Exploring Redox Modulation of Plant UDP-Glucose Pyrophosphorylase
Abstract
:1. Introduction
2. Results
2.1. Redox Modulation of Sugarcane UGPase
2.2. Redox Modulation of Barley UGPase
3. Discussion
3.1. Redox Compounds Affecting Plant UGPase Activity
3.2. Redox and Substrate Affinity of Plant UGPases
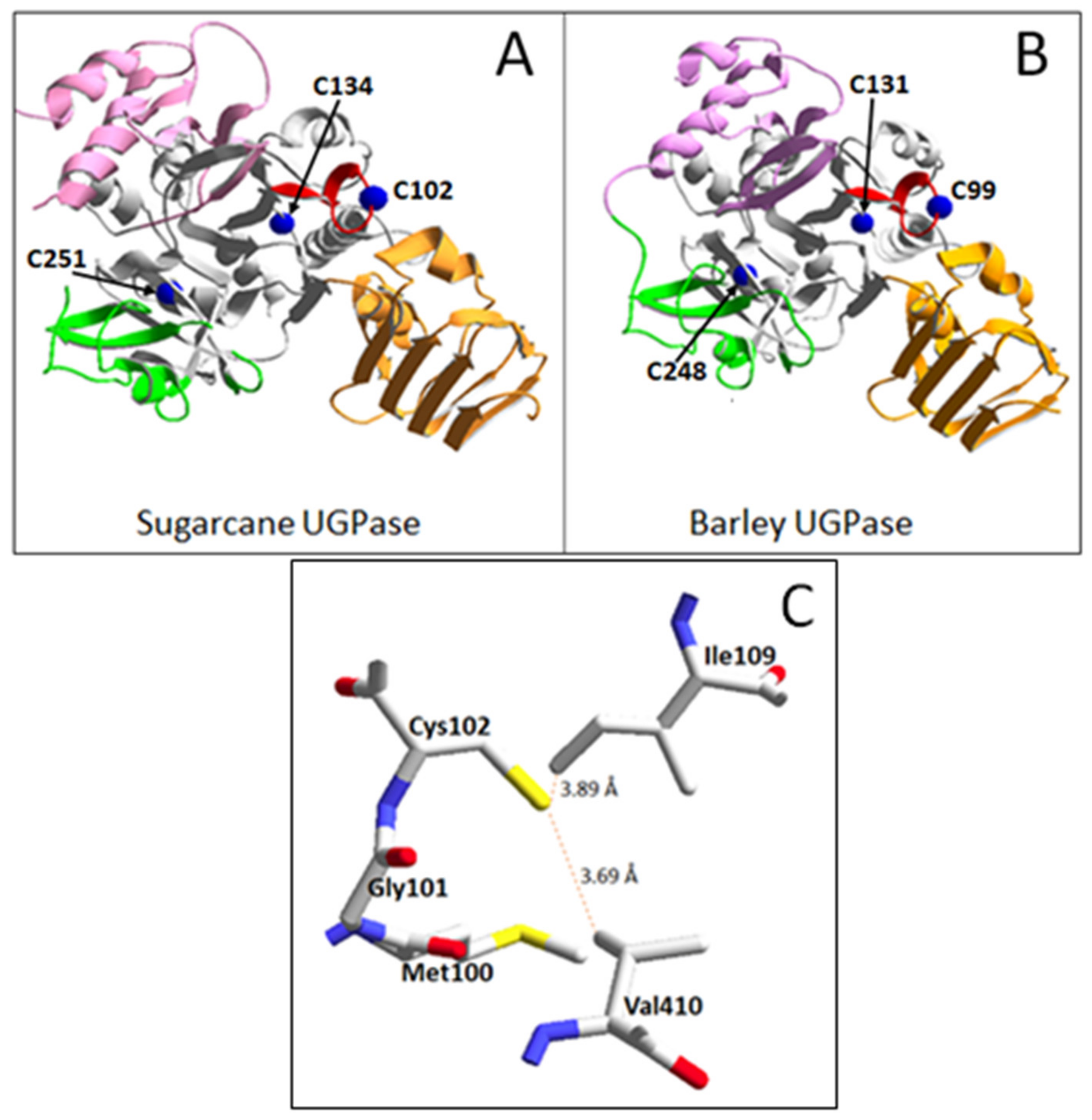
3.3. Structural Basis for Redox Control of Plant UGPases
3.4. Plant UGPase Is under Transcriptional, Posttranslational, and Metabolite Control
4. Materials and Methods
4.1. Materials
4.2. Assays
4.3. Kinetic Studies
4.4. Structural Modeling
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zrenner, R.; Willmitzer, L.; Sonnewald, U. Analysis of the expression of potato uridinediphosphate-glucose pyrophosphorylase and its inhibition by antisense RNA. Planta 1993, 190, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Meng, M.; Geisler, M.; Johansson, H.; Harholt, J.; Scheller, H.V.; Mellerowicz, E.J.; Kleczkowski, L.A. UDP-glucose pyrophosphorylase is not rate-limiting, but is essential in Arabidopsis. Plant Cell Physiol. 2009, 50, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Park, J.I.; Ishimizu, T.; Suwabe, K.; Sudo, K.; Masuko, H.; Hakozaki, H.; Nou, I.S.; Suzuki, G.; Watanabe, M. UDP-glucose pyrophosphorylase is rate limiting in vegetative and reproductive phases in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 981–996. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.A.; Decker, D. Sugar activation for production of nucleotide sugars as substrates for glycosyltransferases in plants. J. Appl. Glycosci. 2015, 62, 25–36. [Google Scholar] [CrossRef]
- Decker, D.; Kleczkowski, L.A. UDP-sugar producing pyrophosphorylases—Distinct and essential enzymes with overlapping substrate specificities, providing de novo precursors for glycosylation reactions. Front. Plant Sci. 2019, 9, 1822. [Google Scholar] [CrossRef] [PubMed]
- Stucki, J.W. The thermodynamic-buffer enzymes. Eur. J. Biochem. 1980, 109, 257–267. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Kleczkowski, L.A. Metabolic systems maintain stable non-equilibrium via thermodynamic buffering. BioEssays 2009, 31, 1091–1099. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Igamberdiev, A.U. Optimization of nucleotide sugar supply for polysaccharide formation via thermodynamic buffering. Biochem. J. 2020, 477, 341–356. [Google Scholar] [CrossRef]
