Intratumoral Restoration of miR-137 Plus Cholesterol Favors Homeostasis of the miR-137/Coactivator p160/AR Axis and Negatively Modulates Tumor Progression in Advanced Prostate Cancer
Abstract
:1. Introduction
2. Results
2.1. Transfection Efficiency with miR-137 Mimic by qPCR
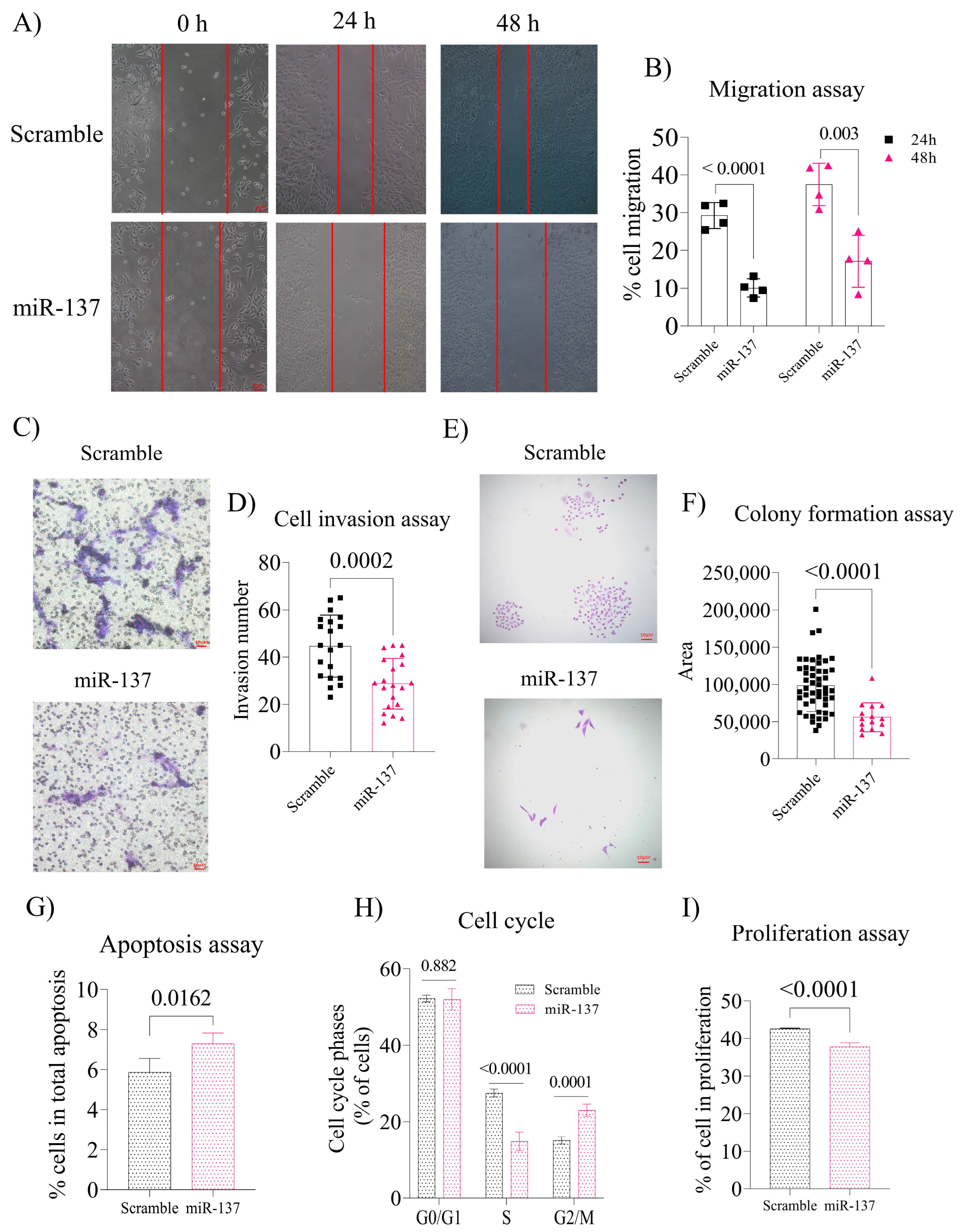
2.2. Exogenous Increase of miR-137 in the PC-3 Cells Negatively Regulates Cell Migration and Invasion and Decreases Colony Formation by Altering the Cell Cycle and Inducing Apoptosis
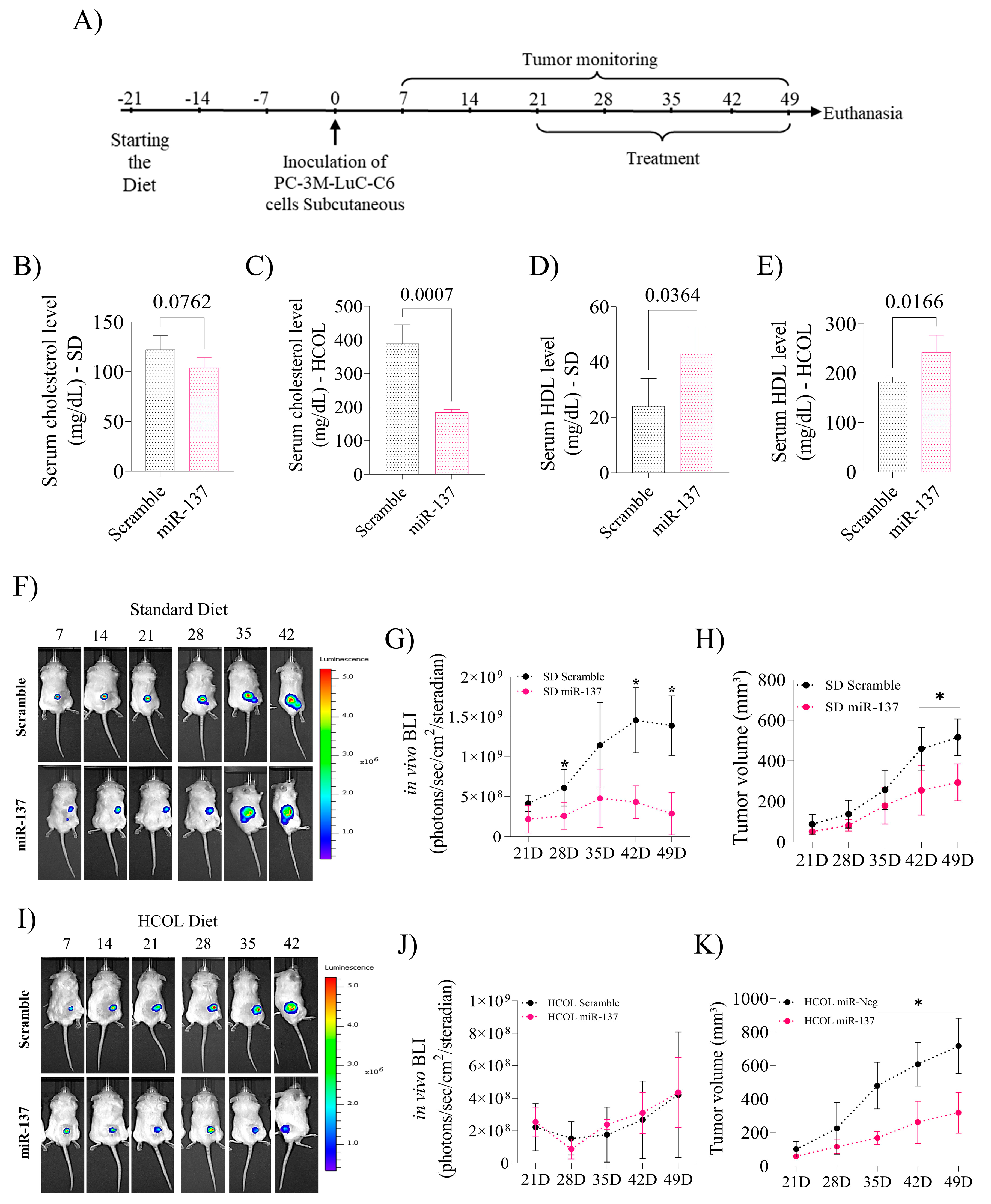
2.3. Induction of miR-137 Expression Attenuates Serum Cholesterol Levels and Favors an Increase in HDL, Reducing Prostate Xenograft Tumor Growth
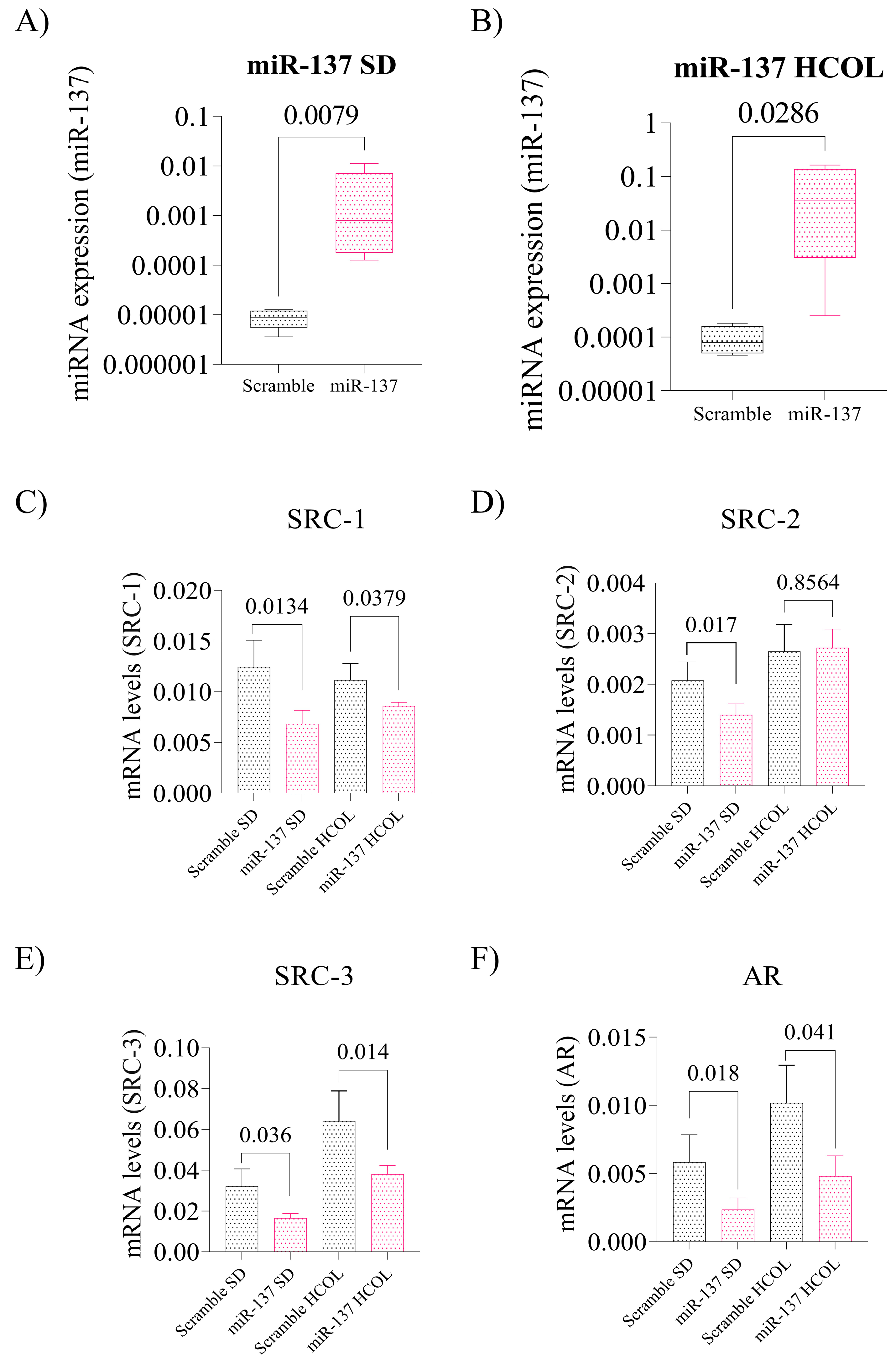
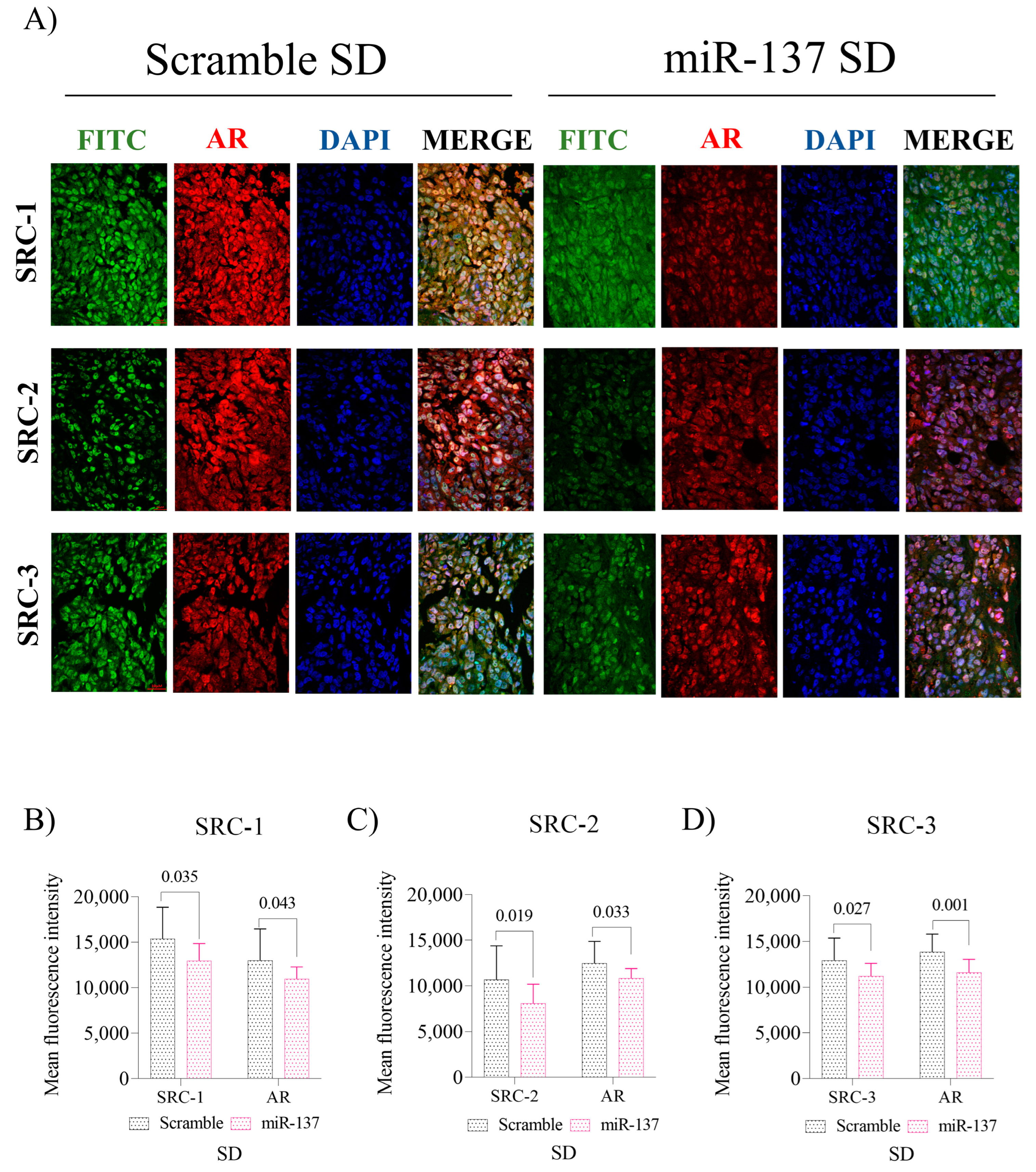
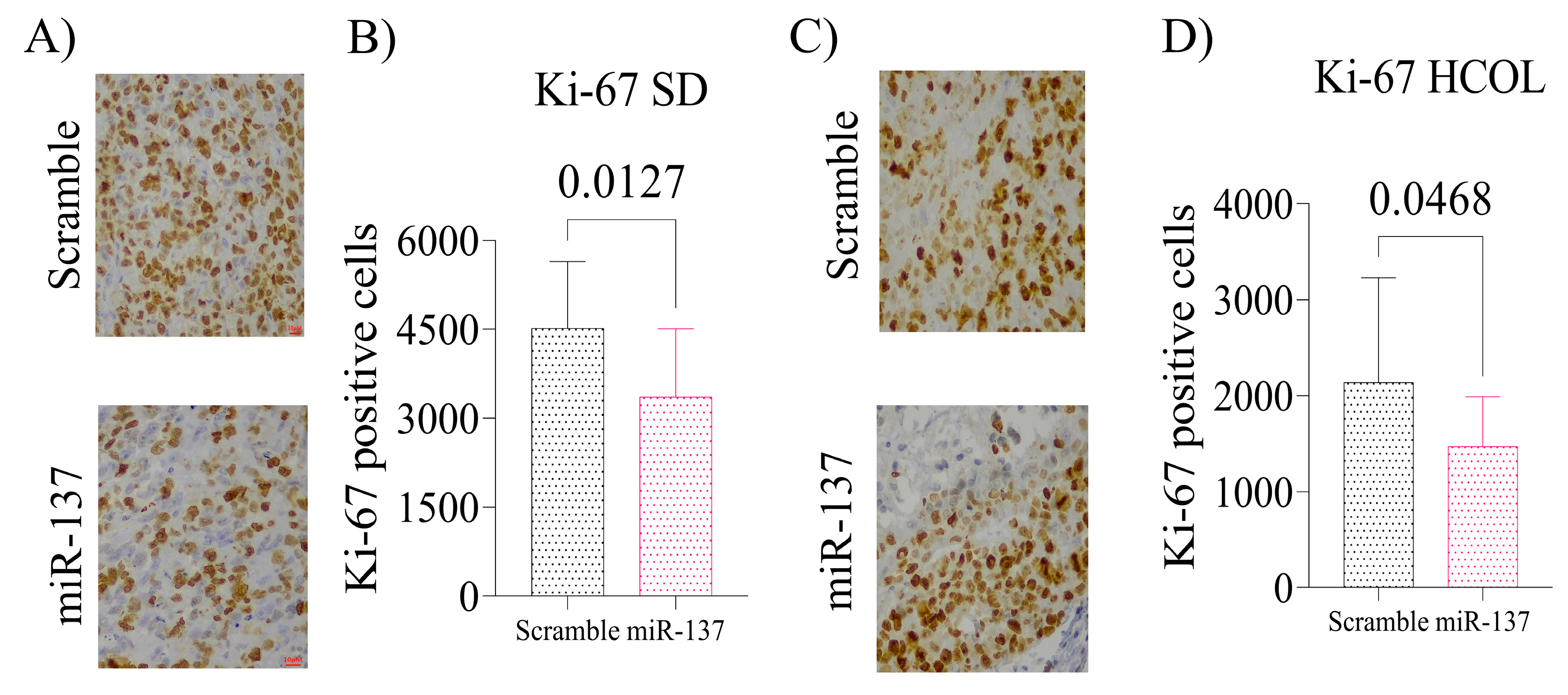
2.4. Intratumoral Reinstatement of the miR-137 Leads to Reduced Expression of p160 Family Proteins and Negatively Impacts AR Expression in Advanced PCa Xenografts, Leading to Reduced Tissue Proliferation
3. Discussion
4. Materials and Methods
4.1. TargetScan, miRDB, and miRmap Databases
4.2. Cell Transfection
4.3. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
4.4. Apoptosis and Cell Cycle Analyses by Flow Cytometry
4.5. Cell Migration Assay
4.6. Colony Formation Assay
4.7. Invasion Assays
4.8. Immunofluorescence In Vitro
4.9. In Vivo Study
4.10. Lipid Profile
4.11. Immunohistochemical Assay
4.12. Immunofluorescence In Vivo
