The Recent Research Progress of NF-κB Signaling on the Proliferation, Migration, Invasion, Immune Escape and Drug Resistance of Glioblastoma
Abstract
:1. Introduction
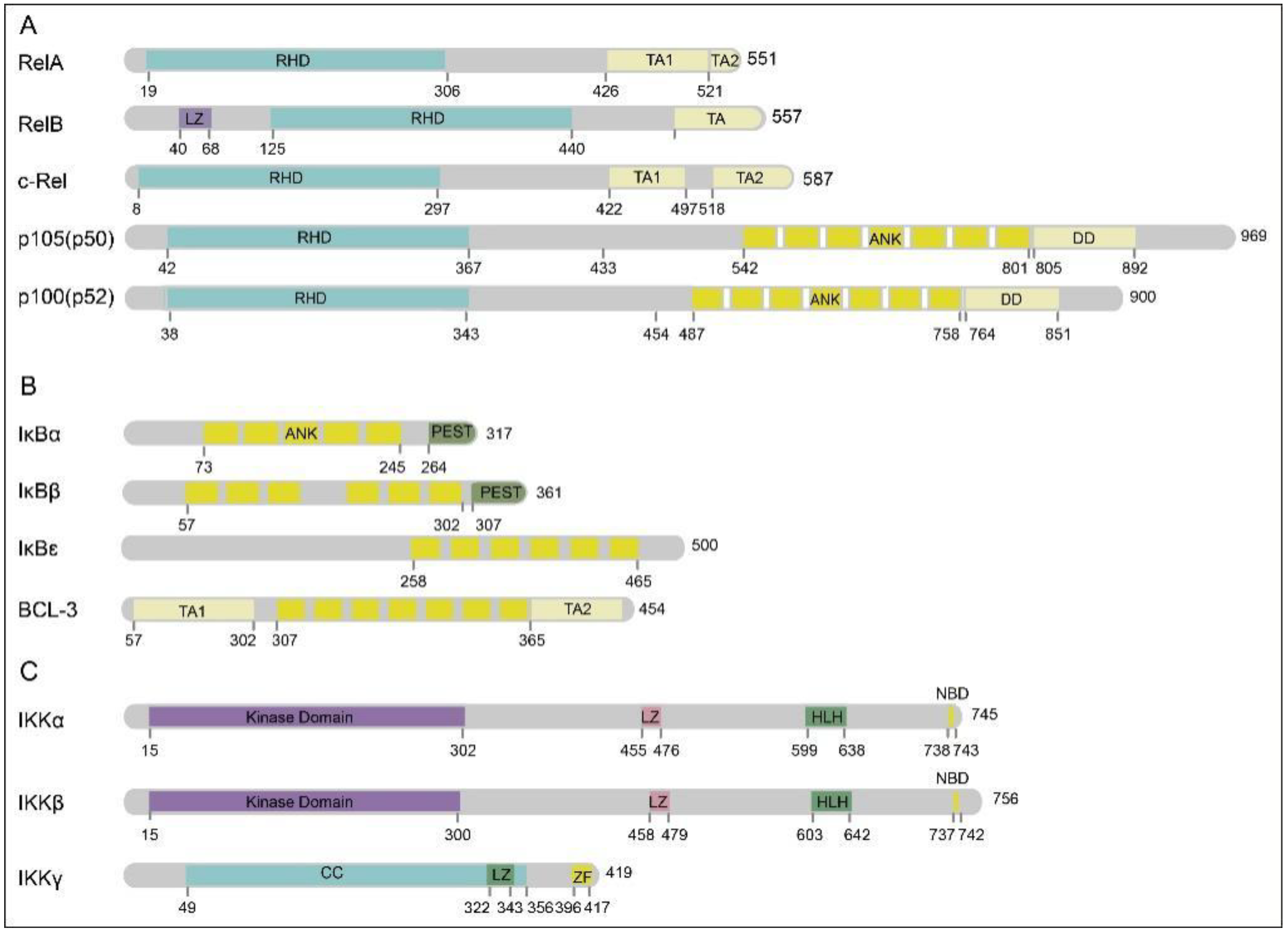
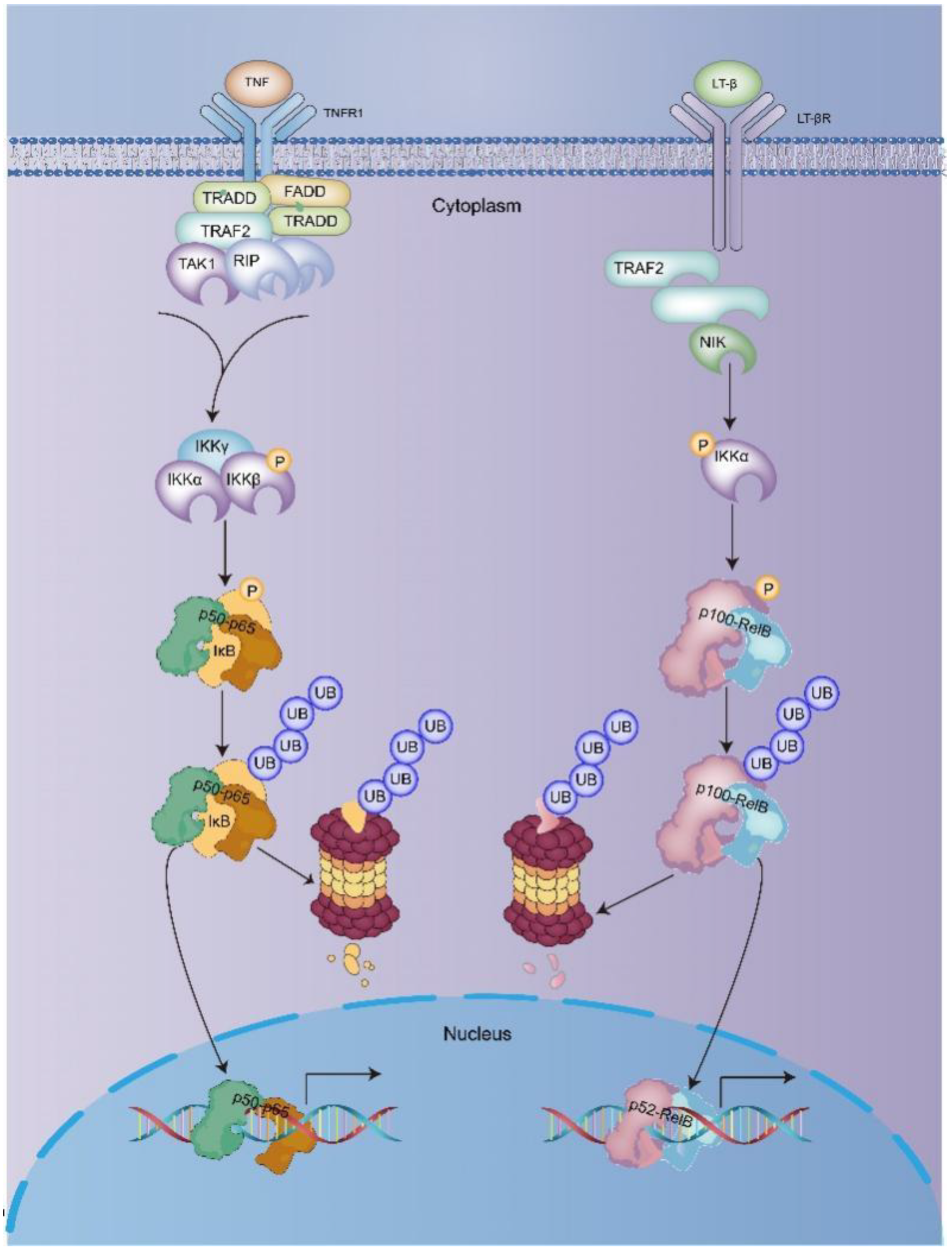
2. NF-κB
3. NF-κB Activation Is Involved in Development and Progression of Glioblastoma
3.1. Inflammation and Proliferation
3.2. Migration and Invasion
3.3. Immune Escape
3.4. Drug Resistance
3.4.1. Promoting Resistance
3.4.2. Inhibiting Resistance
3.5. Apoptosis
4. Summary and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shi, P.; Xu, J.; Xia, F.; Wang, Y.; Ren, J.; Liang, P.; Cui, H. MOXD1 knockdown suppresses the proliferation and tumor growth of glioblastoma cells via ER stress-inducing apoptosis. Cell Death Discov. 2022, 8, 174. [Google Scholar] [CrossRef] [PubMed]
- Michaud, K.; Duffau, H. Surgery of insular and paralimbic diffuse low-grade gliomas: Technical considerations. J. Neuro-Oncol. 2016, 130, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Frosina, G. Recapitulating the Key Advances in the Diagnosis and Prognosis of High-Grade Gliomas: Second Half of 2021 Update. Int. J. Mol. Sci. 2023, 24, 6375. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Min, J.S.; Kim, B.; Chae, U.B.; Yun, J.W.; Choi, M.S.; Kong, I.K.; Chang, K.T.; Lee, D.S. Mitochondrial ROS govern the LPS-induced pro-inflammatory response in microglia cells by regulating MAPK and NF-κB pathways. Neurosci. Lett. 2015, 584, 191–196. [Google Scholar] [CrossRef]
- Mitchell, S.; Vargas, J.; Hoffmann, A. Signaling via the NFκB system. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016, 8, 227–241. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Kadhim, M.M.; Turki Jalil, A.; Obayes, A.M.; Aminov, Z.; Alsaikhan, F.; Ramírez-Coronel, A.A.; Ramaiah, P.; Tayyib, N.A.; Luo, X. Multifaceted role of NF-κB in hepatocellular carcinoma therapy: Molecular landscape, therapeutic compounds and nanomaterial approaches. Environ. Res. 2023, 228, 115767. [Google Scholar] [CrossRef]