- Feingold, D.S.; Avigad, G. Sugar nucleotide transformation in plants. In The Biochemistry of Plants; Stumpf, P.K., Conn, E.E., Eds.; Academic Press: New York, NY, USA, 1980; Volume 3, pp. 101–170. [Google Scholar] [CrossRef]
- Feingold, D.S.; Barber, G.A. Nucleotide sugars. In Methods in Plant Biochemistry; Dey, P.M., Harborne, J.B., Eds.; Academic Press: London, UK, 1990; Volume 2, pp. 39–78. [Google Scholar] [CrossRef]
- Knop, J.K.; Hansen, R.G. Uridine diphosphate glucose pyrophosphorylase. IV. Crystallization and properties of the enzyme from human liver. J. Biol. Chem. 1970, 245, 2499–2504. [Google Scholar] [CrossRef]
- Martínez, L.I.; Piattoni, C.V.; Garay, S.A.; Rodrígues, D.E.; Guerrero, S.A.; Iglesias, A.A. Redox regulation of UDP-glucose pyrophosphorylase from Entamoeba histolytica. Biochimie 2011, 93, 260–268. [Google Scholar] [CrossRef]
- Führing, J.A.; Cramer, J.T.; Schneider, J.; Baruch, P.; Gerardy-Schahn, R.; Fedorov, R. A quaternary mechanism enables the complex biological functions of octameric human UDP-glucose pyrophosphorylase, a key enzyme in cell metabolism. Sci. Rep. 2015, 5, 9618. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Omura, Y.; Tagaya, M.; Fukui, T. UDP-glucose pyrophosphorylase from potato tuber: Purification and characterization. J. Biochem. 1989, 106, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.A. Glucose activation and metabolism through UDP-glucose pyrophosphorylase in plants. Phytochemistry 1994, 37, 1507–1515. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Geisler, M.; Ciereszko, I.; Johansson, H. UDP-glucose pyrophosphorylase. An old protein with new tricks. Plant Physiol. 2004, 134, 912–918. [Google Scholar] [CrossRef] [PubMed]
- Elling, L.; Kula, M.R. Purification of UDP-glucose pyrophosphorylase from germinated barley (malt). J. Biotechnol. 1994, 34, 157–163. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid. Redox Signal. 2009, 11, 861–905. [Google Scholar] [CrossRef] [PubMed]
- Keech, O.; Gardeström, P.; Kleczkowski, L.A.; Rouhier, N. The redox control of photorespiration: From biochemical and physiological aspects to biotechnological considerations. Plant Cell Environ. 2017, 40, 553–569. [Google Scholar] [CrossRef]