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Citron, F.; Armenia, J.; Franchin, G.; Polesel, J.; Talamini, R.; D’Andrea, S.; Sulfaro, S.; Croce, C.M.; Klement, W.; Otasek, D.; et al. An Integrated Approach Identifies Mediators of Local Recurrence in Head and Neck Squamous Carcinoma. Clin. Cancer Res. 2017, 23, 3769–3780. [Google Scholar] [CrossRef] [PubMed]
- Yoon, A.J.; Wang, S.; Kutler, D.I.; Carvajal, R.D.; Dmd, E.P.; Wang, T.; Peters, S.M.; LaRoche, D.; Hernandez, B.Y.; McDowell, B.D.; et al. MicroRNA-based risk scoring system to identify early-stage oral squamous cell carcinoma patients at high-risk for cancer-specific mortality. Head Neck 2020, 42, 1699–1712. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. MicroRNAs in Cancer. Annu. Rev. Pathol. Mech. Dis. 2009, 4, 199–227. [Google Scholar] [CrossRef]
- Suzuki, H.; Maruyama, R.; Yamamoto, E.; Kai, M. DNA methylation and microRNA dysregulation in cancer. Mol. Oncol. 2012, 6, 567–578. [Google Scholar] [CrossRef]
- Nilsson, E.M.; Laursen, K.B.; Whitchurch, J.; McWilliam, A.; Ødum, N.; Persson, J.L.; Heery, D.M.; Gudas, L.J.; Mongan, N.P. MiR137 is an androgen regulated repressor of an extended network of transcriptional coregulators. Oncotarget 2015, 6, 35710–35725. [Google Scholar] [CrossRef]
- Balaguer, F.; Link, A.; Lozano, J.J.; Cuatrecasas, M.; Nagasaka, T.; Boland, C.R.; Goel, A. Epigenetic Silencing of miR-137 Is an Early Event in Colorectal Carcinogenesis. Cancer Res. 2010, 70, 6609–6618. [Google Scholar] [CrossRef]
