Signal Pathways and microRNAs in Osteosarcoma Growth and the Dual Role of Mesenchymal Stem Cells in Oncogenesis
Abstract
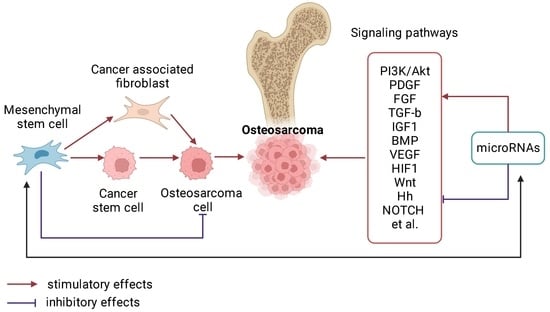
:1. Introduction
2. Molecular Basis of OS Pathogenesis
2.1. The Role of miRNAs in the Pathogenesis of OS
2.2. Signaling Pathways and Regulatory miRNAs in OS
2.2.1. Wnt/β-Catenin
- NKD2
- TCF/LEF
- FOXM1
- E2F1
- E2F5
- E2F3
- RB1-E2F
- NRF2 (NFE2L2)
- YAP
- Rho-ROCK
- AXIN1
- SOX4
- STAT3
- Dkk
- IGF2BP1
- MYC
- CCNE2
2.2.2. Hippo/YAP/TAZ
2.2.3. TGF-β
- SMAD3
2.2.4. Notch
2.2.5. HIF1α
2.2.6. ErbB
- EGFR (epidermal growth factor receptor)
- HER2 (human epidermal growth factor receptor 2)
2.2.7. PI3K/Akt/mTOR
- elF4E (eukaryotic transcription initiation factor 4E)
- HMGB1 (High-Mobility Group Box 1)
2.2.8. RUNX2/CDK4
2.2.9. p53
- FOXP1
- Bak
- Bcl-xL
2.2.10. VEGF-VEGFR-RAS-RAF-MEK-MAPK
2.2.11. PTEN
2.2.12. Hedgehog (Hh)
2.2.13. Additional Signallers in OS
- FOXK1
- NOB1
- TGIF2
- RECK
- KLF10 (Krueppel-like factor 10)
- HDAC1
- HDAC4
- SND1
- Foxp4
- GPX4
3. Heterogeneity of the Cellular Component of the OS Substrate
- Mesenchymal;
- T lymphocytes (Treg, CD4+, CD8+ (highly malignant), NKT, DK, plasmacytoid DK-pDK) [341];
- B lymphocytes (naive B cells, activated B cells, plasma cells);
- Myeloid cells (mast cells, monocytes, M1, M2 macrophages, tumor-associated macrophages (TAM));
- Osteoclasts;
- Endovascular (endothelial, parietal) cells.
3.1. Characteristics of the Tumor Microenvironment and the Behavior of MSCs in OS
3.2. MSC Migration and OS Growth
3.3. Transdifferentiation/Plasticity of MSC
3.3.1. Cancer-Associated Stromal Fibroblasts (CAFs)
3.3.2. Cancer Stem Cells (CSCs) and MSCs
3.4. Transformation of MSCs into OS Cells
3.5. Involvement of MSCs in Metastasis of OS Cells
3.6. MSC and Tumor Cell Death
4. The Potential of MSCs in OS Therapy
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown, H.K.; Schiavone, K.; Gouin, F.; Heymann, M.-F.; Heymann, D. Biology of Bone Sarcomas and New Therapeutic Developments. Calcif. Tissue Int. 2018, 102, 174–195. [Google Scholar] [CrossRef] [PubMed]
- Czarnecka, A.M.; Synoradzki, K.; Firlej, W.; Bartnik, E.; Sobczuk, P.; Fiedorowicz, M.; Grieb, P.; Rutkowski, P. Molecular Biology of Osteosarcoma. Cancers 2020, 12, 2130. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wu, Q.; Gong, X.; Liu, J.; Ma, Y. Osteosarcoma: A Review of Current and Future Therapeutic Approaches. BioMed. Eng. OnLine 2021, 20, 24. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, B.A.; Markel, J.E.; Kleinerman, E.S. Osteosarcoma Overview. Rheumatol. Ther. 2017, 4, 25–43. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, M.; Noirot, C.; Accadbled, F.; de Gauzy, S.J.; Castex, M.P.; Brousset, P.; Gomez-Brouchet, A. Whole-Exome Sequencing in Osteosarcoma Reveals Important Heterogeneity of Genetic Alterations. Ann. Oncol. 2016, 27, 738–744. [Google Scholar] [CrossRef]
- Rickel, K.; Fang, F.; Tao, J. Molecular Genetics of Osteosarcoma. Bone 2017, 102, 69–79. [Google Scholar] [CrossRef]
- Haridy, Y.; Witzmann, F.; Asbach, P.; Schoch, R.R.; Fröbisch, N.; Rothschild, B.M. Triassic Cancer—Osteosarcoma in a 240-Million-Year-Old Stem-Turtle. JAMA Oncol. 2019, 5, 425. [Google Scholar] [CrossRef]
- Menéndez, S.T.; Gallego, B.; Murillo, D.; Rodríguez, A.; Rodríguez, R. Cancer Stem Cells as a Source of Drug Resistance in Bone Sarcomas. J. Clin. Med. 2021, 10, 2621. [Google Scholar] [CrossRef]
- Chellini, L.; Palombo, R.; Riccioni, V.; Paronetto, M.P. Oncogenic Dysregulation of Circulating Noncoding RNAs: Novel Challenges and Opportunities in Sarcoma Diagnosis and Treatment. Cancers 2022, 14, 4677. [Google Scholar] [CrossRef]
- Todosenko, N.; Yurova, K.; Khaziakhmatova, O.; Malashchenko, V.; Khlusov, I.; Litvinova, L. Heparin and Heparin-Based Drug Delivery Systems: Pleiotropic Molecular Effects at Multiple Drug Resistance of Osteosarcoma and Immune Cells. Pharmaceutics 2022, 14, 2181. [Google Scholar] [CrossRef]
- Grünewald, T.G.; Alonso, M.; Avnet, S.; Banito, A.; Burdach, S.; Cidre-Aranaz, F.; Di Pompo, G.; Distel, M.; Dorado-Garcia, H.; Garcia-Castro, J.; et al. Sarcoma Treatment in the Era of Molecular Medicine. EMBO Mol. Med. 2020, 12, e11131. [Google Scholar] [CrossRef]
- Smrke, A.; Anderson, P.M.; Gulia, A.; Gennatas, S.; Huang, P.H.; Jones, R.L. Future Directions in the Treatment of Osteosarcoma. Cells 2021, 10, 172. [Google Scholar] [CrossRef]
- Chang, X.; Ma, Z.; Zhu, G.; Lu, Y.; Yang, J. New Perspective into Mesenchymal Stem Cells: Molecular Mechanisms Regulating Osteosarcoma. J. Bone Oncol. 2021, 29, 100372. [Google Scholar] [CrossRef]
- Harrell, C.R.; Markovic, B.S.; Fellabaum, C.; Arsenijevic, A.; Volarevic, V. Mesenchymal stem cell-based therapy of osteoarthritis: Current knowledge and future perspectives. Biomed. Pharm. 2019, 109, 2318–2326. [Google Scholar] [CrossRef]
- Jiang, W.; Xu, J. Immune modulation by mesenchymal stem cells. Cell Prolif. 2020, 53, e12712. [Google Scholar] [CrossRef]
- Avril, P.; Le Nail, L.-R.; Brennan, M.Á.; Rosset, P.; De Pinieux, G.; Layrolle, P.; Heymann, D.; Perrot, P.; Trichet, V. Mesenchymal Stem Cells Increase Proliferation but Do Not Change Quiescent State of Osteosarcoma Cells: Potential Implications According to the Tumor Resection Status. J. Bone Oncol. 2016, 5, 5–14. [Google Scholar] [CrossRef]
- Perrot, P.; Rousseau, J.; Bouffaut, A.-L.; Rédini, F.; Cassagnau, E.; Deschaseaux, F.; Heymann, M.-F.; Heymann, D.; Duteille, F.; Trichet, V.; et al. Safety Concern between Autologous Fat Graft, Mesenchymal Stem Cell and Osteosarcoma Recurrence. PLoS ONE 2010, 5, e10999. [Google Scholar] [CrossRef]
- Xu, W.; Bian, Z.; Fan, Q.; Li, G.; Tang, T. Human Mesenchymal Stem Cells (HMSCs) Target Osteosarcoma and Promote Its Growth and Pulmonary Metastasis. Cancer Lett. 2009, 281, 32–41. [Google Scholar] [CrossRef]
- Fujiwara, T.; Ozaki, T. Overcoming Therapeutic Resistance of Bone Sarcomas: Overview of the Molecular Mechanisms and Therapeutic Targets for Bone Sarcoma Stem Cells. Stem Cells Int. 2016, 2016, e2603092. [Google Scholar] [CrossRef]
- Hernigou, P.; Flouzat Lachaniette, C.H.; Delambre, J.; Chevallier, N.; Rouard, H. Regenerative Therapy with Mesenchymal Stem Cells at the Site of Malignant Primary Bone Tumour Resection: What Are the Risks of Early or Late Local Recurrence? Int. Orthop. (SICOT) 2014, 38, 1825–1835. [Google Scholar] [CrossRef]
- Litvinova, L.S.; Shupletsova, V.V.; Khaziakhmatova, O.G.; Daminova, A.G.; Kudryavtseva, V.L.; Yurova, K.A.; Malashchenko, V.V.; Todosenko, N.M.; Popova, V.; Litvinov, R.I.; et al. Human Mesenchymal Stem Cells as a Carrier for a Cell-Mediated Drug Delivery. Front. Bioeng. Biotechnol. 2022, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Vulf, M.; Khlusov, I.; Yurova, K.; Todosenko, N.; Komar, A.; Kozlov, I.; Malashchenko, V.; Shunkina, D.; Khaziakhmatova, O.; Litvinova, L. MicroRNA Regulation of Bone Marrow Mesenchymal Stem Cells in the Development of Osteoporosis in Obesity. Front. Biosci.-Sch. 2022, 14, 17. [Google Scholar] [CrossRef] [PubMed]
- Velletri, T.; Xie, N.; Wang, Y.; Huang, Y.; Yang, Q.; Chen, X.; Chen, Q.; Shou, P.; Gan, Y.; Cao, G.; et al. P53 functional abnormality in mesenchymal stem cells promotes osteosarcoma development. Cell Death Dis. 2016, 7, e2015. [Google Scholar] [CrossRef] [PubMed]
- Namløs, H.M.; Meza-Zepeda, L.A.; Barøy, T.; Østensen, I.H.G.; Kresse, S.H.; Kuijjer, M.L.; Serra, M.; Bürger, H.; Cleton-Jansen, A.-M.; Myklebost, O. Modulation of the Osteosarcoma Expression Phenotype by MicroRNAs. PLoS ONE 2012, 7, e48086. [Google Scholar] [CrossRef]
- Sheng, G.; Gao, Y.; Yang, Y.; Wu, H. Osteosarcoma and Metastasis. Front. Oncol. 2021, 11, 780264. [Google Scholar] [CrossRef]
- Lauvrak, S.U.; Munthe, E.; Kresse, S.H.; Stratford, E.W.; Namløs, H.M.; Meza-Zepeda, L.A.; Myklebost, O. Functional Characterisation of Osteosarcoma Cell Lines and Identification of MRNAs and MiRNAs Associated with Aggressive Cancer Phenotypes. Br. J. Cancer 2013, 109, 2228–2236. [Google Scholar] [CrossRef]
- Sikora, M.; Krajewska, K.; Marcinkowska, K.; Raciborska, A.; Wiglusz, R.J.; Śmieszek, A. Comparison of Selected Non-Coding RNAs and Gene Expression Profiles between Common Osteosarcoma Cell Lines. Cancers 2022, 14, 4533. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, G.; Chen, R.; Hua, Y.; Cai, Z. Mesenchymal Stem Cells in the Osteosarcoma Microenvironment: Their Biological Properties, Influence on Tumor Growth, and Therapeutic Implications. Stem Cell Res. Ther. 2018, 9, 22. [Google Scholar] [CrossRef]
- de Azevedo, J.; Fernandes, T.; Fernandes, J.; de Azevedo, J.; Lanza, D.; Bezerra, C.; Andrade, V.; de Araújo, J.; Fernandes, J. Biology and Pathogenesis of Human Osteosarcoma (Review). Oncol. Lett. 2019, 19, 1099–1116. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, N.; Zheng, Y.; Tong, Z.; Yang, T.; Kang, X.; He, Y.; Dong, L. Identification of Key Genes and Pathways in Osteosarcoma by Bioinformatics Analysis. Comput. Math. Methods Med. 2022, 2022, 7549894. [Google Scholar] [CrossRef]
- Alfranca, A.; Martinez-Cruzado, L.; Tornin, J.; Abarrategi, A.; Amaral, T.; de Alava, E.; Menendez, P.; Garcia-Castro, J.; Rodriguez, R. Bone Microenvironment Signals in Osteosarcoma Development. Cell. Mol. Life Sci. 2015, 72, 3097–3113. [Google Scholar] [CrossRef]
- Abarrategi, A.; Tornin, J.; Martinez-Cruzado, L.; Hamilton, A.; Martinez-Campos, E.; Rodrigo, J.P.; González, M.V.; Baldini, N.; Garcia-Castro, J.; Rodriguez, R. Osteosarcoma: Cells-of-Origin, Cancer Stem Cells, and Targeted Therapies. Stem Cells Int. 2016, 2016, 3631764. [Google Scholar] [CrossRef]
- Guan, G.; Zhang, Y.; Lu, Y.; Liu, L.; Shi, D.; Wen, Y.; Yang, L.; Ma, Q.; Liu, T.; Zhu, X.; et al. The HIF-1α/CXCR4 Pathway Supports Hypoxia-Induced Metastasis of Human Osteosarcoma Cells. Cancer Lett. 2015, 357, 254–264. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, T.; Wang, W. Prognostic Significance of Matrix Metalloproteinase 9 Expression in Osteosarcoma: A Meta-Analysis of 16 Studies. Medicine 2018, 97, e13051. [Google Scholar] [CrossRef]
- Ehnman, M.; Chaabane, W.; Haglund, F.; Tsagkozis, P. The Tumor Microenvironment of Pediatric Sarcoma: Mesenchymal Mechanisms Regulating Cell Migration and Metastasis. Curr. Oncol. Rep. 2019, 21, 90. [Google Scholar] [CrossRef]
- Yu, F.-X.; Hu, W.-J.; He, B.; Zheng, Y.-H.; Zhang, Q.-Y.; Chen, L. Bone Marrow Mesenchymal Stem Cells Promote Osteosarcoma Cell Proliferation and Invasion. World J. Surg. Oncol. 2015, 13, 52. [Google Scholar] [CrossRef]
- Xu, S.; Yang, S.; Sun, G.; Huang, W.; Zhang, Y. Transforming Growth Factor-Beta Polymorphisms and Serum Level in the Development of Osteosarcoma. DNA Cell Biol. 2014, 33, 802–806. [Google Scholar] [CrossRef]
- Yang, G.; Yuan, J.; Li, K. EMT Transcription Factors: Implication in Osteosarcoma. Med. Oncol. 2013, 30, 697. [Google Scholar] [CrossRef]
- Katz, L.H.; Li, Y.; Chen, J.-S.; Muñoz, N.M.; Majumdar, A.; Chen, J.; Mishra, L. Targeting TGF-β Signaling in Cancer. Expert Opin. Ther. Targets 2013, 17, 743–760. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, Y.-J.; Zhu, K.; Wang, W.-C. A Systematic Review of Vascular Endothelial Growth Factor Expression as a Biomarker of Prognosis in Patients with Osteosarcoma. Tumor. Biol. 2013, 34, 1895–1899. [Google Scholar] [CrossRef]
- Wang, L.-H.; Tsai, H.-C.; Cheng, Y.-C.; Lin, C.-Y.; Huang, Y.-L.; Tsai, C.-H.; Xu, G.-H.; Wang, S.-W.; Fong, Y.-C.; Tang, C.-H. CTGF Promotes Osteosarcoma Angiogenesis by Regulating MiR-543/Angiopoietin 2 Signaling. Cancer Lett. 2017, 391, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.-H.; Yang, R.; Tsao, Y.-T. Connective Tissue Growth Factor Stimulates Osteosarcoma Cell Migration and Induces Osteosarcoma Metastasis by Upregulating VCAM-1 Expression. Biochem. Pharmacol. 2018, 155, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.-C.; Huang, C.-Y.; Su, H.-L.; Tang, C.-H. CTGF Increases Drug Resistance to Paclitaxel by Upregulating Survivin Expression in Human Osteosarcoma Cells. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2014, 1843, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-C.; Chuang, S.-M.; Hsu, C.-J.; Tsai, C.-H.; Wang, S.-W.; Tang, C.-H. CTGF Increases Vascular Endothelial Growth Factor-Dependent Angiogenesis in Human Synovial Fibroblasts by Increasing MiR-210 Expression. Cell Death Dis. 2014, 5, e1485. [Google Scholar] [CrossRef]
- Heymann, M.-F.; Lézot, F.; Heymann, D. The Contribution of Immune Infiltrates and the Local Microenvironment in the Pathogenesis of Osteosarcoma. Cell. Immunol. 2019, 343, 103711. [Google Scholar] [CrossRef]
- Chang, K.K.; Yoon, C.; Yi, B.C.; Tap, W.D.; Simon, M.C.; Yoon, S.S. Platelet-Derived Growth Factor Receptor-α and -β Promote Cancer Stem Cell Phenotypes in Sarcomas. Oncogenesis 2018, 7, 47. [Google Scholar] [CrossRef]
- Abdeen, A.; Chou, A.J.; Healey, J.H.; Khanna, C.; Osborne, T.S.; Hewitt, S.M.; Kim, M.; Wang, D.; Moody, K.; Gorlick, R. Correlation between Clinical Outcome and Growth Factor Pathway Expression in Osteogenic Sarcoma. Cancer 2009, 115, 5243–5250. [Google Scholar] [CrossRef]
- Zhou, W.; Hao, M.; Du, X.; Chen, K.; Wang, G.; Yang, J. Advances in Targeted Therapy for Osteosarcoma. Discov. Med. 2014, 17, 301–307. [Google Scholar]
- Zhang, J.; Yu, X.-H.; Yan, Y.-G.; Wang, C.; Wang, W.-J. PI3K/Akt Signaling in Osteosarcoma. Clin. Chim. Acta 2015, 444, 182–192. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, M.; Tian, A.; Zhang, X.; Yao, Z.; Ma, X. Aberrant Activation of Wnt/β-Catenin Signaling Drives Proliferation of Bone Sarcoma Cells. Oncotarget 2015, 6, 17570–17583. [Google Scholar] [CrossRef]
- Cai, Z.; Cao, Y.; Luo, Y.; Hu, H.; Ling, H. Signalling Mechanism(s) of Epithelial–Mesenchymal Transition and Cancer Stem Cells in Tumour Therapeutic Resistance. Clin. Chim. Acta 2018, 483, 156–163. [Google Scholar] [CrossRef]
- Iwaya, K.; Ogawa, H.; Kuroda, M.; Izumi, M.; Ishida, T.; Mukai, K. Cytoplasmic and/or Nuclear Staining of Beta-Catenin Is Associated with Lung Metastasis. Clin. Exp. Metastasis 2003, 20, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Kurenbekova, L.; Gao, Y.; Roos, A.; Creighton, C.J.; Rao, P.; Hicks, J.; Man, T.-K.; Lau, C.; Brown, A.M.C.; et al. NKD2, a Negative Regulator of Wnt Signaling, Suppresses Tumor Growth and Metastasis in Osteosarcoma. Oncogene 2015, 34, 5069–5079. [Google Scholar] [CrossRef] [PubMed]