- Ebrecht, A.C.; Asención Diez, M.D.; Piattoni, C.V.; Guerrero, S.A.; Iglesias, A.A. The UDP-glucose pyrophosphorylase from Giardia lamblia is redox regulated and exhibits promiscuity to use galactose-1-phosphate. Biochim. Biophys. Acta 2015, 1850, 88–96. [Google Scholar] [CrossRef]
- Muchut, R.J.; Calloni, R.D.; Herrera, F.E.; Garay, S.A.; Arias, D.G.; Iglesias, A.A.; Guerrero, S.A. Elucidating paramylon and other carbohydrate metabolism in Euglena gracilis: Kinetic characterization, structure and cellular localization of UDP-glucose pyrophosphorylase. Biochimie 2018, 154, 176–186. [Google Scholar] [CrossRef]
- Wong, J.H.; Cai, N.; Balmer, Y.; Tanaka, C.K.; Vensel, W.H.; Hurkman, W.J.; Buchanan, B.B. Thioredoxin targets of developing wheat seeds identified by complementary proteomic approaches. Phytochemistry 2004, 65, 1629–1640. [Google Scholar] [CrossRef]
- Alkhalfioui, F.; Renard, M.; Vensel, W.H.; Wong, J.; Tanaka, C.K.; Hurkman, W.J.; Buchanan, B.B.; Montrichard, F. Thioredoxin-linked proteins are reduced during germination of Medicago truncatula seeds. Plant Physiol. 2007, 144, 1559–1579. [Google Scholar] [CrossRef]
- Collet, J.F.; Messens, J. Structure, function, and mechanism of thioredoxin proteins. Antioxid. Redox Signal. 2010, 13, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Skipsey, M.; Grundy, N.M.; Edwards, R. Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 2005, 138, 2233–2244. [Google Scholar] [CrossRef]
- Wolosiuk, R.A.; Buchanan, B.B. Thioredoxin and glutathione regulate photosynthesis in chloroplasts. Nature 1977, 266, 565–567. [Google Scholar] [CrossRef]
- Lee, K.; Lee, J.; Kim, Y.; Bae, D.; Kang, K.Y.; Yoon, S.C.; Lim, D. Defining the plant disulfide proteome. Electrophoresis 2004, 25, 532–541. [Google Scholar] [CrossRef]
- Fratelli, M.; Gianazza, E.; Ghezzi, P. Redox proteomics: Identification and functional role of glutathionylated proteins. Expert Rev. Proteom. 2004, 1, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Ghezzi, P. Protein glutathionylation in health and disease. Biochim. Biophys. Acta 2013, 1830, 3165–3172. [Google Scholar] [CrossRef]
- Soares, J.S.; Gentile, A.; Scorsato, V.; Lima, A.C.; Kiyota, E.; Dos Santos, M.L.; Piattoni, C.V.; Huber, S.C.; Aparicio, R.; Menossi, M. Oligomerization, membrane association, and in vivo phosphorylation of sugarcane UDP-glucose pyrophosphorylase. J. Biol. Chem. 2014, 289, 33364–33377. [Google Scholar] [CrossRef]