- Eedunuri, V.K.; Rajapakshe, K.; Fiskus, W.; Geng, C.; Chew, S.A.; Foley, C.; Shah, S.S.; Shou, J.; Mohamed, J.S.; Coarfa, C.; et al. miR-137 Targets p160 Steroid Receptor Coactivators SRC1, SRC2, and SRC3 and Inhibits Cell Proliferation. Mol. Endocrinol. 2015, 29, 1170–1183. [Google Scholar] [CrossRef]
- Spencer, T.E.; Jenster, G.; Burcin, M.M.; Allis, C.D.; Zhou, J.; Mizzen, C.A.; McKenna, N.J.; Onate, S.A.; Tsai, S.Y.; Tsai, M.-J.; et al. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 1997, 389, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Ylitalo, E.B.; Thysell, E.; Jernberg, E.; Lundholm, M.; Crnalic, S.; Egevad, L.; Stattin, P.; Widmark, A.; Bergh, A.; Wikström, P. Subgroups of Castration-resistant Prostate Cancer Bone Metastases Defined Through an Inverse Relationship Between Androgen Receptor Activity and Immune Response. Eur. Urol. 2017, 71, 776–787. [Google Scholar] [CrossRef]
- Pimenta, R.; Camargo, J.A.; Candido, P.; Ghazarian, V.; Gonçalves, G.L.; Guimarães, V.R.; Romão, P.; Chiovatto, C.; Mioshi, C.M.; dos Santos, G.A.; et al. Cholesterol Triggers Nuclear Co-Association of Androgen Receptor, p160 Steroid Coactivators, and p300/CBP-Associated Factor Leading to Androgenic Axis Transactivation in Castration-Resistant Prostate Cancer. Cell. Physiol. Biochem. 2022, 56, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chandra, N.C.; Singh, G.; Sankanagoudar, S.; Dogra, P. Interlink between cholesterol & cell cycle in prostate carcinoma. Indian J. Med. Res. 2017, 146, S38–S44. [Google Scholar] [CrossRef]