- Pavitra, E.; Kancharla, J.; Gupta, V.K.; Prasad, K.; Sung, J.Y.; Kim, J.; Tej, M.B.; Choi, R.; Lee, J.H.; Han, Y.K.; et al. The role of NF-κB in breast cancer initiation, growth, metastasis, and resistance to chemotherapy. Biomed. Pharmacother. Biomed. Pharmacother. 2023, 163, 114822. [Google Scholar] [CrossRef]
- Sripathi, S.R.; Hu, M.W.; Turaga, R.C.; Mikeasky, R.; Satyanarayana, G.; Cheng, J.; Duan, Y.; Maruotti, J.; Wahlin, K.J.; Berlinicke, C.A.; et al. IKKβ Inhibition Attenuates Epithelial Mesenchymal Transition of Human Stem Cell-Derived Retinal Pigment Epithelium. Cells 2023, 12, 1155. [Google Scholar] [CrossRef]
- Reicherz, A.; Eltit, F.; Scotland, K.; Almutairi, K.; Bell, R.; Mojtahedzadeh, B.; Cox, M.; Chew, B.; Lange, D. Indwelling stents cause severe inflammation and fibrosis of the ureter via urothelial-mesenchymal transition. Sci. Rep. 2023, 13, 5492. [Google Scholar] [CrossRef]
- Moretti, M.; Bennett, J.; Tornatore, L.; Thotakura, A.K.; Franzoso, G. Cancer: NF-κB regulates energy metabolism. Int. J. Biochem. Cell Biol. 2012, 44, 2238–2243. [Google Scholar] [CrossRef]
- Pflug, K.M.; Sitcheran, R. Targeting NF-κB-Inducing Kinase (NIK) in Immunity, Inflammation, and Cancer. Int. J. Mol. Sci. 2020, 21, 8470. [Google Scholar] [CrossRef]
- Visekruna, A.; Volkov, A.; Steinhoff, U. A key role for NF-κB transcription factor c-Rel in T-lymphocyte-differentiation and effector functions. Clin. Dev. Immunol. 2012, 2012, 239368. [Google Scholar] [CrossRef] [Green Version]
- Pereira, S.G.; Oakley, F. Nuclear factor-kappaB1: Regulation and function. Int. J. Biochem. Cell Biol. 2008, 40, 1425–1430. [Google Scholar] [CrossRef] [PubMed]
- Savinova, O.V.; Hoffmann, A.; Ghosh, G. The Nfkb1 and Nfkb2 proteins p105 and p100 function as the core of high-molecular-weight heterogeneous complexes. Mol. Cell 2009, 34, 591–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neri, A.; Fracchiolla, N.S.; Migliazza, A.; Trecca, D.; Lombardi, L. The involvement of the candidate proto-oncogene NFKB2/lyt-10 in lymphoid malignancies. Leuk. Lymphoma 1996, 23, 3–48. [Google Scholar] [CrossRef]