- Karpinski, S.; Reynolds, H.; Karpinska, B.; Wingsle, G.; Creissen, G.; Mullineaux, P. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 1999, 284, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.A.; Randall, D.D. Light and thiol activation of maize leaf glycerate kinase. The stimulating effect of reduced thioredoxins and ATP. Plant Physiol. 1985, 79, 274–277. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, B.B. The path to thioredoxin and redox regulation in chloroplasts. Annu. Rev. Plant Biol. 2016, 67, 1–24. [Google Scholar] [CrossRef]
- McCoy, J.G.; Bitto, E.; Bingman, C.A.; Wesenberg, G.E.; Bannen, R.M.; Kondrashov, D.A.; Phillips, G.N. Structure and dynamics of UDPglucose pyrophosphorylase from Arabidopsis thaliana with bound UDPglucose and UTP. J. Mol. Biol. 2007, 366, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Cotrim, C.A.; Soares, J.S.M.; Kobe, B.; Menossi, M. Crystal structure and insights into the oligomeric state of UDP-glucose pyrophosphorylase from sugarcane. PLoS ONE 2018, 13, e0193667. [Google Scholar] [CrossRef]
- Martz, F.; Wilczynska, M.; Kleczkowski, L.A. Oligomerization status, with the monomer as active species, defines catalytic efficiency of UDP-glucose pyrophosphorylase. Biochem. J. 2002, 367, 295–300. [Google Scholar] [CrossRef]
- Meng, M.; Fitzek, E.; Gajowniczek, A.; Wilczynska, M.; Kleczkowski, L.A. Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley UDP-glucose pyrophosphorylase. Biochim. Biophys. Acta 2009, 1794, 1734–1742. [Google Scholar] [CrossRef]
- Geisler, M.; Wilczynska, M.; Karpinski, S.; Kleczkowski, L.A. Toward a blueprint for UDP-glucose pyrophosphorylase structure/function properties: Homology-modeling analyses. Plant Mol. Biol. 2004, 56, 783–794. [Google Scholar] [CrossRef]
- Lyublinskaya, O.; Antunes, F. Measuring intracellular concentration of hydrogen peroxide with the use of genetically encoded H2O2 biosensor HyPer. Redox Biol. 2019, 24, 101200. [Google Scholar] [CrossRef]
- Sies, H. Role of metabolic H2O2 generation: Redox signaling and oxidative stress. J. Biol. Chem. 2014, 289, 8735–8741. [Google Scholar] [CrossRef]
- Gechev, T.S.; Hille, J. Hydrogen peroxide as a signal controlling plant programmed cell death. J. Cell Biol. 2005, 168, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Fujita, M. Glutathione in plants: Biosynthesis and physiological role in environmental stress tolerance. Physiol. Mol. Biol. Plants 2017, 23, 249–268. [Google Scholar] [CrossRef] [PubMed]