- Li, X.; Wu, J.B.; Li, Q.; Shigemura, K.; Chung, L.W.; Huang, W.-C. SREBP-2 promotes stem cell-like properties and metastasis by transcriptional activation of c-Myc in prostate cancer. Oncotarget 2016, 7, 12869–12884. [Google Scholar] [CrossRef] [PubMed]
- Pisano, C.; Tucci, M.; Di Stefano, R.F.; Turco, F.; Scagliotti, G.V.; Di Maio, M.; Buttigliero, C. Interactions between androgen receptor signaling and other molecular pathways in prostate cancer progression: Current and future clinical implications. Crit. Rev. Oncol. 2021, 157, 103185. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, T.; Yang, J.C.; Gao, A.C.; Evans, C.P. Targeting molecular resistance in castration-resistant prostate cancer. BMC Med. 2015, 13, 206. [Google Scholar] [CrossRef]
- Varkaris, A.; Katsiampoura, A.D.; Araujo, J.C.; Gallick, G.E.; Corn, P.G. Src signaling pathways in prostate cancer. Cancer Metastasis Rev. 2014, 33, 595–606. [Google Scholar] [CrossRef]
- Shtivelman, E.; Beer, T.M.; Evans, C.P. Molecular pathways and targets in prostate cancer. Oncotarget 2014, 5, 7217–7259. [Google Scholar] [CrossRef]
- Stoykova, G.E.; Schlaepfer, I.R. Lipid Metabolism and Endocrine Resistance in Prostate Cancer, and New Opportunities for Therapy. Int. J. Mol. Sci. 2019, 20, 2626. [Google Scholar] [CrossRef]
- Rane, J.K.; Scaravilli, M.; Ylipää, A.; Pellacani, D.; Mann, V.M.; Simms, M.S.; Nykter, M.; Collins, A.T.; Visakorpi, T.; Maitland, N.J. MicroRNA Expression Profile of Primary Prostate Cancer Stem Cells as a Source of Biomarkers and Therapeutic Targets. Eur. Urol. 2015, 67, 7–10. [Google Scholar] [CrossRef]
