State of the Art of Genomic Technology in Toxicology: A Review
Abstract
:1. Introduction
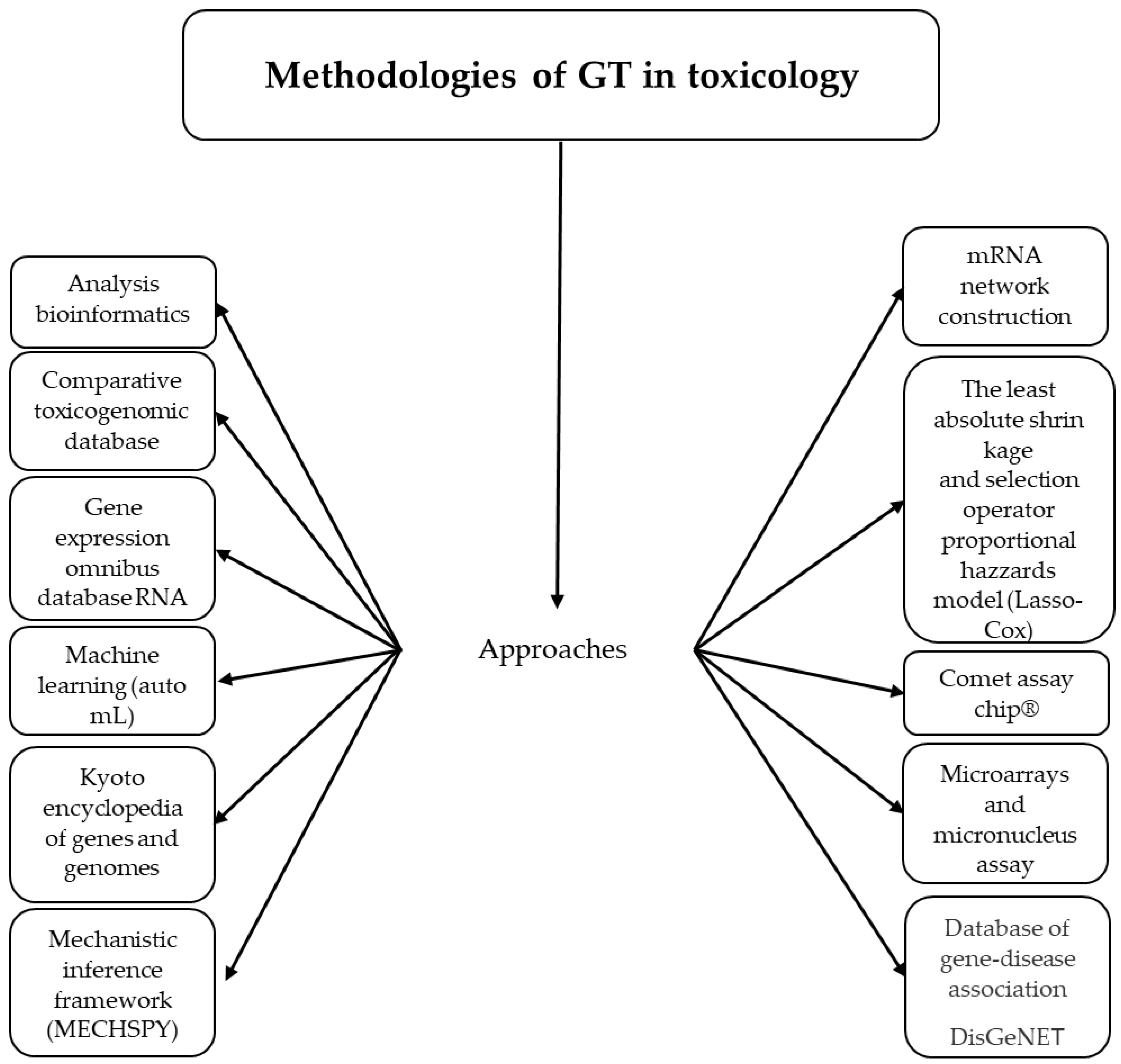
2. Methods
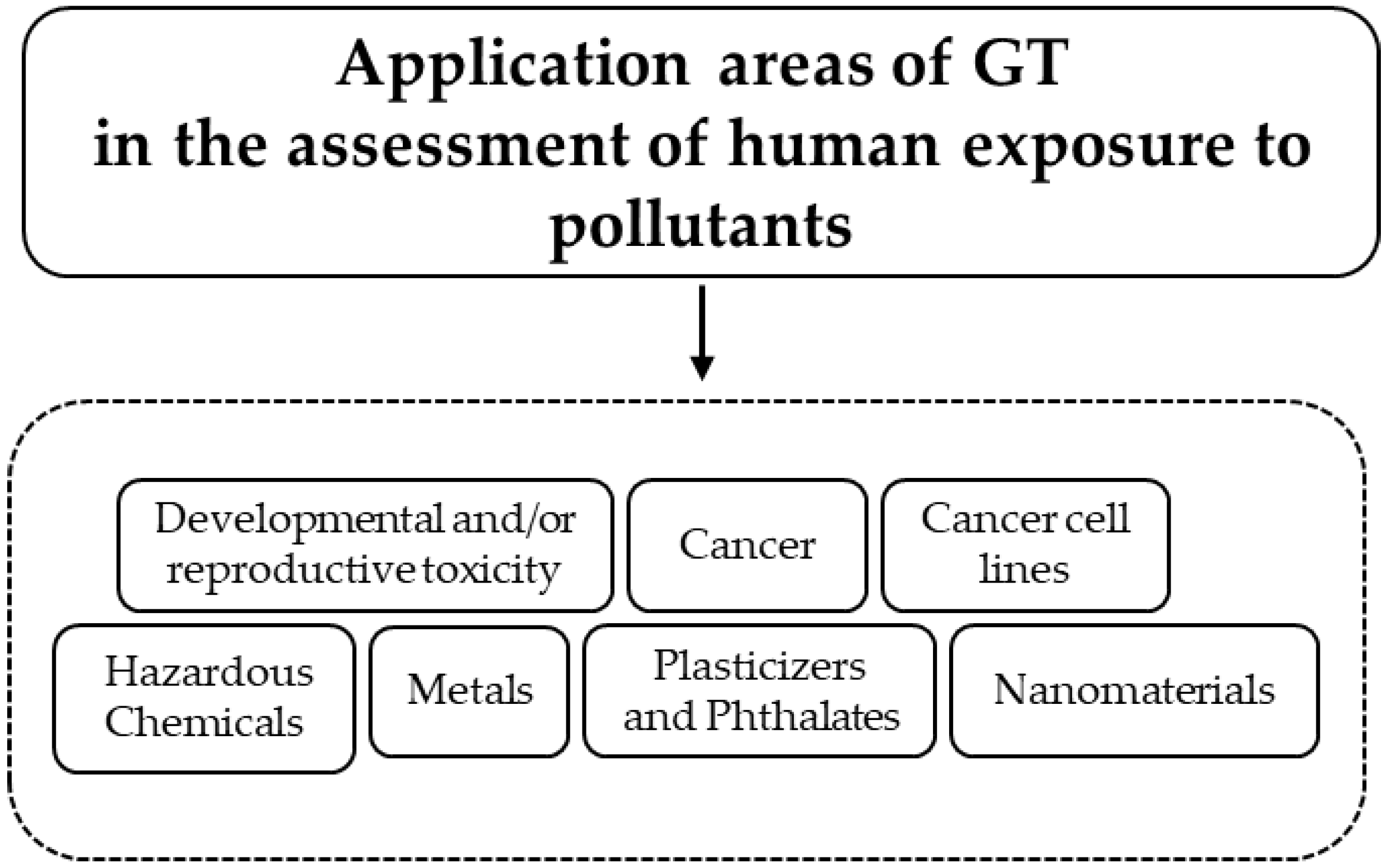
3. Developmental and/or Reproductive Toxicity
4. Cancer
5. Cancer Cell Lines in GT
6. Hazardous Chemicals
7. Metals
8. Plasticizers and Phthalates
9. Nanomaterials
10. Computational Biology/Toxicology
11. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sabbioni, G.; Castaño, A.; Esteban López, M.; Göen, T.; Mol, H.; Riou, M.; Tagne-Fotso, R. Literature review and evaluation of biomarkers, matrices and analytical methods for chemicals selected in the research program Human Biomonitoring for the European Union (HBM4EU). Environ. Int. 2022, 169, 107458. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Tian, Y.; Wang, M.; Li, Y.; Yin, J.; Qu, W.; Yan, C.; Ding, R.; Guan, Y.; Wang, Q. Proteomics unite traditional toxicological assessment methods to evaluate the toxicity of iron oxide nanoparticles. Front. Pharmacol. 2022, 13, 1011065. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, J.S.; Brito, I.M.D.C.; De Oliveira, L.A.; De Barros, L.C.; Almeida, C.; Rossini, B.C.; Sousa, D.L.; Alves, R.S.; Jorge, R.J.B.; Santos, L.D.D. Experimental Bothrops atrox Envenomation: Blood Plasma Proteome Effects after Local Tissue Damage and Perspectives on Thromboinflammation. Toxins 2022, 14, 613. [Google Scholar] [CrossRef]
- Alugubelly, N.; Mohammed, A.N.; Carr, R.L. Persistent proteomic changes in glutamatergic and GABAergic signaling in the amygdala of adolescent rats exposed to chlorpyrifos as juveniles. Neurotoxicology 2021, 85, 234–244. [Google Scholar] [CrossRef]
- LaRocca, J.; Costa, E.; Sriram, S.; Hannas, B.R.; Johnson, K.J. Short-term toxicogenomics as an alternative approach to chronic in vivo studies for derivation of points of departure: A case study in the rat with a triazole fungicide. Regul. Toxicol. Pharmacol. 2020, 113, 104655. [Google Scholar] [CrossRef]
- Granados, J.C.; Falah, K.; Koo, I.; Morgan, E.W.; Perdew, G.H.; Patterson, A.D.; Jamshidi, N.; Nigam, S.K. AHR is a master regulator of diverse pathways in endogenous metabolism. Sci. Rep. 2022, 12, 16625. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Yang, N.; Zhang, Q.; Wang, Z.; Luo, G.; Pang, J. The discovery of combined toxicity effects and mechanisms of hexaconazole and arsenic to mice based on untargeted metabolomics. Ecotoxicol. Environ. Saf. 2021, 226, 112859. [Google Scholar] [CrossRef]
- Wang, Y.; Nong, Y.; Zhang, X.; Mai, T.; Cai, J.; Liu, J.; Lai, K.P.; Zhang, Z. Comparative plasma metabolomic analysis to identify biomarkers for lead-induced cognitive impairment. Chem. Biol. Interact. 2022, 366, 110143. [Google Scholar] [CrossRef]
- Bozack, A.K.; Boileau, P.; Wei, L.; Hubbard, A.E.; Sillé, F.C.M.; Ferreccio, C.; Acevedo, J.; Hou, L.; Ilievski, V.; Steinmaus, C.M.; et al. Exposure to arsenic at different life-stages and DNA methylation meta-analysis in buccal cells and leukocytes. Environ. Health 2021, 20, 79. [Google Scholar] [CrossRef]
- Fitz, N.F.; Barchowsky, A.; Koldamova, R.; Lefterov, I. Genome-wide alteration of histone methylation profiles associated with cognitive changes in response to developmental arsenic exposure in mice. Toxicol. Rep. 2022, 9, 393–403. [Google Scholar] [CrossRef]
- Xiao, L.; Cheng, H.; Cai, H.; Wei, Y.; Zan, G.; Feng, X.; Liu, C.; Li, L.; Huang, L.; Wang, F.; et al. Associations of Heavy Metals with Activities of Daily Living Disability: An Epigenome-Wide View of DNA Methylation and Mediation Analysis. Environ. Health Perspect. 2022, 130, 87009. [Google Scholar] [CrossRef]
- Ao, J.; Liu, Y.; Tang, W.; Zhang, J. Bisphenol S exposure induces intestinal inflammation: An integrated metabolomic and transcriptomic study. Chemosphere 2022, 292, 133510. [Google Scholar] [CrossRef]
- Shukla, V.; Chandrasekaran, B.; Tyagi, A.; Navin, A.K.; Saran, U.; Adam, R.M.; Damodaran, C. A Comprehensive Transcriptomic Analysis of Arsenic-Induced Bladder Carcinogenesis. Cells 2022, 11, 2435. [Google Scholar] [CrossRef] [PubMed]