- Marrs, K.A. The functions and regulation of glutathione S-transferases in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 127–158. [Google Scholar] [CrossRef]
- Lu, S.C. Regulation of glutathione synthesis. Mol. Asp. Med. 2009, 30, 42–59. [Google Scholar] [CrossRef] [PubMed]
- Bohle, F.; Klaus, A.; Tegethof, H.; Schwarzländer, M.; Hochholdinger, F.; Meyer, A.J.; Acosta, I.F.; Müller-Schüssele, S.J. High robustness of cytosolic glutathione redox potential under combined salt and osmotic stress in barley as revealed by the biosensor Grx1-roGFP2. bioRxiv 2022. [Google Scholar] [CrossRef]
- Yan, S.; Tang, Z.; Su, W.; Sun, W. Proteomic analysis of salt stress-responsive proteins in rice roots. Proteomics 2005, 5, 235–244. [Google Scholar] [CrossRef]
- Carpentier, S.C.; Witters, E.; Laukens, K.; Van Onckelen, H.; Swennen, R.; Panis, B. Banana (Musa spp.) as a model to study the meristem proteome: Acclimation to osmotic stress. Proteomics 2007, 7, 92–105. [Google Scholar] [CrossRef]
- Le Gall, H.; Philippe, F.; Domon, J.-M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Decker, D. Effects of magnesium, pyrophosphate and phosphonates on pyrophosphorolytic reaction of UDP-glucose pyrophosphorylase. Plants 2022, 11, 1611. [Google Scholar] [CrossRef] [PubMed]
- Poole, L.B. Formation and functions of protein sulfenic acids. Curr. Protoc. Toxicol. 2003, 17, 1–15. [Google Scholar] [CrossRef]
- Iyer, B.R.; Mahalakshmi, R. Hydrophobic characteristic is energetically preferred for cysteine in a model membrane protein. Biophys. J. 2019, 117, 25–35. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Martz, F.; Wilczynska, M. Factors affecting oligomerization status of UDP-glucose pyrophosphorylase. Phytochemistry 2005, 66, 2815–2821. [Google Scholar] [CrossRef]
- Meng, M.; Wilczynska, M.; Kleczkowski, L.A. Molecular and kinetic characterization of two UDP-glucose pyrophosphorylases, products of distinct genes, from Arabidopsis. Biochim. Biophys. Acta 2008, 1784, 967–972. [Google Scholar] [CrossRef]
- Decker, D.; Meng, M.; Gornicka, A.; Hofer, A.; Wilczynska, M.; Kleczkowski, L.A. Substrate kinetics and substrate effects on the quaternary structure of barley UDP-glucose pyrophosphorylase. Phytochemistry 2012, 79, 39–45. [Google Scholar] [CrossRef]