- Iscaife, A.; Reis, S.T.; Morais, D.R.; Viana, N.I.; da Silva, I.A.; Pimenta, R.; Bordini, A.; Dip, N.; Srougi, M.; Leite, K.R.M. Treating metastatic prostate cancer with microRNA-145. Apoptosis 2018, 23, 388–395. [Google Scholar] [CrossRef]
- Pimenta, R.C.; Viana, N.I.; Amaral, G.Q.; Park, R.; Morais, D.R.; Pontes, J.J.; Guimaraes, V.R.; Camargo, J.A.; Leite, K.R.; Nahas, W.C.; et al. MicroRNA-23b and microRNA-27b plus flutamide treatment enhances apoptosis rate and decreases CCNG1 expression in a castration-resistant prostate cancer cell line. Tumor Biol. 2018, 40, 1010428318803011. [Google Scholar] [CrossRef]
- Leite, K.R.; Tomiyama, A.; Reis, S.; Sousa-Canavez, J.M.; Sanudo, A.; Camara-Lopes, L.H.; Srougi, M. MicroRNA expression profiles in the progression of prostate cancer—From high-grade prostate intraepithelial neoplasia to metastasis. Urol. Oncol. Semin. Orig. Investig. 2013, 31, 796–801. [Google Scholar] [CrossRef] [PubMed]
- Leite, K.R.; Tomiyama, A.; Reis, S.; Sousa-Canavez, J.M.; Sañudo, A.; Dall’Oglio, M.F.; Camara-Lopes, L.H.; Srougi, M. MicroRNA-100 Expression is Independently Related to Biochemical Recurrence of Prostate Cancer. J. Urol. 2011, 185, 1118–1122. [Google Scholar] [CrossRef] [PubMed]
- Katz, B.; Reis, S.T.; Viana, N.I.; Morais, D.R.; Moura, C.M.; Dip, N.; Silva, I.A.; Iscaife, A.; Srougi, M.; Leite, K.R.M. Comprehensive Study of Gene and microRNA Expression Related to Epithelial-Mesenchymal Transition in Prostate Cancer. PLoS ONE 2014, 9, e113700. [Google Scholar] [CrossRef]