- Cunha Nascimento, P.; Alana Bragança Aragão, W.; Oliveira Bittencourt, L.; Dionizio, A.; Buzalaf, M.A.R.; Crespo-Lopez, M.E.; Lima, R.R. Maternal methylmercury exposure changes the proteomic profile of the offspring′s salivary glands: Prospects on translational toxicology. PLoS ONE 2021, 16, e0258969. [Google Scholar] [CrossRef]
- Gao, Y.; Lee, H.; Lee, S.; Kim, K.T. Type 2 Diabetes Induced by Changes in Proteomic Profiling of Zebrafish Chronically Exposed to a Mixture of Organochlorine Pesticides at Low Concentrations. Int. J. Environ. Res. Public Health 2022, 19, 4991. [Google Scholar] [CrossRef]
- Fu, Y.; Bi, Z.; Li, L.; Wadgaonkar, P.; Qiu, Y.; Almutairy, B.; Zhang, W.; Seno, A.; Thakur, C.; Chen, F. Metabolomic dynamics of the arsenic-transformed bronchial epithelial cells and the derived cancer stem-like cells. Int. J. Biol. Sci. 2022, 18, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Quintás, G.; Martínez-Sena, T.; Conde, I.; Pareja Ibars, E.; Kleinjans, J.; Castell, J.V. Metabolomic analysis to discriminate drug-induced liver injury (DILI) phenotypes. Arch. Toxicol. 2021, 95, 3049–3062. [Google Scholar] [CrossRef] [PubMed]
- Tikhodeyev, O.N. Heredity determined by the environment: Lamarckian ideas in modern molecular biology. Sci. Total Environ. 2020, 710, 135521. [Google Scholar] [CrossRef]
- Zhang, X.W.; Li, Q.H.; Xu, Z.D.; Dou, J.J. Mass spectrometry-based metabolomics in health and medical science: A systematic review. RSC Adv. 2020, 10, 3092–3104. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Int. J. Surg. 2021, 372, n71. [Google Scholar] [CrossRef]
- Tung, C.W.; Cheng, H.J.; Wang, C.C.; Wang, S.S.; Lin, P. Leveraging complementary computational models for prioritizing chemicals of developmental and reproductive toxicity concern: An example of food contact materials. Arch. Toxicol. 2020, 94, 485–494. [Google Scholar] [CrossRef]
- Kanmaz, A.G.; İnan, A.H.; Beyan, E.; Budak, A. The effects of threatened abortions on pregnancy outcomes. Ginekol. Pol. 2019, 90, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.M.; Jin, Y.; Loch-Caruso, R.; Padilla, I.Y.; Meeker, J.D.; Bakulski, K.M. Identification of environmental chemicals targeting miscarriage genes and pathways using the comparative toxicogenomics database. Environ. Res. 2020, 184, 109259. [Google Scholar] [CrossRef] [PubMed]
- Haimbaugh, A.; Meyer, D.; Akemann, C.; Gurdziel, K.; Baker, T.R. Comparative Toxicotranscriptomics of Single Cell RNA-Seq and Conventional RNA-Seq in TCDD-Exposed Testicular Tissue. Front. Toxicol. 2022, 4, 821116. [Google Scholar] [CrossRef]
- Wang, Y.; Qian, H. Phthalates and Their Impacts on Human Health. Healthcare 2021, 9, 603. [Google Scholar] [CrossRef]
- Baralić, K.; Bozic, D.; Živančević, K.; Milenković, M.; Javorac, D.; Marić, Đ.; Miljaković, E.A.; Djordjevic, A.B.; Vukomanović, P.; Ćurčić, M.; et al. Integrating in silico with in vivo approach to investigate phthalate and bisphenol A mixture-linked asthma development: Positive probiotic intervention. Food Chem. Toxicol. 2021, 158, 112671. [Google Scholar] [CrossRef]
- Han, Y.; Liang, C.; Yu, Y.; Manthari, R.K.; Cheng, C.; Tan, Y.; Li, X.; Tian, X.; Fu, W.; Yang, J.; et al. Chronic arsenic exposure lowered sperm motility via impairing ultra-microstructure and key proteins expressions of sperm acrosome and flagellum formation during spermiogenesis in male mice. Sci. Total Environ. 2020, 734, 139233. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Huang, Q. Environmental pollutants exposure and male reproductive toxicity: The role of epigenetic modifications. Toxicology 2021, 456, 152780. [Google Scholar] [CrossRef]
- Lu, Z.; Zhao, C.; Yang, J.; Ma, Y.; Qiang, M. Paternal exposure to arsenic and sperm DNA methylation of imprinting gene Meg3 in reproductive-aged men. Environ. Geochem. Health 2022, 45, 3055–3068. [Google Scholar] [CrossRef]
- Deswal, R.; Narwal, V.; Dang, A.; Pundir, C.S. The Prevalence of Polycystic Ovary Syndrome: A Brief Systematic Review. J. Hum. Reprod. Sci. 2020, 13, 261–271. [Google Scholar] [CrossRef]