- Gálvez-Rodríguez, A.; Ferino-Pérez, A.; Rodríguez-Riera, Z.; Rodeiro Guerra, I.; Řeha, D.; Minofar, B.; Jáuregui-Haza, U.J. Explaining the interaction of mangiferin with MMP-9 and NF-ƙβ: A computational study. J. Mol. Model. 2022, 28, 266. [Google Scholar] [CrossRef] [PubMed]
- Knuefermann, P.; Chen, P.; Misra, A.; Shi, S.P.; Abdellatif, M.; Sivasubramanian, N. Myotrophin/V-1, a protein up-regulated in the failing human heart and in postnatal cerebellum, converts NFkappa B p50-p65 heterodimers to p50-p50 and p65-p65 homodimers. J. Biol. Chem. 2002, 277, 23888–23897. [Google Scholar] [CrossRef] [Green Version]
- May, M.J.; Ghosh, S. Rel/NF-kappa B and I kappa B proteins: An overview. Semin. Cancer Biol. 1997, 8, 63–73. [Google Scholar] [CrossRef]
- Gehrke, N.; Wörns, M.A.; Mann, A.; Hövelmeyer, N.; Waisman, A.; Straub, B.K.; Galle, P.R.; Schattenberg, J.M. Hepatocyte Bcl-3 protects from death-receptor mediated apoptosis and subsequent acute liver failure. Cell Death Dis. 2022, 13, 510. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, K.M.; Chen, W.; Marion, J.D.; Bergqvist, S.; Komives, E.A. Exclusivity and Compensation in NFκB Dimer Distributions and IκB Inhibition. Biochemistry 2019, 58, 2555–2563. [Google Scholar] [CrossRef] [PubMed]
- Marienfeld, R.; May, M.J.; Berberich, I.; Serfling, E.; Ghosh, S.; Neumann, M. RelB forms transcriptionally inactive complexes with RelA/p65. J. Biol. Chem. 2003, 278, 19852–19860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoesel, B.; Schmid, J.A. The complexity of NF-κB signaling in inflammation and cancer. Mol. Cancer 2013, 12, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, M.; Yamazaki, S.; Uematsu, S.; Sato, S.; Hemmi, H.; Hoshino, K.; Kaisho, T.; Kuwata, H.; Takeuchi, O.; Takeshige, K.; et al. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature 2004, 430, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Ben-Neriah, Y. Phosphorylation meets ubiquitination: The control of NF-[kappa]B activity. Annu. Rev. Immunol. 2000, 18, 621–663. [Google Scholar] [CrossRef]