- Leite, K.R.M.; Morais, D.R.; Reis, S.T.; Viana, N.; Moura, C.; Florez, M.G.; Silva, I.A.; Dip, N.; Srougi, M. MicroRNA 100: A context dependent miRNA in prostate cancer. Clinics 2013, 68, 797–802. [Google Scholar] [CrossRef] [PubMed]
- de Camargo, J.A.; Lopes, R.E.; Ferreira, G.F.D.; Viana, N.I.; Guimaraes, V.; Leite, K.R.M.; Nahas, W.C.; Srougi, M.; Antunes, A.A.; Reis, S.T. The role of single nucleotide polymorphisms of miRNAs 100 and 146a as prognostic factors for prostate cancer. Int. J. Biol. Markers 2021, 36, 50–56. [Google Scholar] [CrossRef] [PubMed]
- de Moraes, R.P.; Pimenta, R.; Mori, F.N.C.; Dos Santos, G.A.; Viana, N.I.; Guimarães, V.R.; de Camargo, J.A.; Leite, K.R.M.; Srougi, M.; Nahas, W.C.; et al. Tissue expression of MMP-9, TIMP-1, RECK, and miR338-3p in prostate gland: Can it predict cancer? Mol. Biol. Res. Commun. 2021, 10, 149–156. [Google Scholar] [CrossRef]
- Ivanovic, R.F.; Viana, N.I.; Morais, D.R.; Silva, I.A.; Leite, K.R.; Pontes-Junior, J.; Inoue, G.; Nahas, W.C.; Srougi, M.; Reis, S.T. miR-29b enhances prostate cancer cell invasion independently of MMP-2 expression. Cancer Cell Int. 2018, 18, 18. [Google Scholar] [CrossRef]
- Ivanovic, R.F.; Viana, N.I.; Morais, D.R.; Moura, C.; Silva, I.A.; Leite, K.R.; Pontes-Junior, J.; Nahas, W.C.; Srougi, M.; Reis, S.T. miR-618: Possible control over TIMP-1 and its expression in localized prostate cancer. BMC Cancer 2018, 18, 992. [Google Scholar] [CrossRef]