- Guo, Z.; Qiu, H.; Wang, L.; Wang, L.; Wang, C.; Chen, M.; Zuo, Z. Association of serum organochlorine pesticides concentrations with reproductive hormone levels and polycystic ovary syndrome in a Chinese population. Chemosphere 2017, 171, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Lin, X.; Xia, T.; Liu, W.; Tian, X.; Li, M. Identification of Crucial lncRNAs, miRNAs, mRNAs, and Potential Therapeutic Compounds for Polycystic Ovary Syndrome by Bioinformatics Analysis. Biomed. Res. Int. 2020, 2020, 1817094. [Google Scholar] [CrossRef] [PubMed]
- Sakhteman, A.; Failli, M.; Kublbeck, J.; Levonen, A.L.; Fortino, V. A toxicogenomic data space for system-level understanding and prediction of EDC-induced toxicity. Environ. Int. 2021, 156, 106751. [Google Scholar] [CrossRef]
- Kuijpers, T.J.M.; Kleinjans, J.C.S.; Jennen, D.G.J. From multi-omics integration towards novel genomic interaction networks to identify key cancer cell line characteristics. Sci. Rep. 2021, 11, 10542. [Google Scholar] [CrossRef]
- Gong, L.; Luo, Z.; Tang, H.; Tan, X.; Xie, L.; Lei, Y.; He, C.; Ma, J.; Han, S. Integrative, genome-wide association study identifies chemicals associated with common women’s malignancies. Genomics 2020, 112, 5029–5036. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhu, L.; Thakkar, S.; Roberts, R.; Tong, W. Can Transcriptomic Profiles from Cancer Cell Lines Be Used for Toxicity Assessment? Chem. Res. Toxicol. 2020, 33, 271–280. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Health Estimates 2020: Deaths by Cause, Age, Sex, by Country and by Region, 2000–2019: WHO, 2020. Available online: https://www.who.int/data/gho/data/themes/mortality-and-global-health-estimates/ghe-leading-causes-of-death (accessed on 7 March 2023).
- Panaiyadiyan, S.; Quadri, J.A.; Nayak, B.; Pandit, S.; Singh, P.; Seth, A.; Shariff, A. Association of heavy metals and trace elements in renal cell carcinoma: A case-controlled study. Urol. Oncol. 2022, 40, 111.e11–111.e18. [Google Scholar] [CrossRef]
- Meng, M.; Lan, T.; Tian, D.; Qin, Z.; Li, Y.; Li, J.; Cao, H. Integrative Bioinformatics Analysis Demonstrates the Prognostic Value of Chromatin Accessibility Biomarkers in Clear Cell Renal Cell Carcinoma. Front. Oncol. 2021, 11, 814396. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, H.; Li, X.; Liu, Q.; Zhao, Y.; Li, L.; Xu, B.; Hou, Y.; Jin, W. Identification of a Three-RNA Binding Proteins (RBPs) Signature Predicting Prognosis for Breast Cancer. Front. Oncol. 2021, 11, 663556. [Google Scholar] [CrossRef]
- Schyman, P.; Printz, R.L.; Pannala, V.R.; AbdulHameed, M.D.M.; Estes, S.K.; Shiota, C.; Boyd, K.L.; Shiota, M.; Wallqvist, A. Genomics and metabolomics of early-stage thioacetamide-induced liver injury: An interspecies study between guinea pig and rat. Toxicol. Appl. Pharmacol. 2021, 430, 115713. [Google Scholar] [CrossRef]
- Buick, J.K.; Williams, A.; Gagné, R.; Swartz, C.D.; Recio, L.; Ferguson, S.S.; Yauk, C.L. Flow cytometric micronucleus assay and TGx-DDI transcriptomic biomarker analysis of ten genotoxic and non-genotoxic chemicals in human HepaRG™ cells. Genes Environ. 2020, 42, 5. [Google Scholar] [CrossRef] [PubMed]
- Franzosa, J.A.; Bonzo, J.A.; Jack, J.; Baker, N.C.; Kothiya, P.; Witek, R.P.; Hurban, P.; Siferd, S.; Hester, S.; Shah, I.; et al. High-throughput toxicogenomic screening of chemicals in the environment using metabolically competent hepatic cell cultures. NPJ Syst. Biol. Appl. 2021, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Luijten, M.; Wackers, P.F.K.; Rorije, E.; Pennings, J.L.A.; Heusinkveld, H.J. Relevance of In Vitro Transcriptomics for In Vivo Mode of Action Assessment. Chem. Res. Toxicol. 2021, 34, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Kreuzer, K.; Frenzel, F.; Lampen, A.; Braeuning, A.; Böhmert, L. Transcriptomic effect marker patterns of genotoxins—A comparative study with literature data. J. Appl. Toxicol. 2020, 40, 448–457. [Google Scholar] [CrossRef]