- Luke, J.J.; Bao, R.; Sweis, R.F.; Spranger, S.; Gajewski, T.F. WNT/β-Catenin Pathway Activation Correlates with Immune Exclusion across Human Cancers. Clin. Cancer Res. 2019, 25, 3074–3083. [Google Scholar] [CrossRef]
- Lo, W.W.; Pinnaduwage, D.; Gokgoz, N.; Wunder, J.S.; Andrulis, I.L. Aberrant Hedgehog Signaling and Clinical Outcome in Osteosarcoma. Sarcoma 2014, 2014, 261804. [Google Scholar] [CrossRef] [PubMed]
- Bernardini, G.; Geminiani, M.; Gambassi, S.; Orlandini, M.; Petricci, E.; Marzocchi, B.; Laschi, M.; Taddei, M.; Manetti, F.; Santucci, A. Novel Smoothened Antagonists as Anti-Neoplastic Agents for the Treatment of Osteosarcoma. J. Cell Physiol. 2018, 233, 4961–4971. [Google Scholar] [CrossRef]
- Lo, W.W.; Wunder, J.S.; Dickson, B.C.; Campbell, V.; McGovern, K.; Alman, B.A.; Andrulis, I.L. Involvement and Targeted Intervention of Dysregulated Hedgehog Signaling in Osteosarcoma: Hedgehog Signaling in Osteosarcoma. Cancer 2014, 120, 537–547. [Google Scholar] [CrossRef]
- Righi, A.; Gambarotti, M.; Sbaraglia, M.; Sisto, A.; Ferrari, S.; Dei Tos, A.P.; Picci, P. P16 Expression as a Prognostic and Predictive Marker in High-Grade Localized Osteosarcoma of the Extremities: An Analysis of 357 Cases. Hum. Pathol. 2016, 58, 15–23. [Google Scholar] [CrossRef]
- Hingorani, P.; Zhang, W.; Zhang, Z.; Xu, Z.; Wang, W.-L.; Roth, M.E.; Wang, Y.; Gill, J.B.; Harrison, D.J.; Teicher, B.A.; et al. Trastuzumab Deruxtecan, Antibody–Drug Conjugate Targeting HER2, Is Effective in Pediatric Malignancies: A Report by the Pediatric Preclinical Testing Consortium. Mol. Cancer Ther. 2022, 21, 1318–1325. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, W.; Yan, Q.; Luo, Y. Expression of HER-2 in Surgical Specimen and Biopsy as a Biomarker of Metastasis in Patients with Osteosarcoma: A Meta-Analysis. Transl. Cancer Res. 2019, 8, 1129–1136. [Google Scholar] [CrossRef]
- Xie, X.; Li, Y.; Zhu, H.; Kuang, Z.; Chen, D.; Fan, T. Prognostic Significance of β-Catenin Expression in Osteosarcoma: A Meta-Analysis. Front. Oncol. 2020, 10, 402. [Google Scholar] [CrossRef]
- Cersosimo, F.; Lonardi, S.; Bernardini, G.; Telfer, B.; Mandelli, G.E.; Santucci, A.; Vermi, W.; Giurisato, E. Tumor-Associated Macrophages in Osteosarcoma: From Mechanisms to Therapy. Int. J. Mol. Sci. 2020, 21, 5207. [Google Scholar] [CrossRef]
- Yiqi, Z.; Ziyun, L.; Qin, F.; Xingli, W.; Liyu, Y. Identification of 9-Gene Epithelial–Mesenchymal Transition Related Signature of Osteosarcoma by Integrating Multi Cohorts. Technol. Cancer Res. Treat. 2020, 19, 153303382098076. [Google Scholar] [CrossRef]
- Qian, H.; Lei, T.; Hu, Y.; Lei, P. Expression of Lipid-Metabolism Genes Is Correlated With Immune Microenvironment and Predicts Prognosis in Osteosarcoma. Front. Cell Dev. Biol. 2021, 9, 673827. [Google Scholar] [CrossRef]
- Danieau, G.; Morice, S.; Rédini, F.; Verrecchia, F.; Royer, B.B. New Insights about the Wnt/β-Catenin Signaling Pathway in Primary Bone Tumors and Their Microenvironment: A Promising Target to Develop Therapeutic Strategies? Int. J. Mol. Sci. 2019, 20, 3751. [Google Scholar] [CrossRef]
- Katoh, M.; Katoh, M. Molecular Genetics and Targeted Therapy of WNT-Related Human Diseases (Review). Int. J. Mol. Med. 2017, 40, 587–606. [Google Scholar] [CrossRef]
- Robertson, H.; Hayes, J.D.; Sutherland, C. A Partnership with the Proteasome; the Destructive Nature of GSK3. Biochem. Pharmacol. 2018, 147, 77–92. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X. Targeting the Wnt/β-Catenin Signaling Pathway in Cancer. J. Hematol. Oncol. 2020, 13, 165. [Google Scholar] [CrossRef]
- Morice, S.; Danieau, G.; Rédini, F.; Brounais-Le-Royer, B.; Verrecchia, F. Hippo/YAP Signaling Pathway: A Promising Therapeutic Target in Bone Paediatric Cancers? Cancers 2020, 12, 645. [Google Scholar] [CrossRef]
- Kovar, H.; Bierbaumer, L.; Radic-Sarikas, B. The YAP/TAZ Pathway in Osteogenesis and Bone Sarcoma Pathogenesis. Cells 2020, 9, 972. [Google Scholar] [CrossRef]
- Pridgeon, M.G.; Grohar, P.J.; Steensma, M.R.; Williams, B.O. Wnt Signaling in Ewing Sarcoma, Osteosarcoma, and Malignant Peripheral Nerve Sheath Tumors. Curr. Osteoporos. Rep. 2017, 15, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Tomar, V.S.; Patil, V.; Somasundaram, K. Temozolomide Induces Activation of Wnt/β-Catenin Signaling in Glioma Cells via PI3K/Akt Pathway: Implications in Glioma Therapy. Cell Biol. Toxicol. 2020, 36, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.M.; Tao, F.; Roy, A.; Lin, T.; He, X.C.; Chen, S.; Lu, X.; Nemechek, J.; Ruan, L.; Yu, X.; et al. Overcoming Wnt–β-Catenin Dependent Anticancer Therapy Resistance in Leukaemia Stem Cells. Nat. Cell Biol. 2020, 22, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, N.; Kurzrock, R. Targeting the Wnt/Beta-Catenin Pathway in Cancer: Update on Effectors and Inhibitors. Cancer Treat. Rev. 2018, 62, 50–60. [Google Scholar] [CrossRef]
- Zhao, J. The Canonical Wnt-Beta-Catenin Pathway in Development and Chemotherapy of Osteosarcoma. Front. Biosci. 2013, 18, 1384. [Google Scholar] [CrossRef]
- Liu, W.; Zhao, Z.; Wang, Y.; Li, W.; Su, Q.; Jia, Q.; Zhang, J.; Zhang, X.; Shen, J.; Yin, J. Dioscin Inhibits Stem-Cell-like Properties and Tumor Growth of Osteosarcoma through Akt/GSK3/β-Catenin Signaling Pathway. Cell Death Dis. 2018, 9, 343. [Google Scholar] [CrossRef]
- Lu, Y.; Guan, G.-F.; Chen, J.; Hu, B.; Sun, C.; Ma, Q.; Wen, Y.-H.; Qiu, X.-C.; Zhou, Y. Aberrant CXCR4 and β-Catenin Expression in Osteosarcoma Correlates with Patient Survival. Oncol. Lett. 2015, 10, 2123–2129. [Google Scholar] [CrossRef]
- Cai, Y.; Mohseny, A.B.; Karperien, M.; Hogendoorn, P.C.; Zhou, G.; Cleton-Jansen, A.-M. Inactive Wnt/β-Catenin Pathway in Conventional High-Grade Osteosarcoma: Inactive Canonical Wnt Pathway in Osteosarcoma. J. Pathol. 2010, 220, 24–33. [Google Scholar] [CrossRef]
- Du, X.; Yang, J.; Yang, D.; Tian, W.; Zhu, Z. The Genetic Basis for Inactivation of Wnt Pathway in Human Osteosarcoma. BMC Cancer 2014, 14, 450. [Google Scholar] [CrossRef]
- Wang, Y.; He, J.; Zhang, J.; Zhang, N.; Zhou, Y.; Wu, F. Cell Migration Induces Apoptosis in Osteosarcoma Cell via Inhibition of Wnt-β-Catenin Signaling Pathway. Colloids Surf. B Biointerfaces 2023, 223, 113142. [Google Scholar] [CrossRef]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Kahn, M.; Yang, S.X.; Ivy, S.P. Targeting Notch, Hedgehog, and Wnt Pathways in Cancer Stem Cells: Clinical Update. Nat. Rev. Clin. Oncol. 2015, 12, 445–464. [Google Scholar] [CrossRef]
- Martins-Neves, S.R.; Corver, W.E.; Paiva-Oliveira, D.I.; van den Akker, B.E.W.M.; Briaire-de-Bruijn, I.H.; Bovée, J.V.M.G.; Gomes, C.M.F.; Cleton-Jansen, A.-M. Osteosarcoma Stem Cells Have Active Wnt/β-Catenin and Overexpress SOX2 and KLF4: Wnt/β-CATENIN AND SOX2 IN SARCOMA STEM CELLS. J. Cell Physiol. 2016, 231, 876–886. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y.; Wang, N.; Yang, L.; Zhao, W.; Zeng, X. MiR-130b Targets NKD2 and Regulates the Wnt Signaling to Promote Proliferation and Inhibit Apoptosis in Osteosarcoma Cells. Biochem. Biophys. Res. Commun. 2016, 471, 479–485. [Google Scholar] [CrossRef]
- Wang, Q.; Cai, J.; Cai, X.; Chen, L. MiR-346 Regulates Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells by Targeting the Wnt/β-Catenin Pathway. PLoS ONE 2013, 8, e72266. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.; He, M.; Qiao, J.; Sang, Y.; Cheang, L.H.; Gomes, F.C.; Hu, Y.; Li, Z.; Liu, N.; et al. HSP90 Regulates Osteosarcoma Cell Apoptosis by Targeting the P53/TCF-1-mediated Transcriptional Network. J. Cell Physiol. 2020, 235, 3894–3904. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S.; Xu, Y.; Zhang, Y.; Guan, H.; Li, X.; Li, Y.; Wang, Y. Upregulation of MiR-192 Inhibits Cell Growth and Invasion and Induces Cell Apoptosis by Targeting TCF7 in Human Osteosarcoma. Tumor Biol. 2016, 37, 15211–15220. [Google Scholar] [CrossRef]
- Li, R.; Liu, S.; Li, Y.; Tang, Q.; Xie, Y.; Zhai, R. Long Noncoding RNA AFAP1-AS1 Enhances Cell Proliferation and Invasion in Osteosarcoma through Regulating MiR-4695-5p/TCF4-β-catenin Signaling. Mol. Med. Rep. 2018, 18, 1616–1622. [Google Scholar] [CrossRef]
- Zhu, X.; Lu, K.; Cao, L.; Hu, Y.; Yin, Y.; Cai, Y. FoxM1 Is Upregulated in Osteosarcoma and Inhibition of FoxM1 Decreases Osteosarcoma Cell Proliferation, Migration, and Invasion. Cancer Manag. Res. 2020, 12, 9857–9867. [Google Scholar] [CrossRef]
- Halasi, M.; Gartel, A.L. FOX(M1) News—It Is Cancer. Mol. Cancer Ther. 2013, 12, 245–254. [Google Scholar] [CrossRef]
- Gartel, A.L. FOXM1 in Cancer: Interactions and Vulnerabilities. Cancer Res. 2017, 77, 3135–3139. [Google Scholar] [CrossRef]
- Lv, G.-Y.; Miao, J.; Zhang, X.-L. Long Noncoding RNA XIST Promotes Osteosarcoma Progression by Targeting Ras-Related Protein RAP2B via MiR-320b. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2018, 26, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhu, X.; Ke, Y.; Xiao, D.; Liang, C.; Chen, J.; Chang, Y. LncRNA FTX Inhibition Restrains Osteosarcoma Proliferation and Migration via Modulating MiR-320a/TXNRD1. Cancer Biol. Ther. 2020, 21, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, Q.; Shen, W. Circ-FOXM1 Promotes the Proliferation, Migration and EMT Process of Osteosarcoma Cells through FOXM1-Mediated Wnt Pathway Activation. J. Orthop. Surg. Res. 2022, 17, 344. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Zhou, N.; Hu, T.; Zhao, W.; Wu, D.; Wang, S. LncRNA MEG3 Negatively Modified Osteosarcoma Development through Regulation of MiR-361-5p and FoxM1. J. Cell. Physiol. 2019, 234, 13464–13480. [Google Scholar] [CrossRef]
- Sun, Y.; Jia, X.; Wang, M.; Deng, Y. Long Noncoding RNA MIR31HG Abrogates the Availability of Tumor Suppressor MicroRNA-361 for the Growth of Osteosarcoma. Cancer Manag. Res. 2019, 11, 8055–8064. [Google Scholar] [CrossRef]
- Cai, X.; Xiao, W.; Shen, J.; Lian, H.; Lu, Y.; Liu, X.; Gu, J. Thiostrepton and MiR-216b Synergistically Promote Osteosarcoma Cell Cytotoxicity and Apoptosis by Targeting FoxM1. Oncol. Lett. 2020, 20, 391. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Z.; Li, J.; Sang, X. Diallyl Disulfide Suppresses FOXM1-mediated Proliferation and Invasion in Osteosarcoma by Upregulating MiR-134. J. Cell. Biochem. 2019, 120, 7286–7296. [Google Scholar] [CrossRef]
- Sun, C.; Dai, J.; Ma, S.; Pan, Y.; Liu, F.; Wang, Y. MicroRNA-197 Inhibits the Progression of Osteosarcoma through Targeting FOXM1. Minerva Endocrinol. 2020, 45, 153–156. [Google Scholar] [CrossRef]
- Duan, N.; Hu, X.; Yang, X.; Cheng, H.; Zhang, W. MicroRNA-370 Directly Targets FOXM1 to Inhibit Cell Growth and Metastasis in Osteosarcoma Cells. Int. J. Clin. Exp. Pathol. 2015, 8, 10250–10260. [Google Scholar]
- Grant, G.D.; Brooks, L.; Zhang, X.; Mahoney, J.M.; Martyanov, V.; Wood, T.A.; Sherlock, G.; Cheng, C.; Whitfield, M.L. Identification of Cell Cycle–Regulated Genes Periodically Expressed in U2OS Cells and Their Regulation by FOXM1 and E2F Transcription Factors. Mol. Biol. Cell 2013, 24, 3634–3650. [Google Scholar] [CrossRef]
- Xu, J.; Chen, G.; Zhang, Y.; Huang, Z.; Cheng, X.; Gu, H.; Xia, J.; Yin, X. LINC00511 Promotes Osteosarcoma Tumorigenesis and Invasiveness through the MiR-185-3p/E2F1 Axis. BioMed Res. Int. 2020, 2020, 1974506. [Google Scholar] [CrossRef]
- Liu, X.; Wang, H.; Tao, G.-L.; Chu, T.-B.; Wang, Y.-X.; Liu, L. LncRNA-TMPO-AS1 Promotes Apoptosis of Osteosarcoma Cells by Targeting MiR-329 and Regulating E2F1. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 11006–11015. [Google Scholar] [CrossRef]
- Wu, H.; Li, W.; Zhang, M.; Zhu, S.; Zhang, D.; Wang, X. Inhibitory Roles of MiR-320 in Osteosarcoma via Regulating E2F1. J. Cancer Res. Ther. 2016, 12, 68. [Google Scholar] [CrossRef]
- Fang, D.-Z.; Wang, Y.-P.; Liu, J.; Hui, X.-B.; Wang, X.-D.; Chen, X.; Liu, D. MicroRNA-129-3p Suppresses Tumor Growth by Targeting E2F5 in Glioblastoma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 1044–1050. [Google Scholar] [CrossRef]
- Tian, Q.; Gu, Y.; Wang, F.; Zhou, L.; Dai, Z.; Liu, H.; Wu, X.; Wang, X.; Liu, Y.; Chen, S.; et al. Upregulation of MiRNA-154-5p Prevents the Tumorigenesis of Osteosarcoma. Biomed. Pharmacother. 2020, 124, 109884. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, X.; Han, Q.; Huang, Z. Downregulation of Long Non-Coding RNA UCA1 Represses Tumorigenesis and Metastasis of Osteosarcoma via MiR-513b-5p/E2F5 Axis. Anti-Cancer Drugs 2021, 32, 602–613. [Google Scholar] [CrossRef]
- Gao, Y.; Feng, B.; Lu, L.; Han, S.; Chu, X.; Chen, L.; Wang, R. MiRNAs and E2F3: A Complex Network of Reciprocal Regulations in Human Cancers. Oncotarget 2017, 8, 60624–60639. [Google Scholar] [CrossRef]
- Li, S.; Zeng, M.; Yang, L.; Tan, J.; Yang, J.; Guan, H.; Kuang, M.; Li, J. Hsa_circ_0008934 Promotes the Proliferation and Migration of Osteosarcoma Cells by Targeting MiR-145-5p to Enhance E2F3 Expression. Int. J. Biochem. Cell Biol. 2020, 127, 105826. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Z.; Shen, J. Downregulation of Hsa_circ_0000885 Suppressed Osteosarcoma Metastasis and Progression via Regulating E2F3 Expression and Sponging MiR-16-5p. Regen. Ther. 2022, 21, 114–121. [Google Scholar] [CrossRef]
- Wang, L.; Wang, L.; Zhang, X. Knockdown of LncRNA HOXA-AS2 Inhibits Viability, Migration and Invasion of Osteosarcoma Cells by MiR-124-3p/E2F3. OncoTargets Ther. 2019, 12, 10851–10861. [Google Scholar] [CrossRef]
- Dong, D.; Gong, Y.; Zhang, D.; Bao, H.; Gu, G. MiR-874 Suppresses the Proliferation and Metastasis of Osteosarcoma by Targeting E2F3. Tumor Biol. 2016, 37, 6447–6455. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Han, J.; Dong, D.; Wang, N. MicroRNA-152 Suppresses Human Osteosarcoma Cell Proliferation and Invasion by Targeting E2F Transcription Factor 3. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2018, 26, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Qiao, R.H.; Wang, D.M.; Huang, X.W.; Li, B.; Wang, D. UHRF1 Promotes Human Osteosarcoma Cell Invasion by Downregulating the Expression of E-Cadherin in an Rb1-Dependent Manner. Mol. Med. Rep. 2016, 13, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.C.; Benavente, C.A. Chromatin Remodeling Protein HELLS Is Upregulated by Inactivation of the RB-E2F Pathway and Is Nonessential for Osteosarcoma Tumorigenesis. Oncotarget 2018, 9, 32580–32592. [Google Scholar] [CrossRef]
- Jung, O.; Lee, S.Y. Synergistic Anticancer Effects of Timosaponin AIII and Ginsenosides in MG63 Human Osteosarcoma Cells. J. Ginseng Res. 2019, 43, 488–495. [Google Scholar] [CrossRef]
