.; Mosca, F.; et al. Pancreatic islets from type 2 diabetic patients have functional defects and increased apoptosis that are ameliorated by metformin. J. Clin. Endocrinol. Metab. 2004, 89, 5535–5541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curran, M.; Drayson, M.T.; Andrews, R.C.; Zoppi, C.; Barlow, J.P.; Solomon, T.P.J.; Narendran, P. The benefits of physical exercise for the health of the pancreatic β-cell: A review of the evidence. Exp. Physiol. 2020, 105, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Segerstolpe, A.; Palasantza, A.; Eliasson, P.; Andersson, E.-M.; Andréasson, A.-C.; Sun, X.; Picelli, S.; Sabirsh, A.; Clausen, M.; Bjursell, M.K.; et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metabol. 2016, 24, 593–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuraba, H.; Mizukami, H.; Yagihashi, N.; Wada, R.; Hanyu, C.; Yagihashi, S. Reduced beta-cell mass and expression of oxidative stress-related DNA damage in the islet of Japanese type II diabetic patients. Diabetologia 2002, 45, 85–96. [Google Scholar] [CrossRef] [Green Version]
- Meier, J.J.; Breuer, T.G.; Bonadonna, R.C.; Tannapfel, A.; Uhl, W.; Schmidt, W.E.; Schrader, H.; Menge, B.A. Pancreatic diabetes manifests when β cell area declines by approximately 65% in humans. Diabetologia 2012, 55, 1346–1354. [Google Scholar] [CrossRef] [Green Version]
- Bernal-Mizrachi, E.; Cras-Méneur, C.; Ye, B.R.; Johnson, J.D.; Permutt, M.A. Transgenic overexpression of active calcineurin in β-cells results in decreased β-cell mass and hyperglycemia. PLoS ONE 2010, 5, e11969. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, T.; Stewart, S.; Guan, K.-L. The calcium/calmodulin-dependent protein phosphatase calcineurin is the major Elk-1 phosphatase. J. Biol. Chem. 1997, 272, 29415–29418. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.; Karin, M. Stimulation of Elk1 transcriptional activity by mitogen-activated protein kinases is negatively regulated by protein phosphatase 2B (calcineurin). J. Biol. Chem. 1999, 274, 15173–15180. [Google Scholar] [CrossRef] [Green Version]
- Langfermann, D.S.; Schmidt, T.; Rössler, O.G.; Thiel, G. Calcineurin controls gene transcription following stimulation of a Gαq-coupled designer receptor. Exp. Cell Res. 2019, 383, 111553. [Google Scholar] [CrossRef]
- Schaeffer, P.J.; Wende, A.R.; Magee, C.J.; Neilson, J.R.; Leone, T.C.; Chen, F.; Kelly, D.P. Calcineurin and calcium/calmodulin-dependent protein kinase activate distinct metabolic gene regulatory programs in cardiac muscle. J. Biol. Chem. 2004, 279, 39593–39603. [Google Scholar] [CrossRef]
- Baumgärtel, K.; Genoux, D.; Welzl, H.; Tweedie-Cullen, R.Y.; Koshibu, K.; Livingstone-Zatchej, M.; Mamie, C.; Mansuy, I.M. Control of the establishment of aversive memory by calcineurin and Zif268. Nature Neurosci. 2008, 11, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Lam, B.Y.H.; Zhang, W.; Enticknap, N.; Haggis, E.; Cader, M.Z.; Chawla, S. Inverse regulation of plasticity-related immediate early genes by calcineurin in hippocampal neurons. J. Biol. Chem. 2009, 284, 12562–12571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weir, G.C.; Gaglia, J.; Bonner-Weir Joslin, S. Inadequate β-cell mass is essential for the pathogenesis of type 2 diabetes. Lancet Diabetes Endocrinol. 2020, 8, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Poy, M.N.; Hausser, J.; Trajkovski, M.; Braun, M.; Collins, S.; Rorsman, P.; Zavolan, M.; Stoffel, M. miR-375 maintains normal pancreatic α- and β-cell mass. Proc. Natl. Acad. Sci. USA 2009, 106, 5813–5818. [Google Scholar] [CrossRef] [Green Version]
- Cordes, K.R.; Sheehy, N.T.; White, M.P.; Berry, E.C.; Morton, S.U.; Muth, A.N.; Lee, T.-H.; Miano, J.M.; Ivey, K.N.; Srivastava, D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009, 460, 705–710. [Google Scholar] [CrossRef] [Green Version]
- Henley, M.J.; Koehler, A.N. Advances in targeting ‘undruggable’ transcription factors with small molecules. Nature Rev. Drug Discov. 2021, 20, 669–688. [Google Scholar] [CrossRef]
- Li, H.-S.; Israni, D.V.; Gagnon, K.A.; Gan, K.A.; Raymond, M.H.; Sander, J.D.; Roybal, K.T.; Joung, J.K.; Wong, W.W.; Khalil, A.S. Multidimensional control of therapeutic human cell function with synthetic gene circuits. Science 2022, 378, 1227–1234. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thiel, G.; Rössler, O.G. Glucose Homeostasis and Pancreatic Islet Size Are Regulated by the Transcription Factors Elk-1 and Egr-1 and the Protein Phosphatase Calcineurin. Int. J. Mol. Sci. 2023, 24, 815. https://doi.org/10.3390/ijms24010815
Thiel G, Rössler OG. Glucose Homeostasis and Pancreatic Islet Size Are Regulated by the Transcription Factors Elk-1 and Egr-1 and the Protein Phosphatase Calcineurin. International Journal of Molecular Sciences. 2023; 24(1):815. https://doi.org/10.3390/ijms24010815
Chicago/Turabian StyleThiel, Gerald, and Oliver G. Rössler. 2023. "Glucose Homeostasis and Pancreatic Islet Size Are Regulated by the Transcription Factors Elk-1 and Egr-1 and the Protein Phosphatase Calcineurin" International Journal of Molecular Sciences 24, no. 1: 815. https://doi.org/10.3390/ijms24010815