- Gupta, R.; Schrooders, Y.; Hauser, D.; van Herwijnen, M.; Albrecht, W.; Ter Braak, B.; Brecklinghaus, T.; Castell, J.V.; Elenschneider, L.; Escher, S.; et al. Comparing in vitro human liver models to in vivo human liver using RNA-Seq. Arch. Toxicol. 2021, 95, 573–589. [Google Scholar] [CrossRef]
- Saquib, Q.; Al-Salem, A.M.; Siddiqui, M.A.; Ansari, S.M.; Zhang, X.; Al-Khedhairy, A.A. Cyto-Genotoxic and Transcriptomic Alterations in Human Liver Cells by Tris (2-Ethylhexyl) Phosphate (TEHP): A Putative Hepatocarcinogen. Int. J. Mol. Sci. 2022, 23, 3998. [Google Scholar] [CrossRef]
- Perkins, E.J.; Woolard, E.A.; Garcia-Reyero, N. Integration of Adverse Outcome Pathways, Causal Networks and ‘Omics to Support Chemical Hazard Assessment. Front. Toxicol. 2022, 4, 786057. [Google Scholar] [CrossRef]
- Mohanty, B.P.; Mahanty, A.; Mitra, T.; Mohanty, S.; Naik, A.K.; Parija, S.C. Proteomic and transcriptomic changes in rat liver following oral feeding of formaldehyde. Chemosphere 2020, 245, 125599. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, L.; Tagmount, A.; Loguinov, A.; Sobh, A.; Hubbard, A.; McHale, C.M.; Chang, C.J.; Vulpe, C.D.; Zhang, L. Applying genome-wide CRISPR to identify known and novel genes and pathways that modulate formaldehyde toxicity. Chemosphere 2021, 269, 128701. [Google Scholar] [CrossRef]
- Kang, D.S.; Lee, N.; Shin, D.Y.; Jang, Y.J.; Lee, S.H.; Lim, K.M.; Ahn, Y.S.; Lee, C.M.; Seo, Y.R. Network-based integrated analysis for toxic effects of high-concentration formaldehyde inhalation exposure through the toxicogenomic approach. Sci. Rep. 2022, 12, 5645. [Google Scholar] [CrossRef]
- Reinwald, H.; Alvincz, J.; Salinas, G.; Schäfers, C.; Hollert, H.; Eilebrecht, S. Toxicogenomic profiling after sublethal exposure to nerve- and muscle-targeting insecticides reveals cardiac and neuronal developmental effects in zebrafish embryos. Chemosphere 2022, 291, 132746. [Google Scholar] [CrossRef]
- Gust, K.A.; Ji, Q.; Luo, X. Example of Adverse Outcome Pathway Concept Enabling Genome-to-Phenome Discovery in Toxicology. Integr. Comp. Biol. 2020, 60, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ni, M.; Yang, Z.; Chen, X.; Zhang, L.; Chen, J. Bioinformatics analysis and quantitative weight of evidence assessment to map the potential mode of actions of bisphenol A. Environ. Pollut. 2021, 273, 116469. [Google Scholar] [CrossRef] [PubMed]
- Halappanavar, S.; Wu, D.; Boyadzhiev, A.; Solorio-Rodriguez, A.; Williams, A.; Jariyasopit, N.; Saini, A.; Harner, T. Toxicity screening of air extracts representing different source sectors in the Greater Toronto and Hamilton areas: In vitro oxidative stress, pro-inflammatory response, and toxicogenomic analysis. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2021, 872, 503415. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, H.J.; Kim, N.; Song, J.J.; Son, B.S.; Yang, J.H.; Lee, C.M.; Park, M.K.; Seo, Y.R. Toxicogenomic study to identify potential signaling alterations related to nasal inflammatory damages induced by diesel exhaust particles in primary human nasal epithelial cells. Toxicol. Vitr. 2020, 69, 104994. [Google Scholar] [CrossRef] [PubMed]
- Đukić-Ćosić, D.; Baralić, K.; Filipović, T.; Božić, D.; Živančević, K.; Miljaković, E.A.; Đorđević, A.B.; Bulat, Z.; Antonijević, B.; Ćurčić, M. Joint impact of key air pollutants on COVID-19 severity: Prediction based on toxicogenomic data analysis. Arh. Hig. Rada Toksikol. 2022, 73, 119–125. [Google Scholar] [CrossRef]
- Ruiz, P.; Emond, C.; McLanahan, E.D.; Joshi-Barr, S.; Mumtaz, M. Exploring Mechanistic Toxicity of Mixtures Using PBPK Modeling and Computational Systems Biology. Toxicol. Sci. 2020, 174, 38–50. [Google Scholar] [CrossRef]
- Poulsen, S.S.; Bengtson, S.; Williams, A.; Jacobsen, N.R.; Troelsen, J.T.; Halappanavar, S.; Vogel, U. A transcriptomic overview of lung and liver changes one day after pulmonary exposure to graphene and graphene oxide. Toxicol. Appl. Pharmacol. 2021, 410, 115343. [Google Scholar] [CrossRef]