- Reis, S.T.; Timoszczuk, L.S.; Pontes-Junior, J.; Viana, N.; Silva, I.A.; Dip, N.; Srougi, M.; Leite, K.R.M. The role of micro RNAs let7c, 100 and 218 expression and their target RAS, C-MYC, BUB1, RB, SMARCA5, LAMB3 and Ki-67 in prostate cancer. Clinics 2013, 68, 652–657. [Google Scholar] [CrossRef]
- Leite, K.R.; Morais, D.R.; Florez, M.G.; Reis, S.T.; Iscaife, A.; Viana, N.; Moura, C.M.; Silva, I.A.; Katz, B.S.; Pontes, J.; et al. The role of microRNAs 371 and 34a in androgen receptor control influencing prostate cancer behavior. Urol. Oncol. Semin. Orig. Investig. 2015, 33, 267.e15–267.e22. [Google Scholar] [CrossRef]
- Dos Reis, S.T.; Júnior, N.G.D.; Antunes, A.A.; Dall’oglio, M.F.; Dip, N.; Passerotti, C.C.; Rossini, G.A.; Morais, D.R.; Nesrallah, A.J.; Piantino, C.B.; et al. miR-21 may acts as an oncomir by targeting RECK, a matrix metalloproteinase regulator, in prostate cancer. BMC Urol. 2012, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Leite, K.R.; Sousa-Canavez, J.M.; Reis, S.T.; Tomiyama, A.H.; Camara-Lopes, L.H.; Sañudo, A.; Antunes, A.A.; Srougi, M. Change in expression of miR-let7c, miR-100, and miR-218 from high grade localized prostate cancer to metastasis. Urol. Oncol. Semin. Orig. Investig. 2011, 29, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, A.; Likos, E.M.; Weyman, C.M.; Shukla, G.C. Regulation of cholesterol biosynthesis and lipid metabolism: A microRNA management perspective. Steroids 2021, 173, 108878. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef]