- Führing, J.; Damerow, S.; Fedorov, R.; Schneider, J.; Münster-Kühnel, A.K.; Gerardy-Schahn, R. Octamerization is essential for enzymatic function of human UDP-glucose pyrophosphorylase. Glycobiology 2013, 23, 426–437. [Google Scholar] [CrossRef]
- Ciereszko, I.; Johansson, H.; Hurry, V.; Kleczkowski, L.A. Phosphate status affects the gene expression, protein content and enzymatic activity of UDP-glucose pyrophosphorylase in wild-type and pho mutants of Arabidopsis. Planta 2001, 212, 598–605. [Google Scholar] [CrossRef]
- Ciereszko, I.; Johansson, H.; Kleczkowski, L.A. Sucrose and light regulation of a cold-inducible UDP-glucose pyrophosphorylase gene via a hexokinase-independent and abscisic acid-insensitive pathway in Arabidopsis. Biochem. J. 2001, 354, 67–72. [Google Scholar] [CrossRef]
- Repetto, O.; Bestel-Corre, G.; Dumas-Gaudot, E.; Berta, G.; Gianinazzi-Pearson, V.; Gianinazzi, S. Targeted proteomics to identify cadmium-induced protein modifications in Glomus mosseae-inoculated pea roots. New Phytol. 2003, 157, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Ciereszko, I.; Johansson, H.; Kleczkowski, L.A. Interactive effects of phosphate deficiency, sucrose and light/dark conditions on gene expression of UDP-glucose pyrophosphorylase in Arabidopsis. J. Plant Physiol. 2005, 162, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Meng, M.; Geisler, M.; Johansson, H.; Mellerowicz, E.J.; Karpinski, S.; Kleczkowski, L.A. Differential tissue/organ-dependent expression of two sucrose- and cold-responsive genes for UDP-glucose pyrophosphorylase in Populus. Gene 2007, 389, 186–195. [Google Scholar] [CrossRef]
- Vandenabeele, S.; Van Der Kelen, K.; Dat, J.; Gadjev, I.; Boonefaes, T.; Morsa, S.; Rottiers, P.; Slooten, L.; Van Montagu, M.; Zabeau, M.; et al. A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc. Natl. Acad. Sci. USA 2003, 100, 16113–16118. [Google Scholar] [CrossRef]
- Dat, J.F.; Pellinen, R.; Beeckman, T.; Kangasjärvi, J.; Langebartels, C.; Inzé, D.; Van Breusegem, F. Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. Plant J. 2003, 33, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Alexander, R.D.; Morris, P.C. A proteomic analysis of 14-3-3 binding proteins from developing barley grains. Proteomics 2006, 6, 1886–1896. [Google Scholar] [CrossRef] [PubMed]