- Striz, A.; DePina, A.; Jones, R., Jr.; Gao, X.; Yourick, J. Cytotoxic, genotoxic, and toxicogenomic effects of dihydroxyacetone in human primary keratinocytes. Cutan. Ocul. Toxicol. 2021, 40, 232–240. [Google Scholar] [CrossRef]
- Vrijenhoek, N.G.; Wehr, M.M.; Kunnen, S.J.; Wijaya, L.S.; Callegaro, G.; Moné, M.J.; Escher, S.E.; Van de Water, B. Application of high-throughput transcriptomics for mechanism-based biological read-across of short-chain carboxylic acid analogues of valproic acid. Altex 2022, 39, 207–220. [Google Scholar] [CrossRef]
- Oldani, M.; Fabbri, M.; Melchioretto, P.; Callegaro, G.; Fusi, P.; Gribaldo, L.; Forcella, M.; Urani, C. In vitro and bioinformatics mechanistic-based approach for cadmium carcinogenicity understanding. Toxicol. Vitr. 2020, 65, 104757. [Google Scholar] [CrossRef]
- Forcella, M.; Lau, P.; Fabbri, M.; Fusi, P.; Oldani, M.; Melchioretto, P.; Gribaldo, L.; Urani, C. Is Cadmium Toxicity Tissue-Specific? Toxicogenomics Studies Reveal Common and Specific Pathways in Pulmonary, Hepatic, and Neuronal Cell Models. Int. J. Mol. Sci. 2022, 23, 1768. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Fu, Y.; Ai, J.; Zhang, J.; Huang, G.; Deng, Y. Development of predicitve models to distinguish metals from non-metal toxicants, and individual metal from one another. BMC Bioinform. 2020, 21, 239. [Google Scholar] [CrossRef]
- Rehman, M.Y.A.; Briedé, J.J.; van Herwijnen, M.; Krauskopf, J.; Jennen, D.G.J.; Malik, R.N.; Kleinjans, J.C.S. Integrating SNPs-based genetic risk factor with blood epigenomic response of differentially arsenic-exposed rural subjects reveals disease-associated signaling pathways. Environ. Pollut. 2022, 292, 118279. [Google Scholar] [CrossRef] [PubMed]
- Rehman, M.Y.A.; van Herwijnen, M.; Krauskopf, J.; Farooqi, A.; Kleinjans, J.C.S.; Malik, R.N.; Briedé, J.J. Transcriptome responses in blood reveal distinct biological pathways associated with arsenic exposure through drinking water in rural settings of Punjab, Pakistan. Environ. Int. 2020, 135, 105403. [Google Scholar] [CrossRef]
- Pallares, R.M.; Faulkner, D.; An, D.D.; Hébert, S.; Loguinov, A.; Proctor, M.; Villalobos, J.A.; Bjornstad, K.A.; Rosen, C.J.; Vulpe, C.; et al. Genome-wide toxicogenomic study of the lanthanides sheds light on the selective toxicity mechanisms associated with critical materials. Proc. Natl. Acad. Sci. USA 2021, 118, e2025952118. [Google Scholar] [CrossRef] [PubMed]
- Sendra, M.; Pereiro, P.; Figueras, A.; Novoa, B. An integrative toxicogenomic analysis of plastic additives. J. Hazard. Mater. 2021, 409, 124975. [Google Scholar] [CrossRef]
- Nault, R.; Bals, B.; Teymouri, F.; Black, M.B.; Andersen, M.E.; McMullen, P.D.; Krishnan, S.; Kuravadi, N.; Paul, N.; Kumar, S.; et al. A toxicogenomic approach for the risk assessment of the food contaminant acetamide. Toxicol. Appl. Pharmacol. 2020, 388, 114872. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Zhan, H.; Hu, J.; Zhang, T.; Xu, H.; Wong, M.; Xu, B.; Zheng, C. The occurrence, potential toxicity, and toxicity mechanism of bisphenol S, a substitute of bisphenol A: A critical review of recent progress. Ecotoxicol. Environ. Saf. 2019, 173, 192–202. [Google Scholar] [CrossRef]
- Stanic, B.; Kokai, D.; Tesic, B.; Fa, S.; Samardzija Nenadov, D.; Pogrmic-Majkic, K.; Andric, N. Integration of data from the in vitro long-term exposure study on human endothelial cells and the in silico analysis: A case of dibutyl phthalate-induced vascular dysfunction. Toxicol. Lett. 2022, 356, 64–74. [Google Scholar] [CrossRef]
- Ventura, C.; Torres, V.; Vieira, L.; Gomes, B.; Rodrigues, A.S.; Rueff, J.; Penque, D.; Silva, M.J. New “Omics” Approaches as Tools to Explore Mechanistic Nanotoxicology. Adv. Exp. Med. Biol. 2022, 1357, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Rolo, D.; Tavares, A.; Vital, N.; Silva, M.J.; Louro, H. Overview of Adverse Outcome Pathways and Current Applications on Nanomaterials. Adv. Exp. Med. Biol. 2022, 1357, 415–439. [Google Scholar] [CrossRef]