- Vejnar, C.E.; Zdobnov, E.M. MiRmap: Comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 2012, 40, 11673–11683. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pimenta, R.; Mioshi, C.M.; Gonçalves, G.L.; Candido, P.; Camargo, J.A.; Guimarães, V.R.; Chiovatto, C.; Ghazarian, V.; Romão, P.; da Silva, K.S.; et al. Intratumoral Restoration of miR-137 Plus Cholesterol Favors Homeostasis of the miR-137/Coactivator p160/AR Axis and Negatively Modulates Tumor Progression in Advanced Prostate Cancer. Int. J. Mol. Sci. 2023, 24, 9633. https://doi.org/10.3390/ijms24119633
Pimenta R, Mioshi CM, Gonçalves GL, Candido P, Camargo JA, Guimarães VR, Chiovatto C, Ghazarian V, Romão P, da Silva KS, et al. Intratumoral Restoration of miR-137 Plus Cholesterol Favors Homeostasis of the miR-137/Coactivator p160/AR Axis and Negatively Modulates Tumor Progression in Advanced Prostate Cancer. International Journal of Molecular Sciences. 2023; 24(11):9633. https://doi.org/10.3390/ijms24119633
Chicago/Turabian StylePimenta, Ruan, Carolina Mie Mioshi, Guilherme L. Gonçalves, Patrícia Candido, Juliana A. Camargo, Vanessa R. Guimarães, Caroline Chiovatto, Vitória Ghazarian, Poliana Romão, Karina Serafim da Silva, and et al. 2023. "Intratumoral Restoration of miR-137 Plus Cholesterol Favors Homeostasis of the miR-137/Coactivator p160/AR Axis and Negatively Modulates Tumor Progression in Advanced Prostate Cancer" International Journal of Molecular Sciences 24, no. 11: 9633. https://doi.org/10.3390/ijms24119633