- Swatek, K.N.; Graham, K.; Agrawal, G.K.; Thelen, J.J. The 14-3-3 isoforms chi and epsilon differentially bind client proteins from developing Arabidopsis seed. J. Proteome Res. 2011, 10, 4076–4087. [Google Scholar] [CrossRef]
- Chen, R.; Zhao, X.; Shao, Z.; Zhu, L.; He, G. Multiple isoforms of UDP-glucose pyrophosphorylase in rice. Physiol. Plant. 2007, 129, 725–736. [Google Scholar] [CrossRef]
- Komatsu, S.; Yamada, E.; Furukawa, K. Cold stress changes the concanavalin A positive glycosylation pattern of proteins expressed in the basal parts of rice leaf sheaths. Amino Acids 2009, 36, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Silva-Sanchez, C.; Chen, S.; Li, J.; Chourey, P.S. A comparative glycoproteome study of developing endosperm in the hexose-deficient miniature1 (mn1) seed mutant and its wild type Mn1 in maize. Front. Plant Sci. 2014, 5, 63. [Google Scholar] [CrossRef] [PubMed]
- Ferjani, A.; Kawade, K.; Asaoka, M.; Oikawa, A.; Okada, T.; Mochizuki, A.; Maeshima, M.; Hirai, M.Y.; Saito, K.; Tsukaya, H. Pyrophosphate inhibits gluconeogenesis by restricting UDP-glucose formation in vivo. Sci. Rep. 2018, 8, 14696. [Google Scholar] [CrossRef]
- Decker, D.; Öberg, C.; Kleczkowski, L.A. Identification and characterization of inhibitors of UDP-glucose and UDP-sugar pyrophosphorylases for in vivo studies. Plant J. 2017, 90, 1093–1107. [Google Scholar] [CrossRef]
- Decker, D.; Kleczkowski, L.A. Substrate specificity and inhibitor sensitivity of plant UDP-sugar producing pyrophosphorylases. Front. Plant Sci. 2017, 8, 1610. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Geisler, M.; Fitzek, E.; Wilczynska, M. A common structural blueprint for plant UDP-sugar producing pyrophosphorylases. Biochem. J. 2011, 439, 375–379. [Google Scholar] [CrossRef]
- Rose, I.A. The state of magnesium in cells as estimated from the adenylate kinase equilibrium. Proc. Natl. Acad. Sci. USA 1968, 61, 1079–1086. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Kleczkowski, L.A. Pyrophosphate as an alternative energy currency in plants. Biochem. J. 2021, 478, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Igamberdiev, A.U.; Kleczkowski, L.A. Magnesium and cell energetics in plants under anoxia. Biochem. J. 2011, 437, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.A.; Igamberdiev, A.U. Magnesium signaling in plants. Int. J. Mol. Sci. 2021, 22, 1159. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Igamberdiev, A.U. Magnesium and cell energetics: At the junction of metabolism of adenylate and non-adenylate nucleotides. J. Plant Physiol. 2023, 280, 153901. [Google Scholar] [CrossRef]