- Saarimäki, L.A.; Federico, A.; Lynch, I.; Papadiamantis, A.G.; Tsoumanis, A.; Melagraki, G.; Afantitis, A.; Serra, A.; Greco, D. Manually curated transcriptomics data collection for toxicogenomic assessment of engineered nanomaterials. Sci. Data 2021, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Saquib, Q.; Xia, P.; Siddiqui, M.A.; Zhang, J.; Xie, Y.; Faisal, M.; Ansari, S.M.; Alwathnani, H.A.; Alatar, A.A.; Al-Khedhairy, A.A.; et al. High-throughput transcriptomics: An insight on the pathways affected in HepG2 cells exposed to nickel oxide nanoparticles. Chemosphere 2020, 244, 125488. [Google Scholar] [CrossRef]
- Geppert, M.; Himly, M. Iron Oxide Nanoparticles in Bioimaging—An Immune Perspective. Front. Immunol. 2021, 12, 688927. [Google Scholar] [CrossRef] [PubMed]
- Vo, A.H.; Van Vleet, T.R.; Gupta, R.R.; Liguori, M.J.; Rao, M.S. An Overview of Machine Learning and Big Data for Drug Toxicity Evaluation. Chem. Res. Toxicol. 2020, 33, 20–37. [Google Scholar] [CrossRef]
- Lin, Z.; Chou, W.C. Machine Learning and Artificial Intelligence in Toxicological Sciences. Toxicol. Sci. 2022, 189, 7–19. [Google Scholar] [CrossRef]
- Ochoteco Asensio, J.; Verheijen, M.; Caiment, F. Predicting missing proteomics values using machine learning: Filling the gap using transcriptomics and other biological features. Comput. Struct. Biotechnol. J. 2022, 20, 2057–2069. [Google Scholar] [CrossRef]
- Wang, C.C.; Liang, Y.C.; Wang, S.S.; Lin, P.; Tung, C.W. A machine learning-driven approach for prioritizing food contact chemicals of carcinogenic concern based on complementary in silico methods. Food Chem. Toxicol. 2022, 160, 112802. [Google Scholar] [CrossRef]
- Audouze, K.; Taboureau, O. Emerging Bioinformatics Methods and Resources in Drug Toxicology. Methods Mol. Biol. 2022, 2425, 133–146. [Google Scholar] [CrossRef]
- Schyman, P.; Xu, Z.; Desai, V.; Wallqvist, A. TOXPANEL: A Gene-Set Analysis Tool to Assess Liver and Kidney Injuries. Front. Pharmacol. 2021, 12, 601511. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.K.; Eeles, C.; Ho, C.; Beri, G.; Yoo, E.; Tkachuk, D.; Tang, A.; Nijrabi, P.; Smirnov, P.; Seo, H.; et al. ToxicoDB: An integrated database to mine and visualize large-scale toxicogenomic datasets. Nucleic Acids Res. 2020, 48, W455–W462. [Google Scholar] [CrossRef]
- Zare Jeddi, M.; Hopf, N.B.; Viegas, S.; Price, A.B.; Paini, A.; van Thriel, C.; Benfenati, E.; Ndaw, S.; Bessems, J.; Behnisch, P.A.; et al. Towards a systematic use of effect biomarkers in population and occupational biomonitoring. Environ. Int. 2021, 146, 106257. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Plans, J.; Piñero, J.; Souza, T.; Callegaro, G.; Kunnen, S.J.; Sanz, F.; Fernandez-Fuentes, N.; Furlong, L.I.; Guney, E.; Oliva, B. An ensemble learning approach for modeling the systems biology of drug-induced injury. Biol. Direct. 2021, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Krewski, D.; Andersen, M.E.; Tyshenko, M.G.; Krishnan, K.; Hartung, T.; Boekelheide, K.; Wambaugh, J.F.; Jones, D.; Whelan, M.; Thomas, R.; et al. Toxicity testing in the 21st century: Progress in the past decade and future perspectives. Arch. Toxicol. 2020, 94, 1–58. [Google Scholar] [CrossRef]
| Omics Field | Matrix | Aim | Comment | Omics Assessment | Omics Database | Author |
|---|---|---|---|---|---|---|
| Proteomics | Rat spleen. | To evaluate iron oxide nanoparticles toxicity mechanism. | 197 upregulated and 75 downregulated proteins, the AKT/mTOR/TFEB in splenic macrophages, a signaling route promoted autophagy and lysosomal activation. | Tandem mass tag-labeled quantitative proteomics. | Uniprot, geneontology.org, genome.jp. | Han et al., 2022 [2] |
| Proteomics | Bothrops atrox venom. | To measure changes in the plasma proteome of a model of envenomated mice. | B. atrox pathophysiology is caused by direct venom toxicity and indirect mechanisms derived from the tissue inflammatory response to envenomation, such as thromboinflammatory changes in fat metabolism and disturbances in the cell caused by oxidative stress and effects on pathways that regulate gene expression, survival, and cell cycle. | Shotgun proteomics analysis, LC-MS/MS, nanoelectrospray ionization. | UniProtKB/Swiss-Prot, PatternLab, DAVID. | Cavalcante et al., 2022 [3] |