- Zhang, Y.; Jing, X.; Li, Z.; Tian, Q.; Wang, Q.; Chen, X. Investigation of the Role of the MiR17-92 Cluster in BMP9-Induced Osteoblast Lineage Commitment. J. Orthop. Surg. Res. 2021, 16, 652. [Google Scholar] [CrossRef]
- Wu, D.; Zhang, H.; Ji, F.; Ding, W. MicroRNA-17 Promotes Osteosarcoma Cells Proliferation and Migration and Inhibits Apoptosis by Regulating SASH1 Expression. Pathol.-Res. Pract. 2019, 215, 115–120. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.; Wu, W.; Dang, H.; Wang, B. Expression of the Nrf2 and Keap1 Proteins and Their Clinical Significance in Osteosarcoma. Biochem. Biophys. Res. Commun. 2016, 473, 42–46. [Google Scholar] [CrossRef]
- Li, P.-C.; Tu, M.-J.; Ho, P.Y.; Jilek, J.L.; Duan, Z.; Zhang, Q.-Y.; Yu, A.-X.; Yu, A.-M. Bioengineered NRF2-SiRNA Is Effective to Interfere with NRF2 Pathways and Improve Chemosensitivity of Human Cancer Cells. Drug Metab. Dispos. 2018, 46, 2–10. [Google Scholar] [CrossRef]
- Fan, H.-P.; Wang, S.-Y.; Shi, Y.-Y.; Sun, J. MicroRNA-340-5p Inhibits the Malignant Phenotypes of Osteosarcoma by Directly Targeting NRF2 and Deactivating the PI3K/AKT Pathway. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 3661–3669. [Google Scholar] [CrossRef]
- Morice, S.; Mullard, M.; Brion, R.; Dupuy, M.; Renault, S.; Tesfaye, R.; Brounais-Le Royer, B.; Ory, B.; Redini, F.; Verrecchia, F. The YAP/TEAD Axis as a New Therapeutic Target in Osteosarcoma: Effect of Verteporfin and CA3 on Primary Tumor Growth. Cancers 2020, 12, 3847. [Google Scholar] [CrossRef]
- Bouvier, C.; Macagno, N.; Nguyen, Q.; Loundou, A.; Jiguet-Jiglaire, C.; Gentet, J.-C.; Jouve, J.-L.; Rochwerger, A.; Mattei, J.-C.; Bouvard, D.; et al. Prognostic Value of the Hippo Pathway Transcriptional Coactivators YAP/TAZ and Β1-Integrin in Conventional Osteosarcoma. Oncotarget 2016, 7, 64702–64710. [Google Scholar] [CrossRef]
- Morice, S.; Danieau, G.; Tesfaye, R.; Mullard, M.; Brion, R.; Dupuy, M.; Ory, B.; Brounais-Le Royer, B.; Corre, I.; Redini, F.; et al. Involvement of the TGF-β Signaling Pathway in the Development of YAP-Driven Osteosarcoma Lung Metastasis. Front. Oncol. 2021, 11, 765711. [Google Scholar] [CrossRef]
- Shi, P.; Li, Y.; Guo, Q. Circular RNA CircPIP5K1A Contributes to Cancer Stemness of Osteosarcoma by MiR-515-5p/YAP Axis. J. Transl. Med. 2021, 19, 464. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, X.; Zhou, R.; Zhou, Y. TRPS1 and YAP1 Regulate Cell Proliferation and Drug Resistance of Osteosarcoma via Competitively Binding to the Target of CircTADA2A—MiR-129-5p. OncoTargets Ther. 2020, 13, 12397–12407. [Google Scholar] [CrossRef]
- Liu, G.; Huang, K.; Jie, Z.; Wu, Y.; Chen, J.; Chen, Z.; Fang, X.; Shen, S. CircFAT1 Sponges MiR-375 to Promote the Expression of Yes-Associated Protein 1 in Osteosarcoma Cells. Mol. Cancer 2018, 17, 170. [Google Scholar] [CrossRef]
- Yang, C.; Wu, K.; Wang, S.; Wei, G. Long Non-coding RNA XIST Promotes Osteosarcoma Progression by Targeting YAP via MiR-195-5p. J. Cell. Biochem. 2018, 119, 5646–5656. [Google Scholar] [CrossRef]
- Hu, X.-H.; Dai, J.; Shang, H.-L.; Zhao, Z.-X.; Hao, Y.-D. MiR-1285-3p Is a Potential Prognostic Marker in Human Osteosarcoma and Functions as a Tumor Suppressor by Targeting YAP1. Cancer Biomark. 2019, 25, 1–10. [Google Scholar] [CrossRef]
- Guan, H.; Shang, G.; Cui, Y.; Liu, J.; Sun, X.; Cao, W.; Wang, Y.; Li, Y. Long Noncoding RNA APTR Contributes to Osteosarcoma Progression through Repression of MiR-132-3p and Upregulation of Yes-associated Protein 1. J. Cell. Physiol. 2019, 234, 8998–9007. [Google Scholar] [CrossRef]
- Luo, Z.; Wu, G.; Zhang, D.; Liu, J.; Ran, R. MicroRNA-625 Targets Yes-associated Protein 1 to Suppress Cell Proliferation and Invasion of Osteosarcoma. Mol. Med. Rep. 2018, 17, 2005–2011. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, X.; Wei, M. MicroRNA-144 Suppresses Osteosarcoma Growth and Metastasis by Targeting ROCK1 and ROCK2. Oncotarget 2015, 6, 10297–10308. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Choy, E.; Hornicek, F.J.; Yang, S.; Yang, C.; Harmon, D.; Mankin, H.; Duan, Z. ROCK1 as a Potential Therapeutic Target in Osteosarcoma: ROCK1 AND OSTEOSARCOMA. J. Orthop. Res. 2011, 29, 1259–1266. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Poornajaf, Y.; Hussen, B.M.; Abak, A.; Shoorei, H.; Taheri, M.; Sharifi, G. Implication of Non-Coding RNA-Mediated ROCK1 Regulation in Various Diseases. Front. Mol. Biosci. 2022, 9, 986722. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wei, M.; Wang, W. MicroRNA-340 Suppresses Osteosarcoma Tumor Growth and Metastasis by Directly Targeting ROCK1. Biochem. Biophys. Res. Commun. 2013, 437, 653–658. [Google Scholar] [CrossRef]
- Zucchini, C.; Manara, M.C.; Cristalli, C.; Carrabotta, M.; Greco, S.; Pinca, R.S.; Ferrari, C.; Landuzzi, L.; Pasello, M.; Lollini, P.-L.; et al. ROCK2 Deprivation Leads to the Inhibition of Tumor Growth and Metastatic Potential in Osteosarcoma Cells through the Modulation of YAP Activity. J. Exp. Clin. Cancer Res. 2019, 38, 503. [Google Scholar] [CrossRef]
- Cui, S.-Q.; Wang, H. MicroRNA-144 Inhibits the Proliferation, Apoptosis, Invasion, and Migration of Osteosarcoma Cell Line F5M2. Tumor Biol. 2015, 36, 6949–6958. [Google Scholar] [CrossRef]
- Liu, J.L.; Li, J.; Xu, J.J.; Xiao, F.; Cui, P.L.; Qiao, Z.G.; Chen, X.D.; Tao, W.D.; Zhang, X.L. MiR-144 Inhibits Tumor Growth and Metastasis in Osteosarcoma via Dual-Suppressing RhoA/ROCK1 Signaling Pathway. Mol. Pharmacol. 2019, 95, 451–461. [Google Scholar] [CrossRef]
- Cai, H.; Lin, L.; Cai, H.; Tang, M.; Wang, Z. Combined MicroRNA-340 and ROCK1 MRNA Profiling Predicts Tumor Progression and Prognosis in Pediatric Osteosarcoma. Int. J. Mol. Sci. 2014, 15, 560–573. [Google Scholar] [CrossRef]
- Yang, H.; Peng, Z.; Da, Z.; Li, X.; Cheng, Y.; Tan, B.; Xiang, X.; Zheng, H.; Li, Y.; Chen, L.; et al. MicroRNA-148a Acts as a Tumor Suppressor in Osteosarcoma via Targeting Rho-Associated Coiled-Coil Kinase. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 1231–1243. [Google Scholar] [CrossRef]
- Zhou, G. MiR-139 Inhibits Osteosarcoma Cell Proliferation and Invasion by Targeting i ROCK1 i. Front. Biosci. 2019, 24, 1167–1177. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, W.; Fu, Q. MiR-335 Suppresses Migration and Invasion by Targeting ROCK1 in Osteosarcoma Cells. Mol. Cell. Biochem. 2013, 384, 105–111. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, N.; Zeng, X.; Sun, J.; Wang, G.; Xu, H.; Zhao, W. MicroRNA-335 and Its Target Rock1 Synergistically Influence Tumor Progression and Prognosis in Osteosarcoma. Oncol. Lett. 2017, 13, 3057–3065. [Google Scholar] [CrossRef]
- Li, E.; Zhang, J.; Yuan, T.; Ma, B. MiR-145 Inhibits Osteosarcoma Cells Proliferation and Invasion by Targeting ROCK1. Tumor Biol. 2014, 35, 7645–7650. [Google Scholar] [CrossRef]
- Han, C.; Wang, W. MicroRNA-129-5p Suppresses Cell Proliferation, Migration and Invasion via Targeting ROCK1 in Osteosarcoma. Mol. Med. Rep. 2018, 17, 4777–4784. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, D.; Zhu, T.; Yin, R. MiR-214-5p Targets ROCK1 and Suppresses Proliferation and Invasion of Human Osteosarcoma Cells. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 75–81. [Google Scholar] [CrossRef]
- Jiang, R.; Zhang, C.; Liu, G.; Gu, R.; Wu, H. MicroRNA-101 Inhibits Proliferation, Migration and Invasion in Osteosarcoma Cells by Targeting ROCK1. Am. J. Cancer Res. 2017, 7, 88–97. [Google Scholar]
- Li, C.; Ma, D.; Yang, J.; Lin, X.; Chen, B. MiR-202-5p Inhibits the Migration and Invasion of Osteosarcoma Cells by Targeting ROCK1. Oncol. Lett. 2018, 16, 829–834. [Google Scholar] [CrossRef]
- Yang, X.; Liao, H.-Y.; Zhang, H.-H. Roles of MET in Human Cancer. Clin. Chim. Acta 2022, 525, 69–83. [Google Scholar] [CrossRef]
- Kobayashi, E.; Hornicek, F.J.; Duan, Z. MicroRNA Involvement in Osteosarcoma. Sarcoma 2012, 2012, 359739. [Google Scholar] [CrossRef]
- Rothenberger, N.; Stabile, L. Hepatocyte Growth Factor/c-Met Signaling in Head and Neck Cancer and Implications for Treatment. Cancers 2017, 9, 39. [Google Scholar] [CrossRef]
- Refaat, T.; Donnelly, E.D.; Sachdev, S.; Parimi, V.; El Achy, S.; Dalal, P.; Farouk, M.; Berg, N.; Helenowski, I.; Gross, J.P.; et al. C-Met Overexpression in Cervical Cancer, a Prognostic Factor and a Potential Molecular Therapeutic Target. Am. J. Clin. Oncol. 2017, 40, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.; Li, B.; Sun, J.; Sun, L. MiR-454 Is down-Regulated in Osteosarcomas and Suppresses Cell Proliferation and Invasion by Directly Targeting c-Met. Cell Prolif. 2015, 48, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Zvi, Y.; Ugur, E.; Batko, B.; Gill, J.; Roth, M.; Gorlick, R.; Hall, D.; Tingling, J.; Barkauskas, D.A.; Zhang, J.; et al. Prognostic and Therapeutic Utility of Variably Expressed Cell Surface Receptors in Osteosarcoma. Sarcoma 2021, 2021, 8324348. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lu, C.; Wang, J.; Gao, M.; Gao, W. MicroRNA-449b-5p Suppresses Proliferation, Migration, and Invasion of Osteosarcoma by Targeting c-Met. Med. Sci. Monit. 2019, 25, 6236–6243. [Google Scholar] [CrossRef]
- Yan, K.; Gao, J.; Yang, T.; Ma, Q.; Qiu, X.; Fan, Q.; Ma, B. MicroRNA-34a Inhibits the Proliferation and Metastasis of Osteosarcoma Cells Both In Vitro and In Vivo. PLoS ONE 2012, 7, e33778. [Google Scholar] [CrossRef]
- Xie, W.; Xiao, J.; Wang, T.; Zhang, D.; Li, Z. MicroRNA-876-5p Inhibits Cell Proliferation, Migration and Invasion by Targeting C-Met in Osteosarcoma. J. Cell. Mol. Med. 2019, 23, 3293–3301. [Google Scholar] [CrossRef]
- Ji, L.; Lu, B.; Zamponi, R.; Charlat, O.; Aversa, R.; Yang, Z.; Sigoillot, F.; Zhu, X.; Hu, T.; Reece-Hoyes, J.S.; et al. USP7 Inhibits Wnt/β-Catenin Signaling through Promoting Stabilization of Axin. Nat. Commun. 2019, 10, 4184. [Google Scholar] [CrossRef]
- Chen, X.; Zhong, L.; Li, X.; Liu, W.; Zhao, Y.; Li, J. Down-Regulation of MicroRNA-31-5p Inhibits Proliferation and Invasion of Osteosarcoma Cells through Wnt/β-Catenin Signaling Pathway by Enhancing AXIN1. Exp. Mol. Pathol. 2019, 108, 32–41. [Google Scholar] [CrossRef]
- Grimm, D.; Bauer, J.; Wise, P.; Krüger, M.; Simonsen, U.; Wehland, M.; Infanger, M.; Corydon, T.J. The Role of SOX Family Members in Solid Tumours and Metastasis. Semin. Cancer Biol. 2020, 67, 122–153. [Google Scholar] [CrossRef]
- Moreno, C.S. SOX4: The Unappreciated Oncogene. Semin. Cancer Biol. 2020, 67, 57–64. [Google Scholar] [CrossRef]
- Chen, B.; Liu, J.; Qu, J.; Song, Y.; Li, Y.; Pan, S. MicroRNA-25 Suppresses Proliferation, Migration, and Invasion of Osteosarcoma by Targeting SOX4. Tumour Biol. 2017, 39, 101042831770384. [Google Scholar] [CrossRef]
- Pan, L.; Meng, L.; Liang, F.; Cao, L. miR-188 suppresses tumor progression by targeting SOX4 in pediatric osteosarcoma. Mol. Med. Rep. 2018, 18, 441–446. [Google Scholar] [CrossRef]
- Bai, C.J.; Gao, T.; Liu, J.Y.; Li, S.; Wang, X.Y.; Fan, Z.F. SNHG9/miR-214-5p/SOX4 feedback loop regulates osteosarcoma progression. Neoplasma 2022, 69, 1175–1184. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Liu, J.; Wu, Y.; Zhu, Q. MicroRNA-132 Inhibits Cell Growth and Metastasis in Osteosarcoma Cell Lines Possibly by Targeting Sox4. Int. J. Oncol. 2015, 47, 1672–1684. [Google Scholar] [CrossRef]
- Wang, K.; Yan, L.; Lu, F. MiR-363-3p Inhibits Osteosarcoma Cell Proliferation and Invasion via Targeting SOX4. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2019, 27, 157–163. [Google Scholar] [CrossRef]
- Luo, X.-J.; Tang, D.-G.; Gao, T.-L.; Zhang, Y.-L.; Wang, M.; Quan, Z.-X.; Chen, J. MicroRNA-212 Inhibits Osteosarcoma Cells Proliferation and Invasion by Down-Regulation of Sox4. Cell. Physiol. Biochem. 2014, 34, 2180–2188. [Google Scholar] [CrossRef]
- Rongxin, S.; Pengfei, L.; Li, S.; Xiaochen, J.; Yihe, H. MicroRNA-340-5p Suppresses Osteosarcoma Development by down-Regulating the Wnt/β-Catenin Signaling Pathway via Targeting the STAT3 Gene. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 982–991. [Google Scholar] [CrossRef]
- Hu, Y.; Luo, X.; Zhou, J.; Chen, S.; Gong, M.; Deng, Y.; Zhang, H. Piperlongumine Inhibits the Progression of Osteosarcoma by Downregulating the SOCS3/JAK2/STAT3 Pathway via MiR-30d-5p. Life Sci. 2021, 277, 119501. [Google Scholar] [CrossRef]
- Liu, W.; Long, Q.; Zhang, W.; Zeng, D.; Hu, B.; Liu, S.; Chen, L. MiRNA-221-3p Derived from M2-Polarized Tumor-Associated Macrophage Exosomes Aggravates the Growth and Metastasis of Osteosarcoma through SOCS3/JAK2/STAT3 Axis. Aging 2021, 13, 19760–19775. [Google Scholar] [CrossRef]
- Sun, X.; He, Y.; Huang, C.; Ma, T.-T.; Li, J. Distinctive MicroRNA Signature Associated of Neoplasms with the Wnt/β-Catenin Signaling Pathway. Cell. Signal. 2013, 25, 2805–2811. [Google Scholar] [CrossRef]
- Zhang, Z.-C.; Liu, J.-X.; Shao, Z.-W.; Pu, F.-F.; Wang, B.-C.; Wu, Q.; Zhang, Y.-K.; Zeng, X.-L.; Guo, X.-D.; Yang, S.-H.; et al. In Vitro Effect of MicroRNA-107 Targeting Dkk-1 by Regulation of Wnt/β-Catenin Signaling Pathway in Osteosarcoma. Medicine 2017, 96, e7245. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Aireti, A.; Aihaiti, A.; Li, K. Expression of MicroRNA-150 and Its Target Gene IGF2BP1 in Human Osteosarcoma and Their Clinical Implications. Pathol. Oncol. Res. 2019, 25, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Dean, D.C.; Hornicek, F.J.; Spentzos, D.; Hoffman, R.M.; Shi, H.; Duan, Z. Myc Is a Prognostic Biomarker and Potential Therapeutic Target in Osteosarcoma. Ther. Adv. Med. Oncol. 2020, 12, 175883592092205. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Ma, S.; Yang, F.; Chen, X.; Wang, W.; Zhang, J.; Li, Y.; Wang, T.; Shan, L. MiR-193b Exhibits Mutual Interaction with MYC, and Suppresses Growth and Metastasis of Osteosarcoma. Oncol. Rep. 2020, 44, 139–155. [Google Scholar] [CrossRef]
- Matthews, H.K.; Bertoli, C.; de Bruin, R.A.M. Cell Cycle Control in Cancer. Nat. Rev. Mol. Cell Biol. 2022, 23, 74–88. [Google Scholar] [CrossRef]
- Fagundes, R.; Teixeira, L.K. Cyclin E/CDK2: DNA Replication, Replication Stress and Genomic Instability. Front. Cell Dev. Biol. 2021, 9, 774845. [Google Scholar] [CrossRef]
- Tadesse, S.; Anshabo, A.T.; Portman, N.; Lim, E.; Tilley, W.; Caldon, C.E.; Wang, S. Targeting CDK2 in Cancer: Challenges and Opportunities for Therapy. Drug Discov. Today 2020, 25, 406–413. [Google Scholar] [CrossRef]
- Suski, J.M.; Braun, M.; Strmiska, V.; Sicinski, P. Targeting Cell-Cycle Machinery in Cancer. Cancer Cell 2021, 39, 759–778. [Google Scholar] [CrossRef]