- Cakmak, I.; Kirkby, E.A. Role of magnesium in carbon partitioning and alleviating photooxidative damage. Physiol. Plant. 2008, 133, 692–704. [Google Scholar] [CrossRef]
- Kim, G.; Weiss, S.J.; Levine, R.L. Methionine oxidation and reduction in proteins. Biochim. Biophys. Acta 2014, 1840, 901–905. [Google Scholar] [CrossRef]
- Weber, J.; Senior, A.E. Location and properties of pyrophosphate-binding sites in Escherichia coli F1-ATPase. J. Biol. Chem. 1995, 270, 12653–12658. [Google Scholar] [CrossRef]
- Qamer, Z.; Chaudhary, M.T.; Du, X.; Hinze, L.; Azhar, M.T. Review of oxidative stress and antioxidative defense mechanisms in Gossypium hirsutum L. in response to extreme abiotic conditions. J. Cotton Res. 2021, 4, 9. [Google Scholar] [CrossRef]
- Guex, N.; Diemand, A.; Peitsch, M.C. Protein modelling for all. Trends Biochem. Sci. 1999, 24, 364–367. [Google Scholar] [CrossRef]
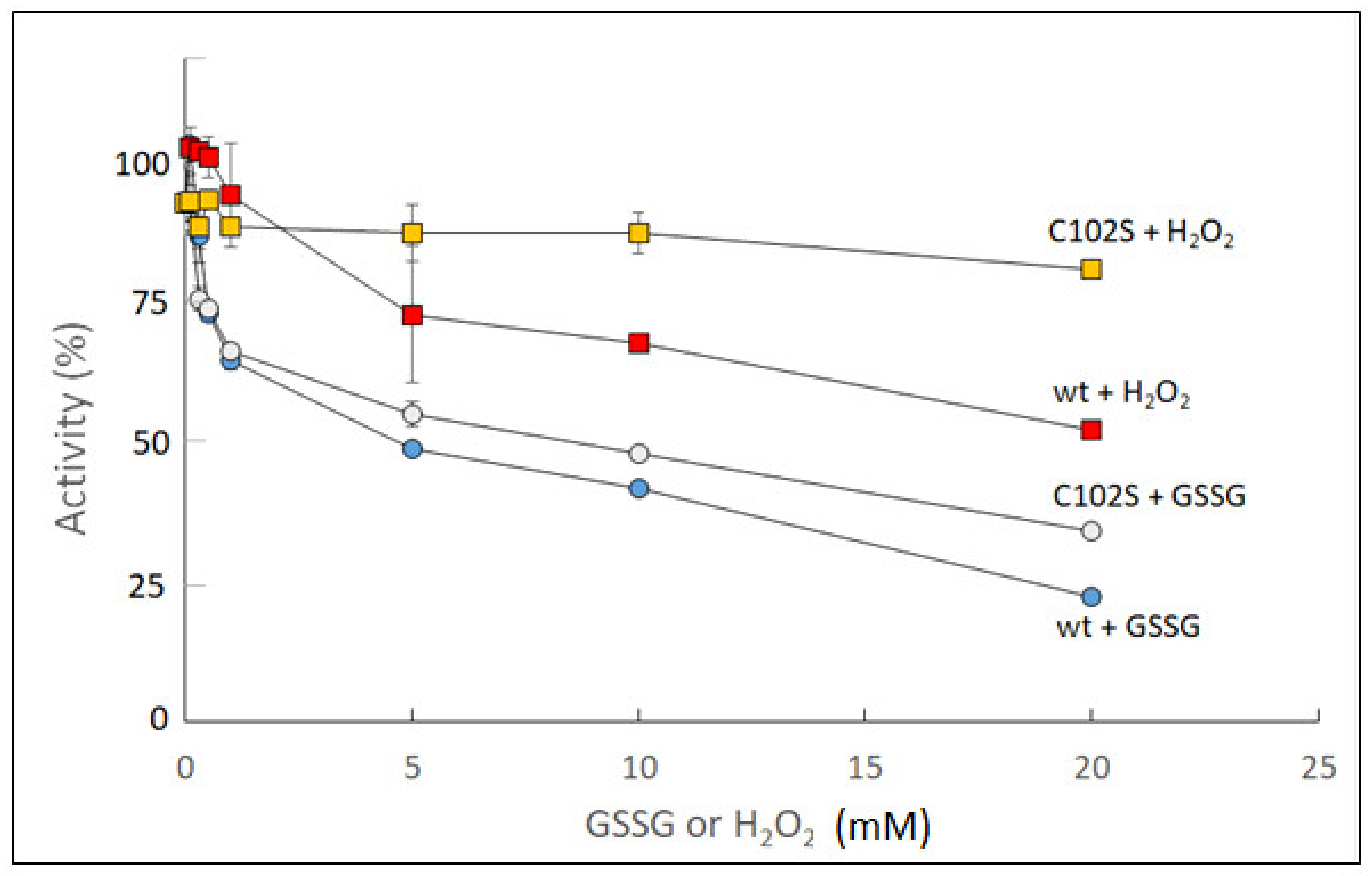
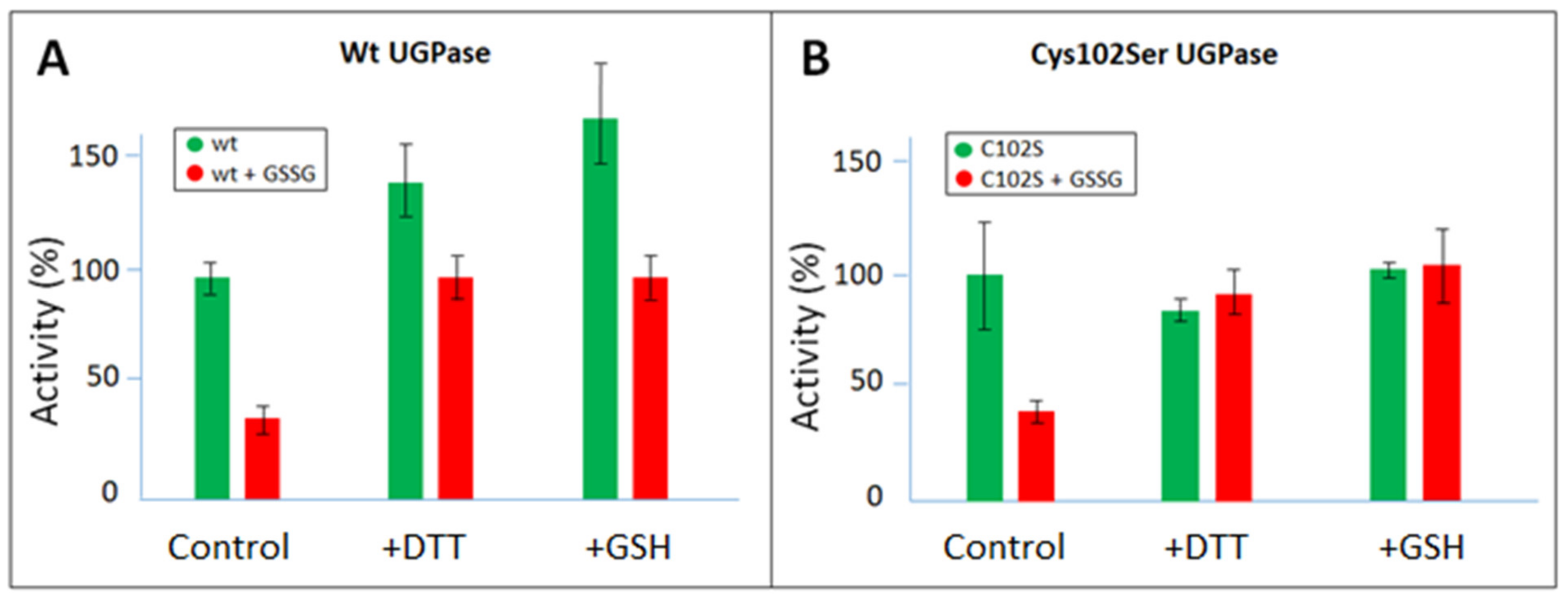
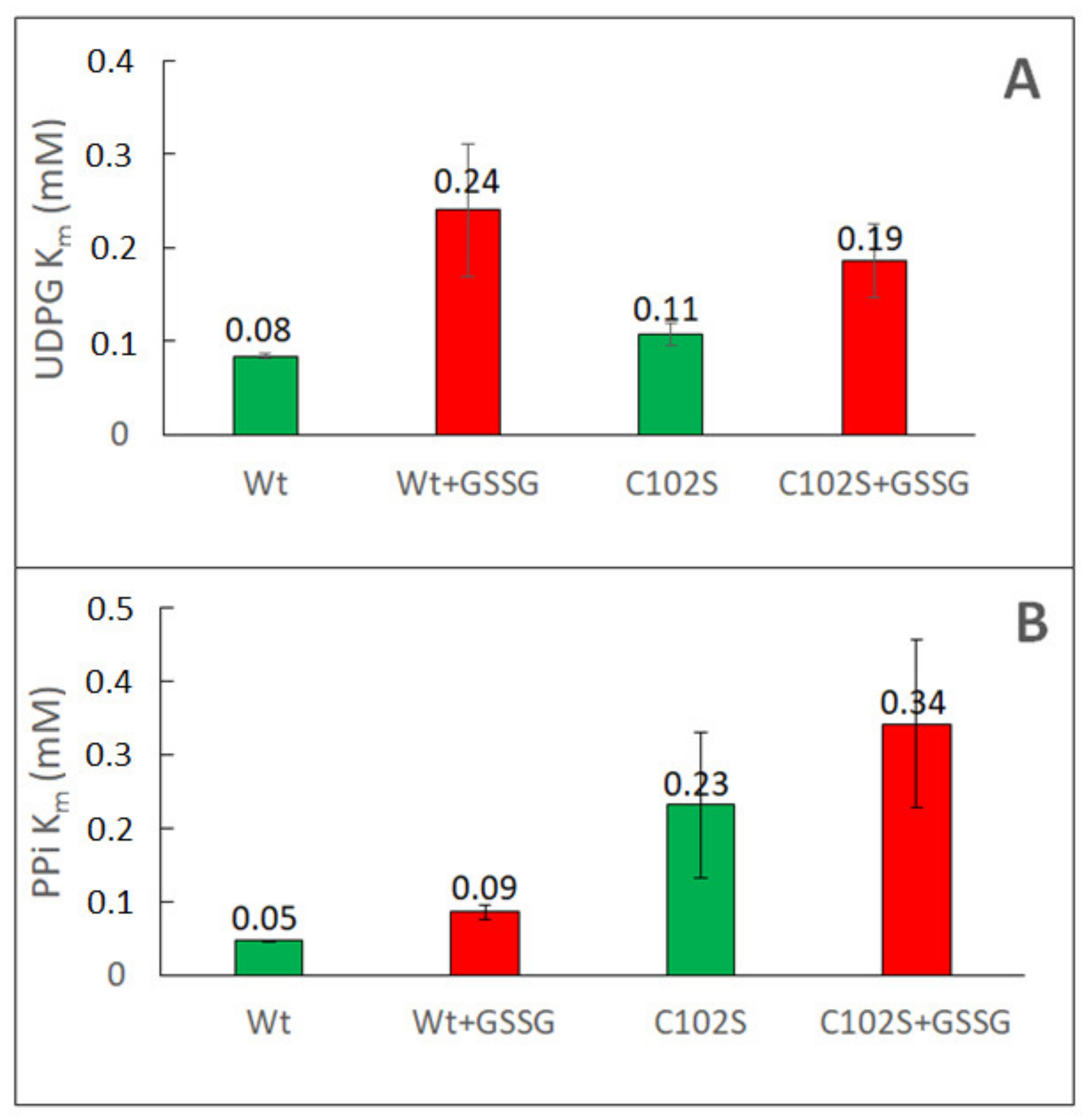
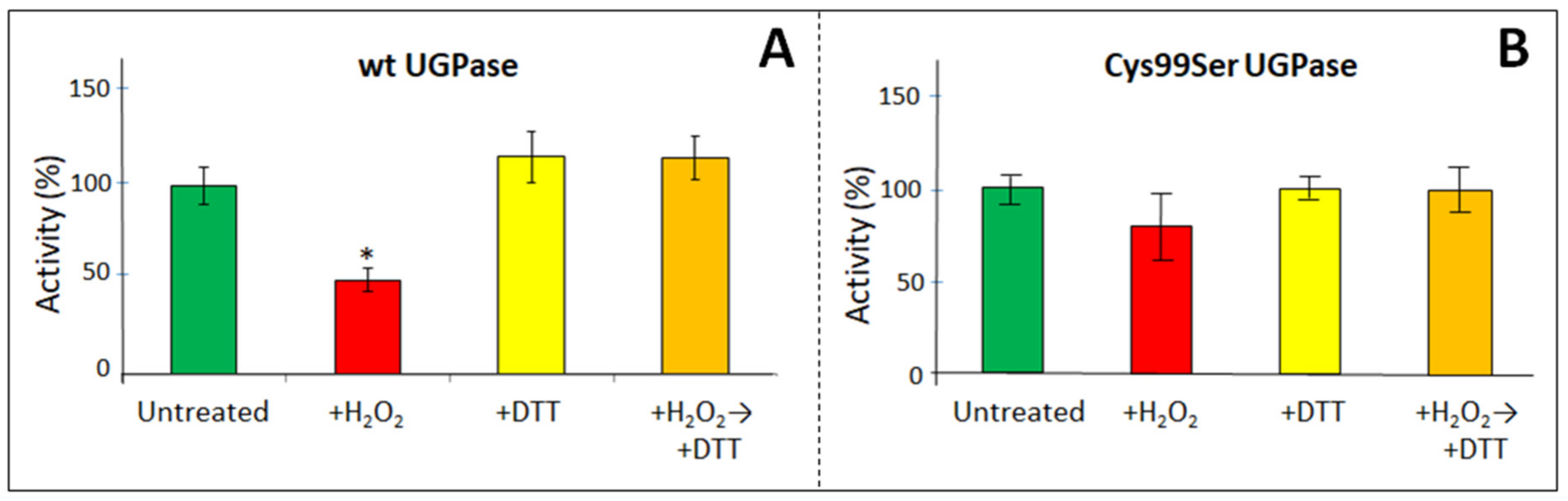
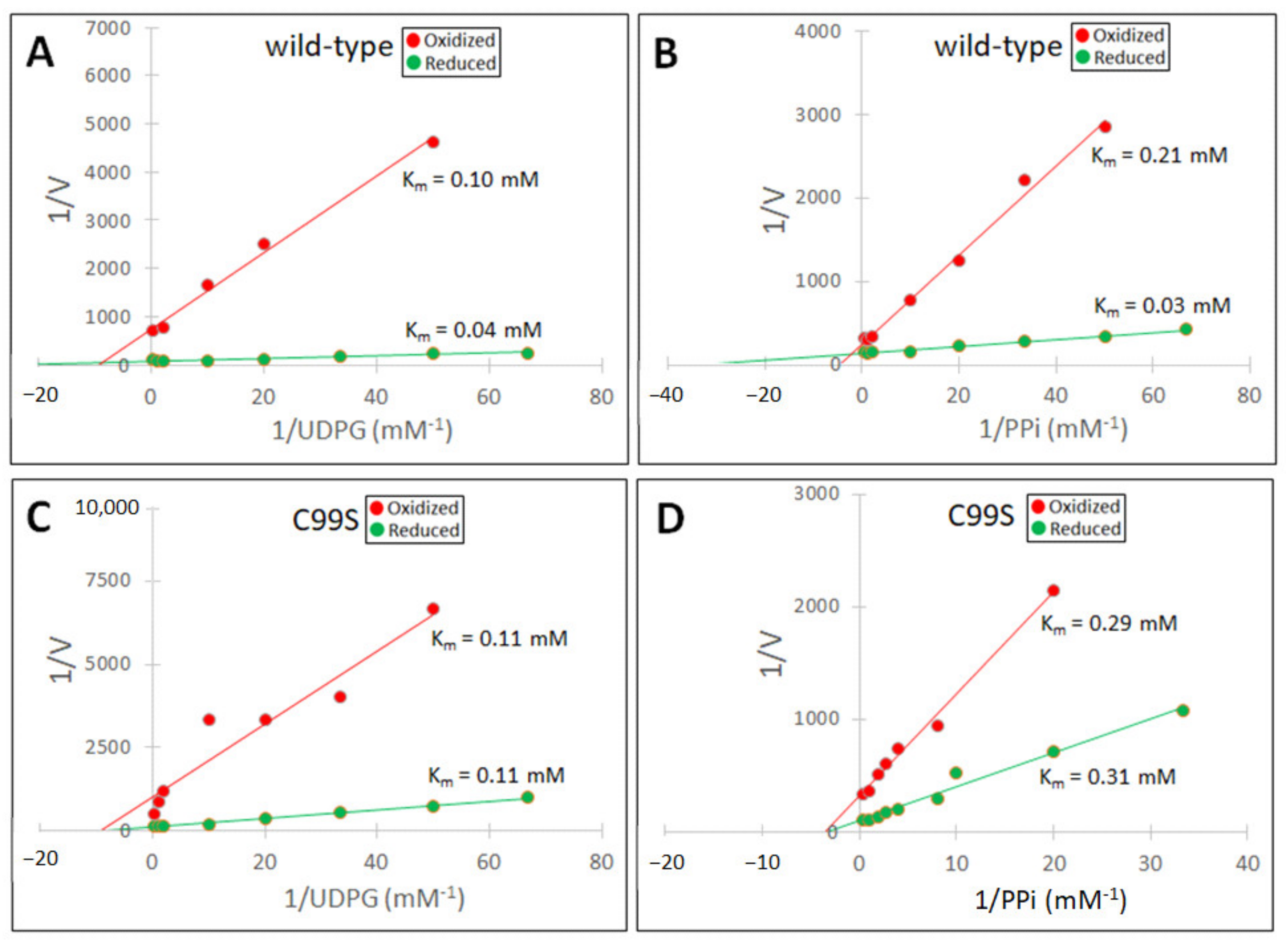
| Sugarcane UGPase | Wild type | Cys102Ser | |||
| Untreated | Oxidized | Untreated | Oxidized | ||
| Km UDPG (mM) | 0.08 | 0.24 | 0.11 | 0.19 | |
| Km PPi (mM) | 0.04 | 0.09 | 0.23 | 0.34 | |
| Barley UGPase | Wild type | Cys99Ser | |||
| Reduced | Oxidized | Reduced | Oxidized | ||
| Km UDPG (mM) | 0.04 | 0.10 | 0.11 | 0.11 | |
| Km PPi (mM) | 0.03 | 0.21 | 0.31 | 0.29 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Decker, D.; Aubert, J.; Wilczynska, M.; Kleczkowski, L.A. Exploring Redox Modulation of Plant UDP-Glucose Pyrophosphorylase. Int. J. Mol. Sci. 2023, 24, 8914. https://doi.org/10.3390/ijms24108914
Decker D, Aubert J, Wilczynska M, Kleczkowski LA. Exploring Redox Modulation of Plant UDP-Glucose Pyrophosphorylase. International Journal of Molecular Sciences. 2023; 24(10):8914. https://doi.org/10.3390/ijms24108914
Chicago/Turabian StyleDecker, Daniel, Juliette Aubert, Malgorzata Wilczynska, and Leszek A. Kleczkowski. 2023. "Exploring Redox Modulation of Plant UDP-Glucose Pyrophosphorylase" International Journal of Molecular Sciences 24, no. 10: 8914. https://doi.org/10.3390/ijms24108914