| Proteomics | Adolescent rat amygdala. | To measure changes in the plasma proteome of a model of envenomated mice. | B. atrox pathophysiology is caused by direct venom toxicity and indirect mechanisms derived from the tissue inflammatory response to envenomation, such as thromboinflammatory changes in fat metabolism and disturbances in the cell caused by oxidative stress and effects on pathways that regulate gene expression, survival, and cell cycle. | LC–MS/MS analysis, electrospray ionization. | Thermo Proteome Discoverer, DAVID. | Alugubelly et al., 2021 [4] |
| Transcriptomics | Rat testis and liver. | To generate apical, hormone, and liver and testis toxicogenomic data following a short-term (14 day) exposure to myclobutanil to compare points of departure from regulatory guideline studies. | Biological effects point of departure from a 14-day study was sensitive apical POD across regulatory guideline studies | Testis and liver transcriptomic as RNA sequencing. | TG-GATES. | LaRocca et al., 2020 [5] |
| Metabolomics | Knockout Ahr−/− mice. | To analyze AHR contributions to metabolism across multiple scales from the organ to the organelle. | AHR interacts in the regulation of 290 metabolites of biochemical pathways from fatty acids, antioxidants, to uremic toxins, which demonstrates different metabolic functions that AHR performs throughout the body. | Global metabolic profiling analysis. | Pubchem, ChemRICH, Gene Expression Omnibus. | Granados et al., 2022 [6] |
| Metabolomics | Mice liver. | To evaluate the combinate toxicity of hexaconazole and Arsenic (As). | As modifies amino-acid-related pathways and metabolites linked to nerve disease, an insufficient supply of energy, and a functional disorder of the liver. | UPLC-MS/MS, electrospray ion, untargeted metabolomics. | BiotreeDB. | Sun et al., 2021 [7] |
| Metabolomics | Human plasma. | To identify plasma metabolite changes under conditions of high Pb concentration and low cognition. | 20 dysregulated metabolites were found, and GO analysis revealed their significance for both normal brain function and the development of neurodegenerative illnesses such as Parkinson’s disease. | LC-MS, untargeted metabolomic analysis. | Online Human Metabolome Database. | Wang et al., 2022 [8] |
| Epigenomics | Buccal and peripheral blood mononuclear cells. | To standardize preprocessing data pipelines and statistical methods to detect As related DNAm signatures. | As exposure has DNA methylation signatures in individual regions and pathways also mechanisms related to As toxicity. | 850 k and 450 k microarray, epigenome-wide association studies. | Kyoto Encyclopedia of Genes and Genomes. | Bozack et al., 2021 [9] |
| Epigenomics | Brain tissue of C57BL/6 wild-type and APP/PS mice. | To evaluate the As epigenomic alterations in pathophysiological neural changes, in particular histone methylation profile manifesting as cognitive decline. | Developmental As exposure affects histone modifications in the brain that persist into adulthood. A potential mechanism of As exposure influencing cognitive function suppressing biological processes related to neuronal development. | Chromatin immunoprecipitation and sequencing. | Database for Annotation, Visualization and Integrated Discovery (DAVID). | Fitz et al., 2022 [10] |
| Epigenomics | Adult whole-blood-derived DNA. | To evaluate potential DNA methylation processes and correlations between heavy metals and impairment of activities of daily living (ADL). | In older people, a higher incidence of ADL impairment was linked to higher exposures to manganese, copper, arsenic, and cadmium. Possible DNA methylation pathways for various heavy metals and possible biomarkers of ADL were revealed by epigenome-wide associations. | Human methylation EPIC bead chip array, EWAS. | Kyoto Encyclopedia of Genes and Genomes. | Xiao et al., 2022 [11] |