- Moon, S.; Kim, W.; Kim, S.; Kim, Y.; Song, Y.; Bilousov, O.; Kim, J.; Lee, T.; Cha, B.; Kim, M.; et al. Phosphorylation by NLK Inhibits YAP -14-3-3-interactions and Induces Its Nuclear Localization. EMBO Rep. 2017, 18, 61–71. [Google Scholar] [CrossRef]
- Zanconato, F.; Forcato, M.; Battilana, G.; Azzolin, L.; Quaranta, E.; Bodega, B.; Rosato, A.; Bicciato, S.; Cordenonsi, M.; Piccolo, S. Genome-Wide Association between YAP/TAZ/TEAD and AP-1 at Enhancers Drives Oncogenic Growth. Nat. Cell Biol. 2015, 17, 1218–1227. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, T.; Jia, Y.; Zou, J.; Wang, X.; Gu, W. LncRNA MALAT1/MiR-181a-5p Affects the Proliferation and Adhesion of Myeloma Cells via Regulation of Hippo-YAP Signaling Pathway. Cell Cycle 2019, 18, 2509–2523. [Google Scholar] [CrossRef]
- Deel, M.D.; Li, J.J.; Crose, L.E.S.; Linardic, C.M. A Review: Molecular Aberrations within Hippo Signaling in Bone and Soft-Tissue Sarcomas. Front. Oncol. 2015, 5, 190. [Google Scholar] [CrossRef]
- Basu-Roy, U.; Bayin, N.S.; Rattanakorn, K.; Han, E.; Placantonakis, D.G.; Mansukhani, A.; Basilico, C. Sox2 Antagonizes the Hippo Pathway to Maintain Stemness in Cancer Cells. Nat. Commun. 2015, 6, 6411. [Google Scholar] [CrossRef]
- Luo, Y.; Liu, W.; Tang, P.; Jiang, D.; Gu, C.; Huang, Y.; Gong, F.; Rong, Y.; Qian, D.; Chen, J.; et al. MiR-624-5p Promoted Tumorigenesis and Metastasis by Suppressing Hippo Signaling through Targeting PTPRB in Osteosarcoma Cells. J. Exp. Clin. Cancer Res. 2019, 38, 488. [Google Scholar] [CrossRef]
- Ma, J.; Huang, K.; Ma, Y.; Zhou, M.; Fan, S. The TAZ–MiR-224–SMAD4 Axis Promotes Tumorigenesis in Osteosarcoma. Cell Death Dis. 2017, 8, e2539. [Google Scholar] [CrossRef]
- Shen, S.; Huang, K.; Wu, Y.; Ma, Y.; Wang, J.; Qin, F.; Ma, J. A MiR-135b-TAZ Positive Feedback Loop Promotes Epithelial–Mesenchymal Transition (EMT) and Tumorigenesis in Osteosarcoma. Cancer Lett. 2017, 407, 32–44. [Google Scholar] [CrossRef]
- Verrecchia, F.; Rédini, F. Transforming Growth Factor-β Signaling Plays a Pivotal Role in the Interplay between Osteosarcoma Cells and Their Microenvironment. Front. Oncol. 2018, 8, 133. [Google Scholar] [CrossRef]
- Ma, K.; Zhang, C.; Li, W. TGF-β Is Associated with Poor Prognosis and Promotes Osteosarcoma Progression via PI3K/Akt Pathway Activation. Cell Cycle 2020, 19, 2327–2339. [Google Scholar] [CrossRef]
- Zhang, J.; Liang, J.H.; Huang, J.G. Bone Morphogenetic Protein 9 Facilitates Osteocarcinoma Cell Apoptosis and Inhibits in Vivo Tumor Growth. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef]
- Li, F.; Li, S.; Cheng, T. TGF-β1 Promotes Osteosarcoma Cell Migration and Invasion through the MiR-143-Versican Pathway. Cell. Physiol. Biochem. 2014, 34, 2169–2179. [Google Scholar] [CrossRef]
- Fu, Y.; Tang, Y.; Wang, J.; Guo, Z. MicroRNA-181c Suppresses the Biological Progression of Osteosarcoma via Targeting SMAD7 and Regulating Transforming Growth Factor-β (TGF-β) Signaling Pathway. Med. Sci. Monit. 2019, 25, 4801–4810. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Guo, J.; Sun, Z.; Lin, C.; Tao, H.; Zhang, Q.; Cui, Y.; Zuo, H.; Lin, Y.; Chen, S.; et al. BMP2-Dependent Gene Regulatory Network Analysis Reveals Klf4 as a Novel Transcription Factor of Osteoblast Differentiation. Cell Death Dis. 2021, 12, 197. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Li, Q.-Q.; Zhai, X.-C.; Qin, L.; Li, H.-B.; Meng, R.; Han, X.-F. MicroRNA-330-5p Promotes the Development of Osteosarcoma by Regulating SPRY2. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8761–8770. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Zhang, N.; Liu, S.; Pang, Z.; Chen, Z. By Targeting TRAF6, MiR-140-3p Inhibits TGF-Β1-Induced Human Osteosarcoma Epithelial-to-Mesenchymal Transition, Migration, and Invasion. Biotechnol. Lett. 2020, 42, 2123–2133. [Google Scholar] [CrossRef]
- Li, C.; Xu, B.; Miu, X.; Deng, Z.; Liao, H.; Hao, L. Inhibition of MiRNA-21 Attenuates the Proliferation and Metastasis of Human Osteosarcoma by Upregulating PTEN. Exp. Ther. Med. 2017, 15, 1036–1040. [Google Scholar] [CrossRef]
- Hu, X.; Li, L.; Lu, Y.; Yu, X.; Chen, H.; Yin, Q.; Zhang, Y. MiRNA-21 Inhibition Inhibits Osteosarcoma Cell Proliferation by Targeting PTEN and Regulating the TGF-β1 Signaling Pathway. Oncol. Lett. 2018, 16, 4337–4342. [Google Scholar] [CrossRef]
- Li, L.-Z.; Wu, Z.-Z.; Lv, Z. The Clinical Significance of MiR-21 in Guiding Chemotherapy for Patients with Osteosarcoma. Pharm. Pers. Med. 2021, 14, 1247–1261. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, M.; Wang, G. Hsa_circ_0051079 Functions as an Oncogene by Regulating MiR-26a-5p/TGF-Β1 in Osteosarcoma. Cell Biosci. 2019, 9, 94. [Google Scholar] [CrossRef]
- Zhang, Q.; Cao, L.; Zou, S.; Feng, Y.; Miao, X.; Huang, L.; Wu, Y. Human Umbilical Cord Mesenchymal Stem Cell-Derived Extracellular Vesicles Carrying MicroRNA-181c-5p Promote BMP2-Induced Repair of Cartilage Injury through Inhibition of SMAD7 Expression. Stem Cells Int. 2022, 2022, 1157498. [Google Scholar] [CrossRef]
- Rypens, C.; Marsan, M.; Van Berckelaer, C.; Billiet, C.; Melis, K.; Lopez, S.P.; van Dam, P.; Devi, G.R.; Finetti, P.; Ueno, N.T.; et al. Inflammatory Breast Cancer Cells Are Characterized by Abrogated TGFβ1-Dependent Cell Motility and SMAD3 Activity. Breast Cancer Res. Treat. 2020, 180, 385–395. [Google Scholar] [CrossRef]
- Saito, M.; Ichikawa, J.; Ando, T.; Schoenecker, J.G.; Ohba, T.; Koyama, K.; Suzuki-Inoue, K.; Haro, H. Platelet-Derived TGF-β Induces Tissue Factor Expression via the Smad3 Pathway in Osteosarcoma Cells. J. Bone Miner. Res. 2018, 33, 2048–2058. [Google Scholar] [CrossRef]
- Gu, Z.; Li, Z.; Xu, R.; Zhu, X.; Hu, R.; Xue, Y.; Xu, W. MiR-16-5p Suppresses Progression and Invasion of Osteosarcoma via Targeting at Smad3. Front. Pharmacol. 2020, 11, 1324. [Google Scholar] [CrossRef]
- Hu, Y.; Liang, D.; Chen, X.; Chen, L.; Bai, J.; Li, H.; Yin, C.; Zhong, W. MiR-671-5p negatively regulates SMAD3 to inhibit migration and invasion of osteosarcoma cells. Nan Fang Yi Ke Da Xue Xue Bao 2021, 41, 1562–1568. [Google Scholar] [CrossRef]
- Liu, M.; Xiusheng, H.; Xiao, X.; Wang, Y. Overexpression of MiR-422a Inhibits Cell Proliferation and Invasion, and Enhances Chemosensitivity in Osteosarcoma Cells. Oncol. Rep. 2016, 36, 3371–3378. [Google Scholar] [CrossRef]
- Wan, G.; Tian, L.; Yu, Y.; Li, F.; Wang, X.; Li, C.; Deng, S.; Yu, X.; Cai, X.; Zuo, Z.; et al. Overexpression of Pofut1 and Activated Notch1 May Be Associated with Poor Prognosis in Breast Cancer. Biochem. Biophys. Res. Commun. 2017, 491, 104–111. [Google Scholar] [CrossRef]
- Zhang, K.; Wu, S.; Wu, H.; Liu, L.; Zhou, J. Effect of the Notch1-Mediated PI3K-Akt-MTOR Pathway in Human Osteosarcoma. Aging 2021, 13, 21090–21101. [Google Scholar] [CrossRef]
- Qin, J.; Wang, R.; Zhao, C.; Wen, J.; Dong, H.; Wang, S.; Li, Y.; Zhao, Y.; Li, J.; Yang, Y.; et al. Notch Signaling Regulates Osteosarcoma Proliferation and Migration through Erk Phosphorylation. Tissue Cell 2019, 59, 51–61. [Google Scholar] [CrossRef]
- Mei, H.; Yu, L.; Ji, P.; Yang, J.; Fang, S.; Guo, W.; Liu, Y.; Chen, X. Doxorubicin Activates the Notch Signaling Pathway in Osteosarcoma. Oncol. Lett. 2015, 9, 2905–2909. [Google Scholar] [CrossRef]
- Xia, R.; Xu, M.; Yang, J.; Ma, X. The Role of Hedgehog and Notch Signaling Pathway in Cancer. Mol. Biomed. 2022, 3, 44. [Google Scholar] [CrossRef]
- Dailey, D.D.; Anfinsen, K.P.; Pfaff, L.E.; Ehrhart, E.; Charles, J.B.; Bønsdorff, T.B.; Thamm, D.H.; Powers, B.E.; Jonasdottir, T.J.; Duval, D.L. HES1, a Target of Notch Signaling, Is Elevated in Canine Osteosarcoma, but Reduced in the Most Aggressive Tumors. BMC Vet. Res. 2013, 9, 130. [Google Scholar] [CrossRef]
- Engin, F.; Bertin, T.; Ma, O.; Jiang, M.M.; Wang, L.; Sutton, R.E.; Donehower, L.A.; Lee, B. Notch Signaling Contributes to the Pathogenesis of Human Osteosarcomas. Hum. Mol. Genet. 2009, 18, 1464–1470. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.P.M. How the NOTCH Pathway Contributes to the Ability of Osteosarcoma Cells to Metastasize. In Pediatric and Adolescent Osteosarcoma; Jaffe, N., Bruland, O.S., Bielack, S., Eds.; Cancer Treatment and Research; Springer: Boston, MA, USA, 2009; Volume 152, pp. 479–496. ISBN 978-1-4419-0283-2. [Google Scholar]
- Tao, J.; Jiang, M.-M.; Jiang, L.; Salvo, J.S.; Zeng, H.-C.; Dawson, B.; Bertin, T.K.; Rao, P.H.; Chen, R.; Donehower, L.A.; et al. Notch Activation as a Driver of Osteogenic Sarcoma. Cancer Cell 2014, 26, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Ryter, S.W.; Mizumura, K.; Choi, A.M.K. The Impact of Autophagy on Cell Death Modalities. Int. J. Cell Biol. 2014, 2014, 502676. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zhang, Y.; Wan, R.; Zhou, J.; Wu, X.; Fan, Q.; He, J.; Tan, W.; Deng, Y. CEMIP Promotes Osteosarcoma Progression and Metastasis Through Activating Notch Signaling Pathway. Front. Oncol. 2022, 12, 919108. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Glassy, M.C.; Abak, A.; Hussen, B.M.; Niazi, V.; Taheri, M. The Interaction between MiRNAs/LncRNAs and Notch Pathway in Human Disorders. Biomed. Pharmacother. 2021, 138, 111496. [Google Scholar] [CrossRef]
- Venkatesh, V.; Nataraj, R.; Thangaraj, G.S.; Karthikeyan, M.; Gnanasekaran, A.; Kaginelli, S.B.; Kuppanna, G.; Kallappa, C.G.; Basalingappa, K.M. Targeting Notch Signalling Pathway of Cancer Stem Cells. Stem Cell Investig. 2018, 5, 5. [Google Scholar] [CrossRef]
- Zanotti, S.; Canalis, E. Notch Signaling and the Skeleton. Endocr. Rev. 2016, 37, 223–253. [Google Scholar] [CrossRef]
- Bae, Y.; Zeng, H.; Chen, Y.; Ketkar, S.; Munivez, E.; Yu, Z.; Gannon, F.H.; Lee, B.H. miRNA-34c Suppresses Osteosarcoma Progression In Vivo by Targeting Notch and E2F. JBMR Plus 2022, 6, e10623. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Tang, J.; Su, X.; Miao, Y. The Study of Mechanism of MiR-34c-5p Targeting FLOT2 to Regulate Proliferation, Migration and Invasion of Osteosarcoma Cells. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3559–3568. [Google Scholar] [CrossRef]
- Won, K.Y.; Kim, Y.W.; Kim, H.-S.; Lee, S.K.; Jung, W.-W.; Park, Y.-K. MicroRNA-199b-5p Is Involved in the Notch Signaling Pathway in Osteosarcoma. Hum. Pathol. 2013, 44, 1648–1655. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, G.; Zhang, Y.; Ma, Y.; Ding, Y.; Xu, N. MiR-199b-5p Promotes Malignant Progression of Osteosarcoma by Regulating HER2. J. Buon 2018, 23, 1816–1824. [Google Scholar]
- Zeng, H.; Zhang, Z.; Dai, X.; Chen, Y.; Ye, J.; Jin, Z. Increased Expression of MicroRNA-199b-5p Associates with Poor Prognosis through Promoting Cell Proliferation, Invasion and Migration Abilities of Human Osteosarcoma. Pathol. Oncol. Res. 2016, 22, 253–260. [Google Scholar] [CrossRef]
- Wang, L.; Hu, K.; Chao, Y.; Wang, X. MicroRNA-1296-5p Suppresses the Proliferation, Migration, and Invasion of Human Osteosarcoma Cells by Targeting NOTCH2. J. Cell. Biochem. 2020, 121, 2038–2046. [Google Scholar] [CrossRef]
- Wang, S.; Ren, T.; Huang, Y.; Bao, X.; Sun, K.; Shen, D.; Guo, W. BMPR2 and HIF1-α Overexpression in Resected Osteosarcoma Correlates with Distant Metastasis and Patient Survival. Chin. J. Cancer Res. 2017, 29, 447–454. [Google Scholar] [CrossRef]
- Ouyang, Y.; Li, H.; Bu, J.; Li, X.; Chen, Z.; Xiao, T. Hypoxia-Inducible Factor-1 Expression Predicts Osteosarcoma Patients’ Survival: A Meta-Analysis. Int. J. Biol. Markers 2016, 31, 229–234. [Google Scholar] [CrossRef]
- Luo, D.; Ren, H.; Zhang, W.; Xian, H.; Lian, K.; Liu, H. Clinicopathological and Prognostic Value of Hypoxia-Inducible Factor-1α in Patients with Bone Tumor: A Systematic Review and Meta-Analysis. J. Orthop. Surg. Res. 2019, 14, 56. [Google Scholar] [CrossRef]
- Li, Z.Q. CircRNA_103801 Accelerates Proliferation of Osteosarcoma Cells by Sponging MiR-338-3p and Regulating HIF-1/Rap1 /PI3K-Akt Pathway. J. Biol. Regul. Homeost. Agents 2021, 35, 1021–1028. [Google Scholar] [CrossRef]
- Xiao, Q.; Wei, Z.; Li, Y.; Zhou, X.; Chen, J.; Wang, T.; Shao, G.; Zhang, M.; Zhang, Z. MiR-186 Functions as a Tumor Suppressor in Osteosarcoma Cells by Suppressing the Malignant Phenotype and Aerobic Glycolysis. Oncol. Rep. 2018, 39, 2703–2710. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, H.; Yao, T.; Gong, H. Advanced Development of ErbB Family-Targeted Therapies in Osteosarcoma Treatment. Investig. New Drugs 2019, 37, 175–183. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, S.-L.; Chen, H.; Shen, R.-K.; Li, X.-D.; Huang, Q.-S.; Wu, C.-Y.; Weng, D.-F.; Lin, J.-H. Clinicopathological and Prognostic Values of ErbB Receptor Family Amplification in Primary Osteosarcoma. Scand. J. Clin. Lab. Investig. 2019, 79, 601–612. [Google Scholar] [CrossRef]
- Tan, J.; Yang, B.; Zhong, H.; Luo, M.; Su, Z.; Xie, C.; Shi, M.; Sun, C.; Lin, L. Circular RNA CircEMB Promotes Osteosarcoma Progression and Metastasis by Sponging MiR-3184-5p and Regulating EGFR Expression. Biomark Res. 2023, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cai, J.; Wang, J.; Xiong, C.; Zhao, J. MiR-143 Inhibits EGFR-Signaling-Dependent Osteosarcoma Invasion. Tumor Biol. 2014, 35, 12743–12748. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lan, M.; Liao, X.; Tang, Z.; Yang, C. Circular RNA Cir-ITCH Promotes Osteosarcoma Migration and Invasion through Cir-ITCH /MiR-7/EGFR Pathway. Technol. Cancer Res. Treat. 2020, 19, 153303381989872. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, G.; Li, B.; Qiu, C.; He, M. MiR-141-3p Is a Key Negative Regulator of the EGFR Pathway in Osteosarcoma. OncoTargets Ther. 2018, 11, 4461–4478. [Google Scholar] [CrossRef] [PubMed]
- Veys, C.; Jammes, M.; Rédini, F.; Poulain, L.; Denoyelle, C.; Legendre, F.; Galera, P. Tumor Suppressive Role of MiR-342-5p and MiR-491-5p in Human Osteosarcoma Cells. Pharmaceuticals 2022, 15, 362. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, F.; Wang, B.; Li, Z.; Zhou, D.; Yang, Q.; Dong, J.; Li, J. HER-2 Expression in Biopsy and Surgical Specimen on Prognosis of Osteosarcoma: A Systematic Review and Meta-Analysis of 16 Studies. Medicine 2016, 95, e3661. [Google Scholar] [CrossRef]
- Khalifa, S.E.; Fathy, Y. HER-2 Immunohistochemical Expression in Bone Sarcomas: A New Hope for Osteosarcoma Patients. Open Access Maced. J. Med. Sci. 2018, 6, 1555–1560. [Google Scholar] [CrossRef]