| Transcriptomic | Human colon mucosal epithelial cells (NCM460). | To examine possible outcomes and underlying processes of exposure to bisphenol S (BPS) on human colon mucosal epithelial cells (NCM460). | The gut–brain axis may become out of balance as a result of exposure to BPS, which could cause the NCM460 cells to produce inflammatory cytokines and the destruction of tight junctions. | A multiomics study. | Ao et al., 2021 [12] | |
| Transcriptomic | The hTERT-immortalized human urothelial cell line (TRT-HU1). | To identify and validate potential candidate biomarkers for the detection, prognosis, and treatment of bladder cancer by demonstrating the gene-expression patterns and transformative changes. | Stem cell activators may be crucial in helping arsenic-exposed cells obtain a survival edge, allowing healthy epithelial cells to reprogram into a cancer stem cell phenotype, and ultimately resulting in malignant transformation. | For high-quality mapping reads to the reference genome, spliced transcripts were aligned to a reference (STAR version 2.7) software. | STAR version 2.7 is free open-source software distributed under GPLv3 license and can be downloaded from http://code.google.com/p/rna-star/ (accessed on 25 April 2023). | Shukla et al., 2022 [13] |
| Proteomic | Parotid, submandibular (SM) sublingual (SL) glands of rat. | To identify and validate potential candidate biomarkers for the detection, prognosis, and treatment of bladder cancer by demonstrating the gene-expression patterns and transformative changes. | Stem cell activators may be crucial in helping arsenic-exposed cells obtain a survival edge, allowing healthy epithelial cells to reprogram into a cancer stem cell phenotype, and ultimately resulting in malignant transformation. | Mass spectrometry. | networkanalyst.ca. | Cunha Nascimento et al., 2021 [14] |
| Proteomic | Blood of zebrafish adults (Danio rerio, AB-wild type). | To research multiplexed quantitative proteomics for zebrafish liver proteome profiling using isobaric tags. | Aldehyde dehydrogenase expression was downregulated, which caused a buildup of aldehydes, which decreased the expression of glyceraldehyde 3-phosphate dehydrogenase and disrupted glucose homeostasis. | Liquid chromatography mass spectrometry. | Gene Ontology (GO) annotation dataset was derived from DAVID (https://david.ncifcrf.gov). | Gao et al., 2022 [15] |
| Metabolomic | BEAS-2B noncancerous human bronchial epithelial cells. | To treat human bronchial epithelial cells with As for 6 to 24 weeks and assess spatiotemporal metabolic patterns of biomolecules, cofactors, and xenobiotics through global untargeted metabolic analysis. | In the cells treated with As3+ for 6 to 13 weeks, a significant suppression of all metabolites in the mitochondrial tricarboxylic acid (TCA) cycle was observed. This thorough metabolomics investigation offers fresh perspectives on the metabolic disruption caused by As and may result in more precise evidence of its role in molecular carcinogenesis. | 584 molecules with known identities are included in the metabolomics collection and are referred to as “biochemicals”. | UCSC genome browser. | Fu et al., 2022 [16] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Recio-Vega, R.; Facio-Campos, R.A.; Hernández-González, S.I.; Olivas-Calderón, E. State of the Art of Genomic Technology in Toxicology: A Review. Int. J. Mol. Sci. 2023, 24, 9618. https://doi.org/10.3390/ijms24119618
Recio-Vega R, Facio-Campos RA, Hernández-González SI, Olivas-Calderón E. State of the Art of Genomic Technology in Toxicology: A Review. International Journal of Molecular Sciences. 2023; 24(11):9618. https://doi.org/10.3390/ijms24119618
Chicago/Turabian StyleRecio-Vega, Rogelio, Rolando Adair Facio-Campos, Sandra Isabel Hernández-González, and Edgar Olivas-Calderón. 2023. "State of the Art of Genomic Technology in Toxicology: A Review" International Journal of Molecular Sciences 24, no. 11: 9618. https://doi.org/10.3390/ijms24119618