- Anderson, P.M.; Subbiah, V.; Trucco, M.M. Current and Future Targeted Alpha Particle Therapies for Osteosarcoma: Radium-223, Actinium-225, and Thorium-227. Front. Med. 2022, 9, 1030094. [Google Scholar] [CrossRef]
- Long, X.-H.; Zhang, G.-M.; Peng, A.-F.; Luo, Q.-F.; Zhang, L.; Wen, H.-C.; Zhou, R.-P.; Gao, S.; Zhou, Y.; Liu, Z.-L. Lapatinib Alters the Malignant Phenotype of Osteosarcoma Cells via Downregulation of the Activity of the HER2-PI3K/AKT-FASN Axis in Vitro. Oncol. Rep. 2014, 31, 328–334. [Google Scholar] [CrossRef]
- Yue, Z.; Guan, X.; Chao, R.; Huang, C.; Li, D.; Yang, P.; Liu, S.; Hasegawa, T.; Guo, J.; Li, M. Diallyl Disulfide Induces Apoptosis and Autophagy in Human Osteosarcoma MG-63 Cells through the PI3K/Akt/MTOR Pathway. Molecules 2019, 24, 2665. [Google Scholar] [CrossRef]
- Jin, R.; Jin, Y.; Tang, Y.; Yang, H.; Zhou, X.; Lei, Z. GPNMB Silencing Suppresses the Proliferation and Metastasis of Osteosarcoma Cells by Blocking the PI3K/Akt/MTOR Signaling Pathway. Oncol. Rep. 2018, 39, 3034–3040. [Google Scholar] [CrossRef]
- Chou, W.-C.; Guo, Z.; Guo, H.; Chen, L.; Zhang, G.; Liang, K.; Xie, L.; Tan, X.; Gibson, S.A.; Rampanelli, E.; et al. AIM2 in Regulatory T Cells Restrains Autoimmune Diseases. Nature 2021, 591, 300–305. [Google Scholar] [CrossRef]
- Zheng, J.; Liu, C.; Shi, J.; Wen, K.; Wang, X. AIM2 Inhibits the Proliferation, Invasion and Migration, and Promotes the Apoptosis of Osteosarcoma Cells by Inactivating the PI3K/AKT/MTOR Signaling Pathway. Mol. Med. Rep. 2021, 25, 53. [Google Scholar] [CrossRef]
- Wang, L.; Tang, B.; Han, H.; Mao, D.; Chen, J.; Zeng, Y.; Xiong, M. MiR-155 Affects Osteosarcoma MG-63 Cell Autophagy Induced by Adriamycin Through Regulating PTEN-PI3K/AKT/MTOR Signaling Pathway. Cancer Biother. Radiopharm. 2018, 33, 32–38. [Google Scholar] [CrossRef]
- Tsubaki, M.; Yamazoe, Y.; Yanae, M.; Satou, T.; Itoh, T.; Kaneko, J.; Kidera, Y.; Moriyama, K.; Nishida, S. Blockade of the Ras/MEK/ERK and Ras/PI3K/Akt Pathways by Statins Reduces the Expression of BFGF, HGF, and TGF-β as Angiogenic Factors in Mouse Osteosarcoma. Cytokine 2011, 54, 100–107. [Google Scholar] [CrossRef]
- Trohatou, O.; Zagoura, D.; Bitsika, V.; Pappa, K.I.; Antsaklis, A.; Anagnou, N.P.; Roubelakis, M.G. Sox2 Suppression by MiR-21 Governs Human Mesenchymal Stem Cell Properties. Stem Cells Transl. Med. 2014, 3, 54–68. [Google Scholar] [CrossRef]
- Pfeffer, S.R.; Yang, C.H.; Pfeffer, L.M. The Role of MiR-21 in Cancer: The Role of MiR-21 in Cancer. Drug Dev. Res. 2015, 76, 270–277. [Google Scholar] [CrossRef]
- Qi, J.; Zhang, R.; Wang, Y. Exosomal MiR-21-5p Derived from Bone Marrow Mesenchymal Stem Cells Promote Osteosarcoma Cell Proliferation and Invasion by Targeting PIK3R1. J. Cell. Mol. Med. 2021, 25, 11016–11030. [Google Scholar] [CrossRef]
- Meng, C.; Zhao, Z.; Bai, R.; Zhao, W.; Wang, Y.; Xue, H.; Sun, L.; Sun, C.; Feng, W.; Guo, S. MicroRNA-22 Mediates the Cisplatin Resistance of Osteosarcoma Cells by Inhibiting Autophagy via the PI3K/Akt/MTOR Pathway. Oncol. Rep. 2020, 43, 1169–1186. [Google Scholar] [CrossRef]
- Ali, M.U.; Ur Rahman, M.S.; Jia, Z.; Jiang, C. Eukaryotic Translation Initiation Factors and Cancer. Tumour Biol. 2017, 39, 101042831770980. [Google Scholar] [CrossRef]
- Bitterman, P.B.; Polunovsky, V.A. EIF4E-Mediated Translational Control of Cancer Incidence. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2015, 1849, 774–780. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Bartish, M.; Gonçalves, C.; Huang, F.; Smith-Voudouris, J.; Krisna, S.S.; Preston, S.E.J.; Emond, A.; Li, V.Z.; Duerr, C.U.; et al. The MNK1/2–EIF4E Axis Supports Immune Suppression and Metastasis in Postpartum Breast Cancer. Cancer Res. 2021, 81, 3876–3889. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, Y.; Zhang, S.; Qiu, L.; Zhang, Y.; Zhou, Y.; Han, J.; Xie, J. Translational Regulation by EIFs and RNA Modifications in Cancer. Genes 2022, 13, 2050. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-I.; Wang, C.-C.; Tai, T.-S.; Hwang, T.-Z.; Yang, C.-C.; Hsu, C.-M.; Su, Y.-C. EIF4E and 4EBP1 Are Prognostic Markers of Head and Neck Squamous Cell Carcinoma Recurrence after Definitive Surgery and Adjuvant Radiotherapy. PLoS ONE 2019, 14, e0225537. [Google Scholar] [CrossRef]
- Qi, N.-N.; Tian, S.; Li, X.; Wang, F.-L.; Liu, B. Up-Regulation of MicroRNA-496 Suppresses Proliferation, Invasion, Migration and in Vivo Tumorigenicity of Human Osteosarcoma Cells by Targeting EIF4E. Biochimie 2019, 163, 1–11. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, H.; Sun, M.; Yin, Z.; Qian, J. High Mobility Group Box 1-Mediated Autophagy Promotes Neuroblastoma Cell Chemoresistance. Oncol. Rep. 2015, 34, 2969–2976. [Google Scholar] [CrossRef]
- Meng, Q.; Zhao, J.; Liu, H.; Zhou, G.; Zhang, W.; Xu, X.; Zheng, M. HMGB1 Promotes Cellular Proliferation and Invasion, Suppresses Cellular Apoptosis in Osteosarcoma. Tumor Biol. 2014, 35, 12265–12274. [Google Scholar] [CrossRef]
- Liu, K.; Huang, J.; Ni, J.; Song, D.; Ding, M.; Wang, J.; Huang, X.; Li, W. MALAT1 Promotes Osteosarcoma Development by Regulation of HMGB1 via MiR-142–3p and MiR-129–5p. Cell Cycle 2017, 16, 578–587. [Google Scholar] [CrossRef]
- Huang, J.; Ni, J.; Liu, K.; Yu, Y.; Xie, M.; Kang, R.; Vernon, P.; Cao, L.; Tang, D. HMGB1 Promotes Drug Resistance in Osteosarcoma. Cancer Res. 2012, 72, 230–238. [Google Scholar] [CrossRef]
- Usman, R.M.; Razzaq, F.; Akbar, A.; Farooqui, A.A.; Iftikhar, A.; Latif, A.; Hassan, H.; Zhao, J.; Carew, J.S.; Nawrocki, S.T.; et al. Role and Mechanism of Autophagy-regulating Factors in Tumorigenesis and Drug Resistance. Asia-Pac. J. Clin. Oncol. 2021, 17, 193–208. [Google Scholar] [CrossRef]
- Liu, Y.-J.; Li, W.; Chang, F.; Liu, J.-N.; Lin, J.-X.; Chen, D.-X. MicroRNA-505 Is Downregulated in Human Osteosarcoma and Regulates Cell Proliferation, Migration and Invasion. Oncol. Rep. 2017, 39, 491–500. [Google Scholar] [CrossRef]
- Li, G.; Liu, F.; Miao, J.; Hu, Y. MiR-505 Inhibits Proliferation of Osteosarcoma via HMGB1. FEBS Open Bio 2020, 10, 1251–1260. [Google Scholar] [CrossRef]
- Lv, S.; Guan, M. MiRNA-1284, a Regulator of HMGB1, Inhibits Cell Proliferation and Migration in Osteosarcoma. Biosci. Rep. 2018, 38, BSR20171675. [Google Scholar] [CrossRef]
- Guo, S.; Bai, R.; Liu, W.; Zhao, A.; Zhao, Z.; Wang, Y.; Wang, Y.; Zhao, W.; Wang, W. MiR-22 Inhibits Osteosarcoma Cell Proliferation and Migration by Targeting HMGB1 and Inhibiting HMGB1-Mediated Autophagy. Tumor Biol. 2014, 35, 7025–7034. [Google Scholar] [CrossRef]
- Diao, Z.; Sun, T.; Zong, Y.; Lin, B.; Xia, Y. Identification of Plasma MicroRNA-22 as a Marker for the Diagnosis, Prognosis, and Chemosensitivity Prediction of Osteosarcoma. J. Int. Med. Res. 2020, 48, 030006052096781. [Google Scholar] [CrossRef]
- Zhu, S.; Wang, X.; Wang, J.; Xi, G.; Liu, Y. Downregulation of MiR-22 Contributes to Epithelial-Mesenchymal Transition in Osteosarcoma by Targeting Twist1. Front. Oncol. 2020, 10, 406. [Google Scholar] [CrossRef]
- Li, Y.; Liu, J.-J.; Zhou, J.-H.; Chen, R.; Cen, C.-Q. LncRNA HULC Induces the Progression of Osteosarcoma by Regulating the MiR-372-3p/HMGB1 Signalling Axis. Mol. Med. 2020, 26, 26. [Google Scholar] [CrossRef]
- Lou, P.; Ding, T.; Zhan, X. Long Noncoding RNA HNF1A-AS1 Regulates Osteosarcoma Advancement Through Modulating the MiR-32-5p/HMGB1 Axis. Cancer Biother. Radiopharm. 2021, 36, 371–381. [Google Scholar] [CrossRef]
- Villanueva, F.; Araya, H.; Briceño, P.; Varela, N.; Stevenson, A.; Jerez, S.; Tempio, F.; Chnaiderman, J.; Perez, C.; Villarroel, M.; et al. The Cancer-related Transcription Factor RUNX2 Modulates Expression and Secretion of the Matricellular Protein Osteopontin in Osteosarcoma Cells to Promote Adhesion to Endothelial Pulmonary Cells and Lung Metastasis. J. Cell. Physiol. 2019, 234, 13659–13679. [Google Scholar] [CrossRef]
- Vega, O.A.; Lucero, C.M.J.; Araya, H.F.; Jerez, S.; Tapia, J.C.; Antonelli, M.; Salazar-Onfray, F.; Las Heras, F.; Thaler, R.; Riester, S.M.; et al. Wnt/Β-Catenin Signaling Activates Expression of the Bone-Related Transcription Factor RUNX2 in Select Human Osteosarcoma Cell Types. J. Cell. Biochem. 2017, 118, 3662–3674. [Google Scholar] [CrossRef]
- Jia, F.; Zhang, Z.; Zhang, X. MicroRNA-338-3p Inhibits Tumor Growth and Metastasis in Osteosarcoma Cells by Targeting RUNX2/CDK4 and Inhibition of MAPK Pathway. J. Cell. Biochem. 2019, 120, 6420–6430. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Huang, L.; Liu, L.; Wang, L.; Lin, W.; Zhu, X.; Su, W.; Lv, C. Knockdown of MicroRNA-203 Reduces Cisplatin Chemo-Sensitivity to Osteosarcoma Cell Lines MG63 and U2OS in Vitro by Targeting RUNX2. J. Chemother. 2021, 33, 328–341. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Sun, W.; Deng, Z.; Zhu, X.; Hu, C.; Cai, L. MiR-302b Suppresses Osteosarcoma Cell Migration and Invasion by Targeting Runx2. Sci. Rep. 2017, 7, 13388. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Ma, J.; Wu, J. MiRNA-218 Inhibits Cell Proliferation, Migration and Invasion by Targeting Runt-Related Transcription Factor 2 (Runx2) in Human Osteosarcoma Cells. Regen. Ther. 2021, 18, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Jacques, C.; Tesfaye, R.; Lavaud, M.; Georges, S.; Baud’huin, M.; Lamoureux, F.; Ory, B. Implication of the P53-Related MiR-34c, -125b, and -203 in the Osteoblastic Differentiation and the Malignant Transformation of Bone Sarcomas. Cells 2020, 9, 810. [Google Scholar] [CrossRef]
- He, C.; Xiong, J.; Xu, X.; Lu, W.; Liu, L.; Xiao, D.; Wang, D. Functional Elucidation of MiR-34 in Osteosarcoma Cells and Primary Tumor Samples. Biochem. Biophys. Res. Commun. 2009, 388, 35–40. [Google Scholar] [CrossRef]
- Li, K.-W.; Wang, S.-H.; Wei, X.; Hou, Y.-Z.; Li, Z.-H. Mechanism of MiR-122-5p Regulating the Activation of PI3K-Akt-MTOR Signaling Pathway on the Cell Proliferation and Apoptosis of Osteosarcoma Cells through Targeting TP53 Gene. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 12655–12666. [Google Scholar] [CrossRef]
- Shi, X.; Wallis, A.M.; Gerard, R.D.; Voelker, K.A.; Grange, R.W.; DePinho, R.A.; Garry, M.G.; Garry, D.J. Foxk1 Promotes Cell Proliferation and Represses Myogenic Differentiation by Regulating Foxo4 and Mef2. J. Cell Sci. 2012, 125, 5329–5337. [Google Scholar] [CrossRef]
- Li, H.; Han, X.; Yang, S.; Wang, Y.; Dong, Y.; Tang, T. FOXP1 Drives Osteosarcoma Development by Repressing P21 and RB Transcription Downstream of P53. Oncogene 2021, 40, 2785–2802. [Google Scholar] [CrossRef]
- Chen, H.; Xiao, Z.; Yu, R.; Wang, Y.; Xu, R.; Zhu, X. MiR-181d-5p-FOXP1 Feedback Loop Modulates the Progression of Osteosarcoma. Biochem. Biophys. Res. Commun. 2018, 503, 1434–1441. [Google Scholar] [CrossRef]
- Peña-Blanco, A.; García-Sáez, A.J. Bax, Bak and beyond—Mitochondrial Performance in Apoptosis. FEBS J. 2018, 285, 416–431. [Google Scholar] [CrossRef]
- Lopez, A.; Reyna, D.E.; Gitego, N.; Kopp, F.; Zhou, H.; Miranda-Roman, M.A.; Nordstrøm, L.U.; Narayanagari, S.-R.; Chi, P.; Vilar, E.; et al. Co-Targeting of BAX and BCL-XL Proteins Broadly Overcomes Resistance to Apoptosis in Cancer. Nat. Commun. 2022, 13, 1199. [Google Scholar] [CrossRef]
- Veys, C.; Benmoussa, A.; Contentin, R.; Duchemin, A.; Brotin, E.; Lafont, J.E.; Saintigny, Y.; Poulain, L.; Denoyelle, C.; Demoor, M.; et al. Tumor Suppressive Role of MiR-342-5p in Human Chondrosarcoma Cells and 3D Organoids. Int. J. Mol. Sci. 2021, 22, 5590. [Google Scholar] [CrossRef]
- Zhang, H.; Cai, X.; Wang, Y.; Tang, H.; Tong, D.; Ji, F. MicroRNA-143, down-Regulated in Osteosarcoma, Promotes Apoptosis and Suppresses Tumorigenicity by Targeting Bcl-2. Oncol. Rep. 2010, 24, 1363–1369. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, J.; Quan, H.; Wang, J.; Liang, Y. MicroRNA-143 Expression Inhibits the Growth and the Invasion of Osteosarcoma. J. Orthop. Surg. Res. 2022, 17, 236. [Google Scholar] [CrossRef]
- Osaki, M.; Takeshita, F.; Sugimoto, Y.; Kosaka, N.; Yamamoto, Y.; Yoshioka, Y.; Kobayashi, E.; Yamada, T.; Kawai, A.; Inoue, T.; et al. MicroRNA-143 Regulates Human Osteosarcoma Metastasis by Regulating Matrix Metalloprotease-13 Expression. Mol. Ther. 2011, 19, 1123–1130. [Google Scholar] [CrossRef]
- Scherr, A.-L.; Mock, A.; Gdynia, G.; Schmitt, N.; Heilig, C.E.; Korell, F.; Rhadakrishnan, P.; Hoffmeister, P.; Metzeler, K.H.; Schulze-Osthoff, K.; et al. Identification of BCL-XL as Highly Active Survival Factor and Promising Therapeutic Target in Colorectal Cancer. Cell Death Dis. 2020, 11, 875. [Google Scholar] [CrossRef]
- Anderson, G.; Carbone, A.; Mazzoccoli, G. Tryptophan Metabolites and Aryl Hydrocarbon Receptor in Severe Acute Respiratory Syndrome, Coronavirus-2 (SARS-CoV-2) Pathophysiology. Int. J. Mol. Sci. 2021, 22, 1597. [Google Scholar] [CrossRef]
- Ji, F.; Zhang, H.; Wang, Y.; Li, M.; Xu, W.; Kang, Y.; Wang, Z.; Wang, Z.; Cheng, P.; Tong, D.; et al. MicroRNA-133a, Downregulated in Osteosarcoma, Suppresses Proliferation and Promotes Apoptosis by Targeting Bcl-XL and Mcl-1. Bone 2013, 56, 220–226. [Google Scholar] [CrossRef]
- Xia, P.; Gu, R.; Zhang, W.; Shao, L.; Li, F.; Wu, C.; Sun, Y. MicroRNA-377 Exerts a Potent Suppressive Role in Osteosarcoma through the Involvement of the Histone Acetyltransferase 1-mediated Wnt Axis. J. Cell. Physiol. 2019, 234, 22787–22798. [Google Scholar] [CrossRef]
- Ferrara, N.; Adamis, A.P. Ten Years of Anti-Vascular Endothelial Growth Factor Therapy. Nat. Rev. Drug Discov. 2016, 15, 385–403. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.-W.; Wu, T.-Y.; Yi, X.; Ren, W.-P.; Zhou, Z.; Sun, Y.; Zhang, C. Prognostic Significance of VEGF Expression in Osteosarcoma: A Meta-Analysis. Tumor Biol. 2014, 35, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Ren, T.; Huang, Y.; Guo, W. Apatinib Inhibits Migration and Invasion as Well as PD-L1 Expression in Osteosarcoma by Targeting STAT3. Biochem. Biophys. Res. Commun. 2018, 495, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Assi, T.; Watson, S.; Samra, B.; Rassy, E.; Le Cesne, A.; Italiano, A.; Mir, O. Targeting the VEGF Pathway in Osteosarcoma. Cells 2021, 10, 1240. [Google Scholar] [CrossRef]
- Qin, Y.; Zhang, B.; Ge, B.-J. MicroRNA-150-5p Inhibits Proliferation and Invasion of Osteosarcoma Cells by down-Regulating VEGFA. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 9265–9273. [Google Scholar] [CrossRef]
- Niu, J.; Sun, Y.; Guo, Q.; Niu, D.; Liu, B. MiR-1 Inhibits Cell Growth, Migration, and Invasion by Targeting VEGFA in Osteosarcoma Cells. Dis. Markers 2016, 2016, 7068986. [Google Scholar] [CrossRef]
- Ma, Z.; Li, K.; Chen, P.; Pan, Q.; Li, X.; Zhao, G. MiR-134, Mediated by IRF1, Suppresses Tumorigenesis and Progression by Targeting VEGFA and MYCN in Osteosarcoma. Anti-Cancer Agents Med. Chem. 2020, 20, 1197–1208. [Google Scholar] [CrossRef]
- Zheng, C.; Tang, F.; Min, L.; Hornicek, F.; Duan, Z.; Tu, C. PTEN in Osteosarcoma: Recent Advances and the Therapeutic Potential. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2020, 1874, 188405. [Google Scholar] [CrossRef]
- Chen, C.; Guo, Y.; Huang, Q.; Wang, B.; Wang, W.; Niu, J.; Lou, J.; Xu, J.; Ren, T.; Huang, Y.; et al. PI3K Inhibitor Impairs Tumor Progression and Enhances Sensitivity to Anlotinib in Anlotinib-Resistant Osteosarcoma. Cancer Lett. 2022, 536, 215660. [Google Scholar] [CrossRef]
- Yuan, H.; Gao, Y. MicroRNA-1908 Is Upregulated in Human Osteosarcoma and Regulates Cell Proliferation and Migration by Repressing PTEN Expression. Oncol. Rep. 2015, 34, 2706–2714. [Google Scholar] [CrossRef]
- Zhu, J.; Liu, F.; Wu, Q.; Liu, X. MiR-221 Increases Osteosarcoma Cell Proliferation, Invasion and Migration Partly through the Downregulation of PTEN. Int. J. Mol. Med. 2015, 36, 1377–1383. [Google Scholar] [CrossRef]
- Zheng, M.; Wu, Y. Piceatannol Suppresses Proliferation and Induces Apoptosis by Regulation of the MicroRNA-21/Phosphatase and Tensin Homolog/Protein Kinase B Signaling Pathway in Osteosarcoma Cells. Mol. Med. Rep. 2020, 22, 3985–3993. [Google Scholar] [CrossRef]
- Vanas, V.; Haigl, B.; Stockhammer, V.; Sutterlüty-Fall, H. MicroRNA-21 Increases Proliferation and Cisplatin Sensitivity of Osteosarcoma-Derived Cells. PLoS ONE 2016, 11, e0161023. [Google Scholar] [CrossRef]
- Ming He, J.; Liu, P.Y.; Wang, J. MicroRNA-17-5p Regulates the Growth, Migration and Invasion of the Human Osteosarcoma Cells by Modulating the Expression of PTEN. J. Buon. 2020, 25, 1028–1034. [Google Scholar]
- Kumar, R.; Fuchs, B. Hedgehog Signaling Inhibitors as Anti-Cancer Agents in Osteosarcoma. Cancers 2015, 7, 784–794. [Google Scholar] [CrossRef]
- Yi, T.; Yi, X.; Lv, X.; Liang, R.; Zan, N.; Su, X. MiR-212 Promotes Proliferation and Inhibits Apoptosis of Osteosarcoma Cells via Regulating Hedgehog Signaling Pathway. J. Buon 2020, 25, 2086–2091. [Google Scholar]
- Zhang, Z.; Zhang, W.; Maoa, J.; Xua, Z.; Fan, M. MiR-186-5p Functions as a Tumor Suppressor in Human Osteosarcoma by Targeting FOXK1. Cell Physiol. Biochem. 2019, 52, 553–564. [Google Scholar] [CrossRef]
- Zhang, Y.; Ni, J.; Zhou, G.; Yuan, J.; Ren, W.; Shan, Y.; Tang, W.; Yu, L.; Zhao, S. Cloning, Expression and Characterization of the Human NOB1 Gene. Mol. Biol. Rep. 2005, 32, 185–189. [Google Scholar] [CrossRef]
- Gao, X.; Wang, J.; Bai, W.; Ji, W.; Wang, L. NOB1 Silencing Inhibits the Growth and Metastasis of Laryngeal Cancer Cells through the Regulation of JNK Signaling Pathway. Oncol. Rep. 2016, 35, 3313–3320. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, F.; Wang, L.; Zhang, Q. MiR-363 Suppresses Cell Migration, Invasion, and Epithelial-Mesenchymal Transition of Osteosarcoma by Binding to NOB1. World J. Surg. Oncol. 2020, 18, 83. [Google Scholar] [CrossRef]
- Liu, K.; Zhang, Y.; Liu, L.; Yuan, Q. MALAT1 Promotes Proliferation, Migration, and Invasion of MG63 Cells by Upregulation of TGIF2 via Negatively Regulating MiR-129. OncoTargets Ther. 2018, 11, 8729–8740. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Diaz, P.C.; Hsiao, T.-H.; Zou, Y.; Sugalski, A.J.; Heim-Hall, J.; Chen, Y.; Langevin, A.-M.; Hung, J.Y. In Silico Functional Analyses and Discovery of Survivalassociated MicroRNA Signatures in Pediatric Osteosarcoma. Oncoscience 2014, 1, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yi, X. MiR-541 Serves as a Prognostic Biomarker of Osteosarcoma and Its Regulatory Effect on Tumor Cell Proliferation, Migration and Invasion by Targeting TGIF2. Diagn. Pathol. 2020, 15, 96. [Google Scholar] [CrossRef]
- Xi, L.; Zhang, Y.; Kong, S.; Liang, W. MiR-34 Inhibits Growth and Promotes Apoptosis of Osteosarcoma in Nude Mice through Targetly Regulating TGIF2 Expression. Biosci. Rep. 2018, 38, BSR20180078. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ge, J.; Ma, T.; Zheng, Y.; Lv, S.; Li, Y.; Liu, S. Promoter Hypermethylation of the Cysteine Protease RECK May Cause Metastasis of Osteosarcoma. Tumor Biol. 2015, 36, 9511–9516. [Google Scholar] [CrossRef] [PubMed]
- Ziyan, W.; Shuhua, Y.; Xiufang, W.; Xiaoyun, L. MicroRNA-21 Is Involved in Osteosarcoma Cell Invasion and Migration. Med. Oncol. 2011, 28, 1469–1474. [Google Scholar] [CrossRef]
- Kang, H.-G.; Kim, H.-S.; Kim, K.-J.; Oh, J.H.; Lee, M.-R.; Seol, S.M.; Han, I. RECK Expression in Osteosarcoma: Correlation with Matrix Metalloproteinases Activation and Tumor Invasiveness. J. Orthop. Res. 2007, 25, 696–702. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, Z.; Wei, H.; Wu, S.; Wang, X.; Xiao, J. Promotion of Tumour Proliferation, Migration and Invasion by MiR-92b in Targeting RECK in Osteosarcoma. Clin. Sci. 2016, 130, 921–930. [Google Scholar] [CrossRef]
- Memon, A.; Lee, W. KLF10 as a Tumor Suppressor Gene and Its TGF-β Signaling. Cancers 2018, 10, 161. [Google Scholar] [CrossRef]
- Wang, L.; Du, Z.-G.; Huang, H.; Li, F.-S.; Li, G.-S.; Xu, S.-N. Circ-0003998 Promotes Cell Proliferative Ability and Invasiveness by Binding to MiR-197-3p in Osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10638–10646. [Google Scholar] [CrossRef]
- Huang, J.; Xu, Y.; Lin, F. The Inhibition of MicroRNA-326 by SP1/HDAC1 Contributes to Proliferation and Metastasis of Osteosarcoma through Promoting SMO Expression. J. Cell. Mol. Med. 2020, 24, 10876–10888. [Google Scholar] [CrossRef]
- Zeng, H.; Zhang, J.; Du, Y.; Wang, J.; Ren, Y.; Li, M.; Li, H.; Cai, Z.; Chu, Q.; Yang, C. Crosstalk between ATF4 and MTA1/HDAC1 Promotes Osteosarcoma Progression. Oncotarget 2016, 7, 7329–7342. [Google Scholar] [CrossRef]
- Cao, L.; Wang, J.; Wang, Q. MiR-326 Is a Diagnostic Biomarker and Regulates Cell Survival and Apoptosis by Targeting Bcl-2 in Osteosarcoma. Biomed. Pharmacother. 2016, 84, 828–835. [Google Scholar] [CrossRef]
- Liu, T.; Hou, L.; Zhao, Y.; Huang, Y. Epigenetic Silencing of HDAC1 by MiR-449a Upregulates Runx2 and Promotes Osteoblast Differentiation. Int. J. Mol. Med. 2015, 35, 238–246. [Google Scholar] [CrossRef]
- Lu, D.-G.; Tang, Q.-L.; Wei, J.-H.; He, F.-Y.; Lu, L.; Tang, Y.-J. Targeting EZH2 by MicroRNA-449a Inhibits Osteosarcoma Cell Proliferation, Invasion and Migration via Regulation of PI3K/AKT Signaling Pathway and Epithelial-Mesenchymal Transition. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1656–1665. [Google Scholar] [CrossRef]
- Kushwaha, P.; Khedgikar, V.; Sharma, D.; Yuen, T.; Gautam, J.; Ahmad, N.; Karvande, A.; Mishra, P.R.; Trivedi, P.K.; Sun, L.; et al. MicroRNA 874-3p Exerts Skeletal Anabolic Effects Epigenetically during Weaning by Suppressing Hdac1 Expression. J. Biol. Chem. 2016, 291, 3959–3966. [Google Scholar] [CrossRef]
- Wen, Y.; Li, B.; He, M.; Teng, S.; Sun, Y.; Wang, G. CircHIPK3 Promotes Proliferation and Migration and Invasion via Regulation of MiR-637/HDAC4 Signaling in Osteosarcoma Cells. Oncol. Rep. 2020, 45, 169–179. [Google Scholar] [CrossRef]
- Song, B.; Wang, Y.; Titmus, M.A.; Botchkina, G.; Formentini, A.; Kornmann, M.; Ju, J. Molecular Mechanism of Chemoresistance by MiR-215 in Osteosarcoma and Colon Cancer Cells. Mol. Cancer 2010, 9, 96. [Google Scholar] [CrossRef]
- Cui, X.; Zhao, C.; Yao, X.; Qian, B.; Su, C.; Ren, Y.; Yao, Z.; Gao, X.; Yang, J. SND1 Acts as an Anti-Apoptotic Factor via Regulating the Expression of LncRNA UCA1 in Hepatocellular Carcinoma. RNA Biol. 2018, 15, 1364–1375. [Google Scholar] [CrossRef]
- Bilbao-Aldaiturriaga, N.; Gutierrez-Camino, A.; Martin-Guerrero, I.; Pombar-Gomez, M.; Zalacain-Diez, M.; Patiño-Garcia, A.; Lopez-Lopez, E.; Garcia-Orad, A. Polymorphisms in MiRNA Processing Genes and Their Role in Osteosarcoma Risk: MiRNA-Related SNPs and Osteosarcoma Risk. Pediatr. Blood Cancer 2015, 62, 766–769. [Google Scholar] [CrossRef]
- Huang, Y.-Z.; Zhang, J.; Shen, J.-J.; Zhao, T.-X.; Xu, Y.-J. MiRNA-296-5p Functions as a Potential Tumor Suppressor in Human Osteosarcoma by Targeting SND1. Chin. Med. J. 2021, 134, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Wiehagen, K.R.; Corbo-Rodgers, E.; Li, S.; Staub, E.S.; Hunter, C.A.; Morrisey, E.E.; Maltzman, J.S. Foxp4 Is Dispensable for T Cell Development, but Required for Robust Recall Responses. PLoS ONE 2012, 7, e42273. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Ding, H.; He, E.; Chen, J.; Li, M. Up-Regulation of MicroRNA-491-5p Suppresses Cell Proliferation and Promotes Apoptosis by Targeting FOXP4 in Human Osteosarcoma. Cell Prolif. 2017, 50, e12308. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Qin, D.; Zhao, C.; Chai, L.; Yu, Z.; Wang, W.; Tong, L.; Lv, L.; Wang, Y.; Rehwinkel, J.; et al. Redox Homeostasis Maintained by GPX4 Facilitates STING Activation. Nat. Immunol. 2020, 21, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Chen, L.; Wang, C.; Zhang, L.; Xu, W. MicroRNA-1287-5p Promotes Ferroptosis of Osteosarcoma Cells through Inhibiting GPX4. Free Radic. Res. 2021, 55, 1119–1129. [Google Scholar] [CrossRef]
- Corre, I.; Verrecchia, F.; Crenn, V.; Redini, F.; Trichet, V. The Osteosarcoma Microenvironment: A Complex but Targetable Ecosystem. Cells 2020, 9, 976. [Google Scholar] [CrossRef]
- Cascini, C.; Chiodoni, C. The Immune Landscape of Osteosarcoma: Implications for Prognosis and Treatment Response. Cells 2021, 10, 1668. [Google Scholar] [CrossRef]
- Luo, Z.-W.; Liu, P.-P.; Wang, Z.-X.; Chen, C.-Y.; Xie, H. Macrophages in Osteosarcoma Immune Microenvironment: Implications for Immunotherapy. Front. Oncol. 2020, 10, 586580. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, T.; Gong, H.; Jiang, R.; Zhou, W.; Sun, H.; Huang, R.; Wang, Y.; Wu, Z.; Xu, W.; et al. A Novel Molecular Classification Method for Osteosarcoma Based on Tumor Cell Differentiation Trajectories. Bone Res. 2023, 11, 1. [Google Scholar] [CrossRef]
- Huang, R.; Wang, X.; Yin, X.; Zhou, Y.; Sun, J.; Yin, Z.; Zhu, Z. Combining Bulk RNA-Sequencing and Single-Cell RNA-Sequencing Data to Reveal the Immune Microenvironment and Metabolic Pattern of Osteosarcoma. Front. Genet. 2022, 13, 976990. [Google Scholar] [CrossRef]
- Pietrovito, L.; Leo, A.; Gori, V.; Lulli, M.; Parri, M.; Becherucci, V.; Piccini, L.; Bambi, F.; Taddei, M.L.; Chiarugi, P. Bone Marrow-derived Mesenchymal Stem Cells Promote Invasiveness and Transendothelial Migration of Osteosarcoma Cells via a Mesenchymal to Amoeboid Transition. Mol. Oncol. 2018, 12, 659–676. [Google Scholar] [CrossRef]
- Wang, Y.; Chu, Y.; Yue, B.; Ma, X.; Zhang, G.; Xiang, H.; Liu, Y.; Wang, T.; Wu, X.; Chen, B. Adipose-Derived Mesenchymal Stem Cells Promote Osteosarcoma Proliferation and Metastasis by Activating the STAT3 Pathway. Oncotarget 2017, 8, 23803–23816. [Google Scholar] [CrossRef]
- Wang, Y.; Chu, Y.; Li, K.; Zhang, G.; Guo, Z.; Wu, X.; Qiu, C.; Li, Y.; Wan, X.; Sui, J.; et al. Exosomes Secreted by Adipose-Derived Mesenchymal Stem Cells Foster Metastasis and Osteosarcoma Proliferation by Increasing COLGALT2 Expression. Front. Cell Dev. Biol. 2020, 8, 353. [Google Scholar] [CrossRef]
- Ullah, M.; Liu, D.D.; Thakor, A.S. Mesenchymal Stromal Cell Homing: Mechanisms and Strategies for Improvement. IScience 2019, 15, 421–438. [Google Scholar] [CrossRef]
- Costa, A.; Kieffer, Y.; Scholer-Dahirel, A.; Pelon, F.; Bourachot, B.; Cardon, M.; Sirven, P.; Magagna, I.; Fuhrmann, L.; Bernard, C.; et al. Fibroblast Heterogeneity and Immunosuppressive Environment in Human Breast Cancer. Cancer Cell 2018, 33, 463–479. [Google Scholar] [CrossRef]
- Cortini, M.; Massa, A.; Avnet, S.; Bonuccelli, G.; Baldini, N. Tumor-Activated Mesenchymal Stromal Cells Promote Osteosarcoma Stemness and Migratory Potential via IL-6 Secretion. PLoS ONE 2016, 11, e0166500. [Google Scholar] [CrossRef]
- Jiang, K.; Li, J.; Wang, L.; Zhang, Q.; Ge, J.; Guo, Y.; Wang, B.; Huang, Y.; Yang, T.; Hao, D.; et al. SDF-1/CXCR4 axis facilitates myelpid-derived suppressor cells accumulation in osteosarcoma microenvironment and blunts the response to anti-PD-1 therapy. Int. Immunopharmacol. 2019, 75, 105818. [Google Scholar] [CrossRef]
- Ling, Z.; Yang, C.; Tan, J.; Dou, C.; Chen, Y. Beyond immunosupressive effects: Dual roles of mieloid-derived supressor cells in bone-related diseases. Cell. Mol. Life Sci. 2021, 78, 7161–7183. [Google Scholar] [CrossRef]
- Cipriani, P.; Ruscitti, P.; Di Benedetto, P.; Carubbi, F.; Liakouli, V.; Berardicurti, O.; Ciccia, F.; Triolo, G.; Giacomelli, R. Mesenchymal Stromal Cells and Rheumatic Diseases: New Tools from Pathogenesis to Regenerative Therapies. Cytotherapy 2015, 17, 832–849. [Google Scholar] [CrossRef]
- Sarhadi, V.K.; Daddali, R.; Seppänen-Kaijansinkko, R. Mesenchymal Stem Cells and Extracellular Vesicles in Osteosarcoma Pathogenesis and Therapy. Int. J. Mol. Sci. 2021, 22, 11035. [Google Scholar] [CrossRef]
- Batlle, R.; Andrés, E.; Gonzalez, L.; Llonch, E.; Igea, A.; Gutierrez-Prat, N.; Berenguer-Llergo, A.; Nebreda, A.R. Regulation of Tumor Angiogenesis and Mesenchymal–Endothelial Transition by P38α through TGF-β and JNK Signaling. Nat. Commun. 2019, 10, 3071. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Zhu, B.; Huang, G.; Zeng, Q.; Wang, C. Microvesicles Derived from Human Bone Marrow Mesenchymal Stem Cells Promote U2OS Cell Growth under Hypoxia: The Role of PI3K/AKT and HIF-1α. Hum. Cell 2019, 32, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Vallabhaneni, K.C.; Hassler, M.-Y.; Abraham, A.; Whitt, J.; Mo, Y.-Y.; Atfi, A.; Pochampally, R. Mesenchymal Stem/Stromal Cells under Stress Increase Osteosarcoma Migration and Apoptosis Resistance via Extracellular Vesicle Mediated Communication. PLoS ONE 2016, 11, e0166027. [Google Scholar] [CrossRef] [PubMed]
- Benslimane-Ahmim, Z.; Pereira, J.; Lokajczyk, A.; Dizier, B.; Galy-Fauroux, I.; Fischer, A.-M.; Heymann, D.; Boisson-Vidal, C. Osteoprotegerin Regulates Cancer Cell Migration through SDF-1/CXCR4 Axis and Promotes Tumour Development by Increasing Neovascularization. Cancer Lett. 2017, 395, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Tang, H.; Zhang, Y.; Zhang, Z.; Huang, P.; Zhu, J. Bone Marrow-derived Mesenchymal Stem Cell-derived Exosomal MicroRNA-208a Promotes Osteosarcoma Cell Proliferation, Migration, and Invasion. J. Cell. Physiol. 2020, 235, 4734–4745. [Google Scholar] [CrossRef]
- Xu, Z.; Zhou, X.; Wu, J.; Cui, X.; Wang, M.; Wang, X.; Gao, Z. Mesenchymal Stem Cell-Derived Exosomes Carrying MicroRNA-150 Suppresses the Proliferation and Migration of Osteosarcoma Cells via Targeting IGF2BP1. Transl. Cancer Res. TCR 2020, 9, 5323–5335. [Google Scholar] [CrossRef]
- Avnet, S.; Di Pompo, G.; Chano, T.; Errani, C.; Ibrahim-Hashim, A.; Gillies, R.J.; Donati, D.M.; Baldini, N. Cancer-Associated Mesenchymal Stroma Fosters the Stemness of Osteosarcoma Cells in Response to Intratumoral Acidosis via NF-ΚB Activation: Tumor Acidic Microenvironment Fosters Osteosarcoma Stemness via Mesenchymal Stroma. Int. J. Cancer 2017, 140, 1331–1345. [Google Scholar] [CrossRef]
- Zhu, T.; Han, J.; Yang, L.; Cai, Z.; Sun, W.; Hua, Y.; Xu, J. Immune Microenvironment in Osteosarcoma: Components, Therapeutic Strategies and Clinical Applications. Front. Immunol. 2022, 13, 907550. [Google Scholar] [CrossRef]
- Melzer, C.; von der Ohe, J.; Lehnert, H.; Ungefroren, H.; Hass, R. Cancer Stem Cell Niche Models and Contribution by Mesenchymal Stroma/Stem Cells. Mol. Cancer 2017, 16, 28. [Google Scholar] [CrossRef]
- Tian, B.; Du, X.; Zheng, S.; Zhang, Y. The Role of Tumor Microenvironment in Regulating the Plasticity of Osteosarcoma Cells. Int. J. Mol. Sci. 2022, 23, 16155. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, H.; Zheng, J.; Yu, P.; Xu, L.; Jiang, P.; Gao, J.; Wang, H.; Zhang, Y. Transforming Growth Factor Β1 Signal Is Crucial for Dedifferentiation of Cancer Cells to Cancer Stem Cells in Osteosarcoma. Stem Cells 2013, 31, 433–446. [Google Scholar] [CrossRef]
- Galbo, P.M.; Zang, X.; Zheng, D. Molecular Features of Cancer-Associated Fibroblast Subtypes and Their Implication on Cancer Pathogenesis, Prognosis, and Immunotherapy Resistance. Clin. Cancer Res. 2021, 27, 2636–2647. [Google Scholar] [CrossRef]
- Labernadie, A.; Kato, T.; Brugués, A.; Serra-Picamal, X.; Derzsi, S.; Arwert, E.; Weston, A.; González-Tarragó, V.; Elosegui-Artola, A.; Albertazzi, L.; et al. A Mechanically Active Heterotypic E-Cadherin/N-Cadherin Adhesion Enables Fibroblasts to Drive Cancer Cell Invasion. Nat. Cell Biol. 2017, 19, 224–237. [Google Scholar] [CrossRef]
- García-Silva, S.; Peinado, H. Melanosomes Foster a Tumour Niche by Activating CAFs. Nat. Cell Biol. 2016, 18, 911–913. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, X.; Sun, W.; Yue, S.; Yang, J.; Li, J.; Ma, B.; Wang, J.; Yang, X.; Pu, M.; et al. Loss of Exosomal MiR-320a from Cancer-Associated Fibroblasts Contributes to HCC Proliferation and Metastasis. Cancer Lett. 2017, 397, 33–42. [Google Scholar] [CrossRef]
- Liu, T.; Han, C.; Wang, S.; Fang, P.; Ma, Z.; Xu, L.; Yin, R. Cancer-Associated Fibroblasts: An Emerging Target of Anti-Cancer Immunotherapy. J. Hematol. Oncol. 2019, 12, 86. [Google Scholar] [CrossRef]
- Zhu, H.; Guo, S.; Zhang, Y.; Yin, J.; Yin, W.; Tao, S.; Wang, Y.; Zhang, C. Proton-Sensing GPCR-YAP Signalling Promotes Cancer-Associated Fibroblast Activation of Mesenchymal Stem Cells. Int. J. Biol. Sci. 2016, 12, 389–396. [Google Scholar] [CrossRef]
- Yang, L.; Achreja, A.; Yeung, T.-L.; Mangala, L.S.; Jiang, D.; Han, C.; Baddour, J.; Marini, J.C.; Ni, J.; Nakahara, R.; et al. Targeting Stromal Glutamine Synthetase in Tumors Disrupts Tumor Microenvironment-Regulated Cancer Cell Growth. Cell Metab. 2016, 24, 685–700. [Google Scholar] [CrossRef]
- Nurmik, M.; Ullmann, P.; Rodriguez, F.; Haan, S.; Letellier, E. In Search of Definitions: Cancer-associated Fibroblasts and Their Markers. Int. J. Cancer 2020, 146, 895–905. [Google Scholar] [CrossRef]
- Wang, J.-W.; Wu, X.-F.; Gu, X.-J.; Jiang, X.-H. Exosomal MiR-1228 From Cancer-Associated Fibroblasts Promotes Cell Migration and Invasion of Osteosarcoma by Directly Targeting SCAI. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2019, 27, 979–986. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, D.; Yang, Q.; Lv, X.; Huang, W.; Zhou, Z.; Wang, Y.; Zhang, Z.; Yuan, T.; Ding, X.; et al. Single-Cell RNA Landscape of Intratumoral Heterogeneity and Immunosuppressive Microenvironment in Advanced Osteosarcoma. Nat. Commun. 2020, 11, 6322. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Feng, W.; Dai, Y.; Bao, M.; Yuan, Z.; He, M.; Qin, Z.; Liao, S.; He, J.; Huang, Q.; et al. Single-Cell Transcriptomics Reveals the Complexity of the Tumor Microenvironment of Treatment-Naive Osteosarcoma. Front. Oncol. 2021, 11, 709210. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Huang, K.; Guo, W.; Zhou, C.; Wang, G.; Zhao, Q. Conditioned Medium of the Osteosarcoma Cell Line U2OS Induces HBMSCs to Exhibit Characteristics of Carcinoma-Associated Fibroblasts via Activation of IL-6/STAT3 Signalling. J. Biochem. 2020, 168, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-M.; Wang, W.; Qiu, E.-D. Osteosarcoma Cells Induce Differentiation of Mesenchymal Stem Cells into Cancer Associated Fibroblasts through Notch and Akt Signaling Pathway. Int. J. Clin. Exp. Pathol. 2017, 10, 8479–8486. [Google Scholar] [PubMed]
- Murdocca, M.; De Masi, C.; Pucci, S.; Mango, R.; Novelli, G.; Di Natale, C.; Sangiuolo, F. LOX-1 and Cancer: An Indissoluble Liaison. Cancer Gene Ther. 2021, 28, 1088–1098. [Google Scholar] [CrossRef]
- Wen, B.; Xu, L.-Y.; Li, E.-M. LOXL2 in Cancer: Regulation, Downstream Effectors and Novel Roles. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2020, 1874, 188435. [Google Scholar] [CrossRef]
- Huang, X.; Wang, L.; Guo, H.; Zhang, W.; Shao, Z. Single-Cell Transcriptomics Reveals the Regulative Roles of Cancer Associated Fibroblasts in Tumor Immune Microenvironment of Recurrent Osteosarcoma. Theranostics 2022, 12, 5877–5887. [Google Scholar] [CrossRef]
- Kim, H.-S.; Kim, H.J.; Lee, M.R.; Han, I. EMMPRIN Expression Is Associated with Metastatic Progression in Osteosarcoma. BMC Cancer 2021, 21, 1059. [Google Scholar] [CrossRef]
- Zeng, C.; Wen, M.; Liu, X. Fibroblast Activation Protein in Osteosarcoma Cells Promotes Angiogenesis via AKT and ERK Signaling Pathways. Oncol. Lett. 2018, 15, 6029–6035. [Google Scholar] [CrossRef]
- Chen, Z.; Yang, H.; Zhang, Q.; Hu, Q.; Zhao, Z. Chelerythrine Inhibits Stemness of Cancer Stem-Like Cells of Osteosarcoma and PI3K/AKT/mTOR Signal. J. Oncol. 2022, 2022, 6435431. [Google Scholar] [CrossRef]
- Nallasamy, P.; Nimmakayala, R.K.; Parte, S.; Are, A.C.; Batra, S.K.; Ponnusamy, M.P. Tumor Microenvironment Enriches the Stemness Features: The Architectural Event of Therapy Resistance and Metastasis. Mol. Cancer 2022, 21, 225. [Google Scholar] [CrossRef]
- Xu, A.; Qian, C.; Lin, J.; Yu, W.; Jin, J.; Liu, B.; Tao, H. Cell Differentiation Trajectory-Associated Molecular Classification of Osteosarcoma. Genes 2021, 12, 1685. [Google Scholar] [CrossRef]
- Brown, H.K.; Tellez-Gabriel, M.; Heymann, D. Cancer Stem Cells in Osteosarcoma. Cancer Lett. 2017, 386, 189–195. [Google Scholar] [CrossRef]
- Salerno, M.; Avnet, S.; Bonuccelli, G.; Eramo, A.; De Maria, R.; Gambarotti, M.; Gamberi, G.; Baldini, N. Sphere-Forming Cell Subsets with Cancer Stem Cell Properties in Human Musculoskeletal Sarcomas. Int. J. Oncol. 2013, 43, 95–102. [Google Scholar] [CrossRef]
- Gibbs, C.P.; Kukekov, V.G.; Reith, J.D.; Tchigrinova, O.; Suslov, O.N.; Scott, E.W.; Ghivizzani, S.C.; Ignatova, T.N.; Steindler, D.A. Stem-Like Cells in Bone Sarcomas: Implications for Tumorigenesis. Neoplasia 2005, 7, 967–976. [Google Scholar] [CrossRef]
- Jubelin, C.; Muñoz-Garcia, J.; Cochonneau, D.; Moranton, E.; Heymann, M.-F.; Heymann, D. Biological Evidence of Cancer Stem-like Cells and Recurrent Disease in Osteosarcoma. Cancer Drug Resist. 2022, 5, 184. [Google Scholar] [CrossRef]
- Hammad, M.; Cornejo, Y.R.; Batalla-Covello, J.; Majid, A.A.; Burke, C.; Liu, Z.; Yuan, Y.-C.; Li, M.; Dellinger, T.H.; Lu, J.; et al. Neural Stem Cells Improve the Delivery of Oncolytic Chimeric Orthopoxvirus in a Metastatic Ovarian Cancer Model. Mol. Ther.-Oncolytics 2020, 18, 326–334. [Google Scholar] [CrossRef]
- Pennati, A.; Riggio, E.; Marano, G.; Biganzoli, E. Autologous Fat Grafting after Sarcoma Surgery: Evaluation of Oncological Safety. J. Plast. Reconstr. Aesthetic Surg. 2018, 71, 1723–1729. [Google Scholar] [CrossRef]
- Honoki, K. Possible Involvement of Stem-like Populations with Elevated ALDH1 in Sarcomas for Chemotherapeutic Drug Resistance. Oncol. Rep. 2010, 24, 501–505. [Google Scholar] [CrossRef]
- Tian, J.; Li, X.; Si, M.; Liu, T.; Li, J. CD271+ Osteosarcoma Cells Display Stem-Like Properties. PLoS ONE 2014, 9, e98549. [Google Scholar] [CrossRef]
- Sun, D.-X.; Liao, G.-J.; Liu, K.-G.; Jian, H. Endosialin-Expressing Bone Sarcoma Stem-like Cells Are Highly Tumor-Initiating and Invasive. Mol. Med. Rep. 2015, 12, 5665–5670. [Google Scholar] [CrossRef]
- Oshimori, N.; Guo, Y.; Taniguchi, S. An Emerging Role for Cellular Crosstalk in the Cancer Stem Cell Niche. J. Pathol. 2021, 254, 384–394. [Google Scholar] [CrossRef] [PubMed]
- Brown, H.K.; Tellez-Gabriel, M.; Cartron, P.-F.; Vallette, F.M.; Heymann, M.-F.; Heymann, D. Characterization of Circulating Tumor Cells as a Reflection of the Tumor Heterogeneity: Myth or Reality? Drug Discov. Today 2019, 24, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Vallette, F.M.; Olivier, C.; Lézot, F.; Oliver, L.; Cochonneau, D.; Lalier, L.; Cartron, P.-F.; Heymann, D. Dormant, Quiescent, Tolerant and Persister Cells: Four Synonyms for the Same Target in Cancer. Biochem. Pharmacol. 2019, 162, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabières, C. Tumour Microenvironment: Informing on Minimal Residual Disease in Solid Tumours. Nat. Rev. Clin. Oncol. 2017, 14, 325–326. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, A.S.; Agarwal, N.; Wood, B.M.; Porretta, C.; Ruiz, B.; Pochampally, R.R.; Iwakuma, T. CD117 and Stro-1 Identify Osteosarcoma Tumor-Initiating Cells Associated with Metastasis and Drug Resistance. Cancer Res. 2010, 70, 4602–4612. [Google Scholar] [CrossRef]
- Brune, J.C.; Tormin, A.; Johansson, M.C.; Rissler, P.; Brosjö, O.; Löfvenberg, R.; von Steyern, F.V.; Mertens, F.; Rydholm, A.; Scheding, S. Mesenchymal Stromal Cells from Primary Osteosarcoma Are Non-Malignant and Strikingly Similar to Their Bone Marrow Counterparts. Int. J. Cancer 2011, 129, 319–330. [Google Scholar] [CrossRef]
- Mohseny, A.B.; Szuhai, K.; Romeo, S.; Buddingh, E.P.; Briaire-de Bruijn, I.; de Jong, D.; van Pel, M.; Cleton-Jansen, A.-M.; Hogendoorn, P.C. Osteosarcoma Originates from Mesenchymal Stem Cells in Consequence of Aneuploidization and Genomic Loss of Cdkn2: Mesenchymal Stem Cells-Based Model to Study Osteosarcoma. J. Pathol. 2009, 219, 294–305. [Google Scholar] [CrossRef]
- Wang, J.-Y.; Wu, P.-K.; Chen, P.C.-H.; Lee, C.-W.; Chen, W.-M.; Hung, S.-C. Generation of Osteosarcomas from a Combination of Rb Silencing and C-Myc Overexpression in Human Mesenchymal Stem Cells. Stem Cells Transl. Med. 2017, 6, 512–526. [Google Scholar] [CrossRef]
- Gambera, S.; Abarrategi, A.; Rodríguez-Milla, M.A.; Mulero, F.; Menéndez, S.T.; Rodriguez, R.; Navarro, S.; García-Castro, J. Role of Activator Protein-1 Complex on the Phenotype of Human Osteosarcomas Generated from Mesenchymal Stem Cells. Stem Cells 2018, 36, 1487–1500. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, S.; Zhao, R.C. The Roles of Mesenchymal Stem Cells in Tumor Inflammatory Microenvironment. J. Hematol. Oncol. 2014, 7, 14. [Google Scholar] [CrossRef]
- Kundu, B.; Bastos, A.R.F.; Brancato, V.; Cerqueira, M.T.; Oliveira, J.M.; Correlo, V.M.; Reis, R.L.; Kundu, S.C. Mechanical Property of Hydrogels and the Presence of Adipose Stem Cells in Tumor Stroma Affect Spheroid Formation in the 3D Osteosarcoma Model. ACS Appl. Mater. Interfaces 2019, 11, 14548–14559. [Google Scholar] [CrossRef]
- Aanstoos, M.E.; Regan, D.P.; Rose, R.J.; Chubb, L.S.; Ehrhart, N.P. Do Mesenchymal Stromal Cells Influence Microscopic Residual or Metastatic Osteosarcoma in a Murine Model? Clin. Orthop. Relat. Res. 2016, 474, 707–715. [Google Scholar] [CrossRef]
- Liu, J.; Yang, L.; Fu, Q.; Liu, S. Emerging Roles and Potential Biological Value of CircRNA in Osteosarcoma. Front. Oncol. 2020, 10, 552236. [Google Scholar] [CrossRef]
- Du, L.; Han, X.; Tu, B.; Wang, M.; Qiao, H.; Zhang, S.; Fan, Q.; Tang, T. CXCR1/Akt Signaling Activation Induced by Mesenchymal Stem Cell-Derived IL-8 Promotes Osteosarcoma Cell Anoikis Resistance and Pulmonary Metastasis. Cell Death Dis. 2018, 9, 714. [Google Scholar] [CrossRef]
- Gauthaman, K.; Yee, F.C.; Cheyyatraivendran, S.; Biswas, A.; Choolani, M.; Bongso, A. Human Umbilical Cord Wharton’s Jelly Stem Cell (HWJSC) Extracts Inhibit Cancer Cell Growth in Vitro. J. Cell Biochem. 2012, 113, 2027–2039. [Google Scholar] [CrossRef]
- Sanchez, C.; Oskowitz, A.; Pochampally, R.R. Epigenetic Reprogramming of IGF1 and Leptin Genes by Serum Deprivation in Multipotential Mesenchymal Stromal Cells. Stem Cells 2009, 27, 375–382. [Google Scholar] [CrossRef]
- Lee, S.-W.; Jeon, T.J.; Biswal, S. Effect of Local Treatment with Adipose Tissue-Derived Mesenchymal Stem Cells in the Early Tumorigenesis of Osteosarcoma. Oncol. Rep. 2015, 33, 1381–1387. [Google Scholar] [CrossRef]
- Toh, W.S.; Lai, R.C.; Zhang, B.; Lim, S.K. MSC Exosome Works through a Protein-Based Mechanism of Action. Biochem. Soc. Trans. 2018, 46, 843–853. [Google Scholar] [CrossRef]
- Milane, L.; Singh, A.; Mattheolabakis, G.; Suresh, M.; Amiji, M.M. Exosome Mediated Communication within the Tumor Microenvironment. J. Control. Release 2015, 219, 278–294. [Google Scholar] [CrossRef]
- Baglio, S.R.; Lagerweij, T.; Pérez-Lanzón, M.; Ho, X.D.; Léveillé, N.; Melo, S.A.; Cleton-Jansen, A.-M.; Jordanova, E.S.; Roncuzzi, L.; Greco, M.; et al. Blocking Tumor-Educated MSC Paracrine Activity Halts Osteosarcoma Progression. Clin. Cancer Res. 2017, 23, 3721–3733. [Google Scholar] [CrossRef] [PubMed]
- Klopp, A.H.; Gupta, A.; Spaeth, E.; Andreeff, M.; Marini, F. Concise Review: Dissecting a Discrepancy in the Literature: Do Mesenchymal Stem Cells Support or Suppress Tumor Growth? Stem Cells 2011, 29, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, J.; Ren, T.; Huang, Y.; Liang, X.; Yu, Y.; Wang, W.; Niu, J.; Guo, W. Bone Marrow Mesenchymal Stem Cell-Derived Exosomal MiR-206 Inhibits Osteosarcoma Progression by Targeting TRA2B. Cancer Lett. 2020, 490, 54–65. [Google Scholar] [CrossRef]
- NguyenThai, Q.-A.; Sharma, N.; Luong, D.H.; Sodhi, S.S.; Kim, J.-H.; Kim, N.; Oh, S.-J.; Jeong, D.K. Targeted Inhibition of Osteosarcoma Tumor Growth by Bone Marrow-Derived Mesenchymal Stem Cells Expressing Cytosine Deaminase/5-Fluorocytosine in Tumor-Bearing Mice: Anti-Tumor Activity of CD/5FC-MSCs. J. Gene Med. 2015, 17, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, X.; Jiang, N.; Xie, X.; Liu, N.; Liu, J.; Shen, J.; Peng, T. Exosome-transmitted Linc00852 Associated with Receptor Tyrosine Kinase AXL Dysregulates the Proliferation and Invasion of Osteosarcoma. Cancer Med. 2020, 9, 6354–6366. [Google Scholar] [CrossRef]
- Zhang, H.; Du, Y.; Xin, P.; Man, X. The LINC00852/MiR-29a-3p/JARID2 Axis Regulates the Proliferation and Invasion of Prostate Cancer Cell. BMC Cancer 2022, 22, 1269. [Google Scholar] [CrossRef]
- Ge, X.; Liu, W.; Zhao, W.; Feng, S.; Duan, A.; Ji, C.; Shen, K.; Liu, W.; Zhou, J.; Jiang, D.; et al. Exosomal Transfer of LCP1 Promotes Osteosarcoma Cell Tumorigenesis and Metastasis by Activating the JAK2/STAT3 Signaling Pathway. Mol. Ther.-Nucleic Acids 2020, 21, 900–915. [Google Scholar] [CrossRef]
- Zhang, H.; Song, J.; Zhou, X. Long Noncoding RNA P53 Upregulated Regulator of P53 Levels Promotes Osteogenic Differentiation in Osteoporosis Progression Through Sponging MiR-135a-5p. J. Biomater. Tissue Eng. 2022, 12, 2085–2091. [Google Scholar] [CrossRef]
- Zhang, K.; Dong, C.; Chen, M.; Yang, T.; Wang, X.; Gao, Y.; Wang, L.; Wen, Y.; Chen, G.; Wang, X.; et al. Extracellular Vesicle-Mediated Delivery of MiR-101 Inhibits Lung Metastasis in Osteosarcoma. Theranostics 2020, 10, 411–425. [Google Scholar] [CrossRef]
- Mei, X.; Zhang, B.; Zhao, M.; Lu, Q. An Update on Epigenetic Regulation in Autoimmune Diseases. J. Transl. Autoimmun. 2022, 5, 100176. [Google Scholar] [CrossRef]
- Mazumdar, A.; Urdinez, J.; Boro, A.; Migliavacca, J.; Arlt, M.J.E.; Muff, R.; Fuchs, B.; Snedeker, J.G.; Gvozdenovic, A. Osteosarcoma-Derived Extracellular Vesicles Induce Lung Fibroblast Reprogramming. Int. J. Mol. Sci. 2020, 21, 5451. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Wang, L.; Wu, C.; Huang, L.; Ruan, Y.; Xue, W. Tumor-Derived Exosomes Induced M2 Macrophage Polarization and Promoted the Metastasis of Osteosarcoma Cells Through Tim-3. Arch. Med. Res. 2021, 52, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Gomes de Morais, A.L.; Cerdá, S.; de Miguel, M. New Checkpoint Inhibitors on the Road: Targeting TIM-3 in Solid Tumors. Curr. Oncol. Rep. 2022, 24, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Z.; Yang, X.; Lu, W.; Chen, Y.; Lin, Y.; Wang, J.; Lin, S.; Yun, J.-P. H3K27 Acetylation Activated-COL6A1 Promotes Osteosarcoma Lung Metastasis by Repressing STAT1 and Activating Pulmonary Cancer-Associated Fibroblasts. Theranostics 2021, 11, 1473–1492. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, Y.; Wang, L.; Zou, M.; Sun, Y.; Sun, H.; Guo, Q.; Peng, X. Mycoplasma Gallisepticum Escapes the Host Immune Response via Gga-MiR-365-3p/SOCS5/STATs Axis. Vet. Res. 2022, 53, 103. [Google Scholar] [CrossRef]
- Qiao, B.; Shui, W.; Cai, L.; Guo, S.; Jiang, D. Human Mesenchymal Stem Cells as Delivery Of & nbsp;Osteoprotegerin Gene: Homing and Therapeutic Effect for Osteosarcoma. Drug Des. Dev. Ther. 2015, 9, 969. [Google Scholar] [CrossRef]
- Grisendi, G.; Spano, C.; D’souza, N.; Rasini, V.; Veronesi, E.; Prapa, M.; Petrachi, T.; Piccinno, S.; Rossignoli, F.; Burns, J.S.; et al. Mesenchymal Progenitors Expressing TRAIL Induce Apoptosis in Sarcomas. Stem Cells 2015, 33, 859–869. [Google Scholar] [CrossRef]
- Gamie, Z.; Kapriniotis, K.; Papanikolaou, D.; Haagensen, E.; Da Conceicao Ribeiro, R.; Dalgarno, K.; Krippner-Heidenreich, A.; Gerrand, C.; Tsiridis, E.; Rankin, K.S. TNF-Related Apoptosis-Inducing Ligand (TRAIL) for Bone Sarcoma Treatment: Pre-Clinical and Clinical Data. Cancer Lett. 2017, 409, 66–80. [Google Scholar] [CrossRef]
- Lenna, S.; Bellotti, C.; Duchi, S.; Martella, E.; Columbaro, M.; Dozza, B.; Ballestri, M.; Guerrini, A.; Sotgiu, G.; Frisoni, T.; et al. Mesenchymal Stromal Cells Mediated Delivery of Photoactive Nanoparticles Inhibits Osteosarcoma Growth in Vitro and in a Murine in Vivo Ectopic Model. J. Exp. Clin. Cancer Res. 2020, 39, 40. [Google Scholar] [CrossRef]
- Duchi, S.; Sotgiu, G.; Lucarelli, E.; Ballestri, M.; Dozza, B.; Santi, S.; Guerrini, A.; Dambruoso, P.; Giannini, S.; Donati, D.; et al. Mesenchymal Stem Cells as Delivery Vehicle of Porphyrin Loaded Nanoparticles: Effective Photoinduced in Vitro Killing of Osteosarcoma. J. Control. Release 2013, 168, 225–237. [Google Scholar] [CrossRef]
- Morales-Molina, A.; Gambera, S.; Leo, A.; García-Castro, J. Combination Immunotherapy Using G-CSF and Oncolytic Virotherapy Reduces Tumor Growth in Osteosarcoma. J. ImmunoTherapy Cancer 2021, 9, e001703. [Google Scholar] [CrossRef]
- Wei, H.; Chen, F.; Chen, J.; Lin, H.; Wang, S.; Wang, Y.; Wu, C.; Lin, J.; Zhong, G. Mesenchymal Stem Cell Derived Exosomes as Nanodrug Carrier of Doxorubicin for Targeted Osteosarcoma Therapy via SDF1-CXCR4 Axis. Int. J. Nanomed. 2022, 17, 3483–3495. [Google Scholar] [CrossRef]
- Wei, H.; Chen, J.; Wang, S.; Fu, F.; Zhu, X.; Wu, C.; Liu, Z.; Zhong, G.; Lin, J. A Nanodrug Consisting Of Doxorubicin And Exosome Derived From Mesenchymal Stem Cells For Osteosarcoma Treatment In Vitro. Int. J. Nanomed. 2019, 14, 8603–8610. [Google Scholar] [CrossRef]
- Shimbo, K.; Miyaki, S.; Ishitobi, H.; Kato, Y.; Kubo, T.; Shimose, S.; Ochi, M. Exosome-Formed Synthetic MicroRNA-143 Is Transferred to Osteosarcoma Cells and Inhibits Their Migration. Biochem. Biophys. Res. Commun. 2014, 445, 381–387. [Google Scholar] [CrossRef]
- Fei, D.; Sui, G.; Lu, Y.; Tan, L.; Dongxu, Z.; Zhang, K. The Long Non-Coding RNA-ROR Promotes Osteosarcoma Progression by Targeting MiR-206. J. Cell. Mol. Med. 2019, 23, 1865–1872. [Google Scholar] [CrossRef]
- Xu, X.; Qiu, B.; Yi, P.; Li, H. Overexpression of MiR-206 in Osteosarcoma and Its Associated Molecular Mechanisms as Assessed through TCGA and GEO Databases. Oncol. Lett. 2020, 19, 1751–1758. [Google Scholar] [CrossRef]
- Keremu, A.; Aila, P.; Tusun, A.; Abulikemu, M.; Zou, X. Extracellular Vesicles from Bone Mesenchymal Stem Cells Transport MicroRNA-206 into Osteosarcoma Cells and Target NRSN2 to Block the ERK1/2-Bcl-XL Signaling Pathway. Eur. J. Histochem. 2022, 66, 3394. [Google Scholar] [CrossRef]
- Xu, Z.; Zeng, S.; Gong, Z.; Yan, Y. Exosome-Based Immunotherapy: A Promising Approach for Cancer Treatment. Mol. Cancer 2020, 19, 160. [Google Scholar] [CrossRef]
- Khlusov, I.; Yurova, K.; Shupletsova, V.; Khaziakhmatova, O.; Malashchenko, V.; Kudryavtseva, V.; Khlusova, M.; Sukhorukov, G.; Litvinova, L. Microcapsule-Based Dose-Dependent Regulation of the Lifespan and Behavior of Adipose-Derived MSCs as a Cell-Mediated Delivery System: In Vitro Study. Int. J. Mol. Sci. 2022, 24, 292. [Google Scholar] [CrossRef]



| Effects | Signal Path | Component of the Signal Path | Activation (↑)/Inactivation (↓) of the Signal Path Component in OS | miRs Regulating Components of the Signaling Pathway | High (↑)/Low (↓) miRs Expression Levels in OS | Influence of miRs on Components of Signaling Pathways (Suppression ↓/Mediated Activation ↑) | References |
|---|---|---|---|---|---|---|---|
| Proliferation, migration, invasion | NKD2 | ↓ | miR-130b | ↑ | ↓ | [83] | |
| GSK-3β | ↓ | miR-346 | ↑ | ↓ | [84] | ||
| TCF-4 | ↑ | miR-4695-5p | ↓ | ↓ | [87] | ||
| TCF-7 | ↑ | miR-192 | ↓ | ↓ | [87] | ||
| FOXM1 | ↑ | miR-320a/b | ↓ | ↓ | [91,92,93] | ||
| FOXM1 | ↑ | miR-361 miR-361-5p | ↓ | ↓ | [94,95] | ||
| FOXM1 | ↑ | miR-216b | ↓ | ↓ | [96] | ||
| Apoptosis | Bax/Bcl-2 | ↓/↑ | miR-216b | ↓ | ↑/↓ | [96] | |
| FOXM1 | ↑ | miR-134 | ↓ | ↓ | [97,98] | ||
| Cell cycle | E2F1 | ↑ | miR-185-3p | ↓ | ↓ | [101] | |
| EMT | E2F1 | ↑ | miR-211-5p | ↓ | ↓ | [101] | |
| E2F1 | ↑ | miR-329 | ↓ | ↓ | [102] | ||
| E2F1 | ↑ | miR-320 | ↓ | ↓ | [103] | ||
| E2F3 | ↑ | miR-145-5p | ↑ | ↑ | [108] | ||
| E2F3 | ↑ | miR-16-5p | ↓ | ↓ | [109] | ||
| E2F3 | ↑ | miR-124-3p | ↓ | ↓ | [110] | ||
| E2F3 | ↑ | miR-874 | ↓ | ↓ | [111] | ||
| E2F3 | ↑ | miR-152 | ↓ | ↓ | [112] | ||
| E2F5 | ↑ | miR-154-5p | ↓ | ↓ | [105] | ||
| E2F5 | ↑ | miR-513b-5p | ↓ | ↓ | [106] | ||
| Cell cycle | RB1 | ↓ | miR-17 | ↑ | ↓ | [116,117] | |
| Chemoresistance, EMT | NRF2 | ↑ | miR-340-5p | ↓ | ↓ | [120] | |
| Migration, invasion, stemness | YAP | ↑ | miR-515-5p | ↓ | ↓ | [124] | |
| YAP | ↑ | miR-129-5p | ↓ | ↓ | [125] | ||
| YAP | ↑ | miR-375 | ↓ | ↓ | [126] | ||
| YAP | ↑ | miR-195-5p | ↓ | ↓ | [127] | ||
| YAP | ↑ | miR-1285-3p | ↓ | ↓ | [128] | ||
| YAP | ↑ | miR-132-3p | ↓ | ↓ | [129] | ||
| YAP | ↑ | miR-625 | ↓ | ↓ | [130] | ||
| ROCK | ↑ | miR-144 | ↓ | ↓ | [131,137] | ||
| ROCK1 | ↑ | miR-340 | ↓ | ↓ | [134] | ||
| ROCK1 | ↑ | miR-148a | ↓ | ↓ | [139] | ||
| ROCK1 | ↑ | miR-139 | ↓ | ↓ | [140] | ||
| ROCK1 | ↑ | miR-335 | ↓ | ↓ | [141,142] | ||
| ROCK1 | ↑ | miR-145 | ↓ | ↓ | [143] | ||
| ROCK1 | ↑ | miR-129-5p | ↓ | ↓ | [144] | ||
| ROCK1 | ↑ | miR-214-5p | ↓ | ↓ | [145] | ||
| ROCK1 | ↑ | miR-101 | ↓ | ↓ | [146] | ||
| ROCK1 | ↑ | miR-202-5p | ↓ | ↓ | [147] | ||
| Proliferation, migration, invasion | c-Met | ↑ | miR-449b-5p | ↓ | ↓ | [154] | |
| c-Met | ↑ | miR-454 | ↓ | ↓ | [152] | ||
| c-Met | ↑ | miR-34a | ↓ | ↓ | [155] | ||
| c-Met | ↑ | miR-876-5p | ↓ | ↓ | [156] | ||
| AXIN1 | ↑ | miR-31-5p | ↓ | ↓ | [158] | ||
| Proliferation, migration, apoptosis, EMT | PI3K, Wnt, TGFβ | SOX4 | ↑ | miR-25 | ↓ | ↓ | [161] |
| SOX4 | ↑ | miR-132 | ↓ | ↓ | [164] | ||
| SOX4 | ↑ | miR-363-3p | ↓ | ↓ | [165] | ||
| SOX4 | ↑ | miR-212 | ↓ | ↓ | [166] | ||
| Proliferation | STAT3 | ↑ | miR-340-5p | ↓ | ↓ | [167] | |
| STAT3 | ↑ | miR-30d-5p | ↑ | ↑ | [168] | ||
| STAT3 | ↑ | miR-221-3p | ↑ | ↑ | [169] | ||
| Dkk-1 | ↑ | miR-107 | ↓ | ↓ | [171] | ||
| IGF2BP1 | ↑ | miR-150 | ↓ | ↓ | [172] | ||
| MYC | ↑ | miR-193b | ↓ | ↓ | [174] | ||
| Cell cycle | PI3K, Wnt, TGFβ | CCNE2 | ↑ | miR-34c | ↓ | ↓ | [178] |
| Hippo | PTPRB | ↓ | miR-624-5p | ↑ | ↓ | [184] | |
| TGFβ | SMAD4 | ↓ | miR-224 | ↑ | ↓ | [185] | |
| SMAD7 | ↑ | miR-181c | ↓ | ↓ | [191] | ||
| TRAF6 | ↑ | miR-140-3p | ↓ | ↓ | [194] | ||
| PTEN and TGF-β1 | ↓ | miR-21 | ↓ | ↑ | [195] | ||
| TGF-β1 | miR-26a-5p | ↓ | [198] | ||||
| Migration, invasion | SMAD3 | ↑ | miR-671-5p | ↓ | ↓ | [203] | |
| SMAD3 | ↑ | miR-16-5p | ↓ | ↓ | [202] | ||
| TGFβ2 | ↑ | miR-422a | ↓ | ↓ | [204] | ||
| Metastasis | Notch | NOTCH1, JAG1, HEY2 | ↑ | miR-34c | ↓ | ↓ | [219] |
| Notch | ↑ | miR-199b-5p | ↓ | ↓ | [221,223] | ||
| Notch2 | ↑ | miR-1296-5p | ↓ | ↓ | [224] | ||
| HIF-1/ Rap1/ PI3K-Akt | HIF1α | ↑ | miR-338-3p | ↓ | ↓ | [228] | |
| HIF1α | ↑ | miR-186 | ↓ | ↓ | [229] | ||
| Migration, invasion | ErbB | EGFR | ↑ | miR-7 | ↓ | ↓ | [234] |
| EGFR | ↑ | miR-3184-5p | ↓ | ↓ | [232] | ||
| EGFR (ERK/MAPK) | ↑ | miR-143 | ↓ | ↓ | [233] | ||
| EGFR | ↑ | miR-141-3p | ↓ | ↓ | [235] | ||
| EGFR | ↑ | miR-491-5p | ↓ | ↓ | [236] | ||
| Metastasis | ErbB | HER2 | ↑ | miR-199b-5p | ↑ | ↑ | [222] |
| PI3K/Akt/mTOR | PI3KR1 | ↓ | miR-21-5p | ↑ | ↑ | [249] | |
| NRF2 | ↑ | miR-340-5p | ↓ | ↓ | [120] | ||
| PI3K, Akt, mTOR | ↑ | miR-22 | ↓ | ↓ | [250] | ||
| elF4E | ↑ | miR-496 | ↓ | ↓ | [256] | ||
| Oncogenesis, angiogenesis, proliferation, metastasis, chemoresistance | PI3K/Akt/mTOR | HMGB1 | ↑ | miR-505 | ↓ | ↓ | [262,263] |
| HMGB1 | ↑ | miR-1284 | ↓ | ↓ | [264] | ||
| HMGB1 | ↑ | miR-22 | ↓ | ↓ | [265,266] | ||
| Twist1 | ↑ | miR-22 | ↓ | ↓ | [267] | ||
| HMGB1 | ↑ | miR-372-3p | ↓ | ↓ | [268] | ||
| HMGB1 | ↑ | miR-142-3p | ↓ | ↓ | [259] | ||
| HMGB1 | ↑ | miR-129-3p | ↓ | ↓ | [259] | ||
| HMGB1 | ↑ | miR-32-5p | ↓ | ↓ | [259] | ||
| RUNX2 | ↑ | miR-338-3p | ↓ | ↓ | [272] | ||
| CDK4 | ↑ | miR-338-3p | ↓ | ↓ | [272] | ||
| MAPK | ↑ | miR-338-3p | ↓ | ↓ | [272] | ||
| Chemoresistance | RUNX2 | ↑ | miR-203 | ↓ | ↓ | [273] | |
| RUNX2 | ↑ | miR-320b | ↓ | ↓ | [274] | ||
| RUNX2 | ↑ | miR-218 | ↓ | ↓ | [275] | ||
| Apoptosis | p53 | CDK6, E2F3, Cyclin E2, BCL2 | ↑ | miR-34 | ↓ | ↓ | [9] |
| p53 | ↓ | miR-122-5p | ↓ | ↑ | [278] | ||
| PI3K/Akt/mTOR | PI3K/Akt/mTOR | ↑ | miR-122-5p | ↓ | ↓ | [278] | |
| Proliferation, migration, invasion | PI3K/Akt/mTOR | FOXP1 | ↑ | miR-181d-5p | ↓ | ↓ | [281] |
| Apoptosis | Bak | ↓ | miR-491-5p | ↓ | ↑ | [284] | |
| pro-caspase-9 | ↑ | miR-342-5p | ↓ | ↓ | [236,284] | ||
| BCL-2 | ↓ | miR-143 | ↓ | ↑ | [285] | ||
| Bcl-xL | ↑ | miR-133a | ↓ | ↓ | [290] | ||
| VEGF | VEGF | ↑ | miR-150-5p | ↓ | ↓ | [296] | |
| VEGF | ↑ | miR-1 | ↓ | ↓ | [297] | ||
| VEGF | ↑ | miR-134 | ↓ | ↓ | [298] | ||
| PI3K/Akt/mTOR | PTEN | ↓ | miR-1908 | ↑ | ↓ | [301] | |
| PTEN | ↓ | miR-221 | ↑ | ↓ | [302] | ||
| PTEN | ↓ | miR-21 | ↑ | ↓ | [306] | ||
| PTEN | ↓ | miR-187 | ↑ | ↓ | [305] | ||
| PTEN | ↓ | miR-17 | ↑ | ↓ | [305] | ||
| Proliferation | Hh | Hh | ↑ | miR-212 | ↑ | ↑ | [307] |
| Proliferation, cell cycle, EMT | FOXK1 | ↑ | miR-186-5p | ↓ | ↓ | [308] | |
| Proliferation, migration, invasion | NOB1 | ↑ | miR-363 | ↓ | ↓ | [311] | |
| TGIF2 | ↑ | miR-541 | ↓ | ↓ | [314] | ||
| TGIF2 | ↑ | miR-34 | ↓ | ↓ | [315] | ||
| TGIF2 | ↑ | miR-129 | ↓ | ↓ | [312] | ||
| Metastasis | RECK | ↑ | miR-21 | ↓ | ↓ | [317,318] | |
| RECK | ↑ | miR-92b | ↓ | ↓ | [319] | ||
| Antiproliferative effect, proapoptotic | KLF10 | ↓ | miR-197-3p | ↑ | ↓ | [321] | |
| Metastasis | HDAC1 | ↑ | miR-326 | ↓ | ↓ | [322] | |
| HDAC1 | ↑ | miR-449a | ↓ | ↓ | [325] | ||
| Proliferation, migration, invasion | HDAC4 | ↑ | miR-637 | ↓ | ↓ | [328] | |
| Development, metastasis | SND1 | ↑ | miR-296-5p | ↓ | ↓ | [332] | |
| Proliferation | FOXP4 | ↑ | miR-491-5p | ↓ | ↓ | [334] | |
| Cellular redox homeostasis | GPX4 | ↑ | miR-1287-5p | ↓ | ↓ | [336] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Todosenko, N.; Khlusov, I.; Yurova, K.; Khaziakhmatova, O.; Litvinova, L. Signal Pathways and microRNAs in Osteosarcoma Growth and the Dual Role of Mesenchymal Stem Cells in Oncogenesis. Int. J. Mol. Sci. 2023, 24, 8993. https://doi.org/10.3390/ijms24108993
Todosenko N, Khlusov I, Yurova K, Khaziakhmatova O, Litvinova L. Signal Pathways and microRNAs in Osteosarcoma Growth and the Dual Role of Mesenchymal Stem Cells in Oncogenesis. International Journal of Molecular Sciences. 2023; 24(10):8993. https://doi.org/10.3390/ijms24108993
Chicago/Turabian StyleTodosenko, Natalia, Igor Khlusov, Kristina Yurova, Olga Khaziakhmatova, and Larisa Litvinova. 2023. "Signal Pathways and microRNAs in Osteosarcoma Growth and the Dual Role of Mesenchymal Stem Cells in Oncogenesis" International Journal of Molecular Sciences 24, no. 10: 8993. https://doi.org/10.3390/ijms24108993