- Roh, Y.S.; Song, J.; Seki, E. TAK1 regulates hepatic cell survival and carcinogenesis. J. Gastroenterol. 2014, 49, 185–194. [Google Scholar] [CrossRef] [Green Version]
- Aashaq, S.; Batool, A.; Andrabi, K.I. TAK1 mediates convergence of cellular signals for death and survival. Apoptosis Int. J. Program. Cell Death 2019, 24, 3–20. [Google Scholar] [CrossRef]
- Che, W.; Lerner-Marmarosh, N.; Huang, Q.; Osawa, M.; Ohta, S.; Yoshizumi, M.; Glassman, M.; Lee, J.D.; Yan, C.; Berk, B.C.; et al. Insulin-like growth factor-1 enhances inflammatory responses in endothelial cells: Role of Gab1 and MEKK3 in TNF-alpha-induced c-Jun and NF-kappaB activation and adhesion molecule expression. Circ. Res. 2002, 90, 1222–1230. [Google Scholar] [CrossRef] [Green Version]
- Lei, C.Q.; Wu, X.; Zhong, X.; Jiang, L.; Zhong, B.; Shu, H.B. USP19 Inhibits TNF-α- and IL-1β-Triggered NF-κB Activation by Deubiquitinating TAK1. J. Immunol. 2019, 203, 259–268. [Google Scholar] [CrossRef]
- Vendel, A.C.; Calemine-Fenaux, J.; Izrael-Tomasevic, A.; Chauhan, V.; Arnott, D.; Eaton, D.L. B and T lymphocyte attenuator regulates B cell receptor signaling by targeting Syk and BLNK. J. Immunol. 2009, 182, 1509–1517. [Google Scholar] [CrossRef] [Green Version]
- Heyninck, K.; Beyaert, R. Crosstalk between NF-kappaB-activating and apoptosis-inducing proteins of the TNF-receptor complex. Mol. Cell Biol. Res. Commun. MCBRC 2001, 4, 259–265. [Google Scholar] [CrossRef]
- Verstrepen, L.; Bekaert, T.; Chau, T.L.; Tavernier, J.; Chariot, A.; Beyaert, R. TLR-4, IL-1R and TNF-R signaling to NF-kappaB: Variations on a common theme. Cell. Mol. Life Sci. CMLS 2008, 65, 2964–2978. [Google Scholar] [CrossRef] [PubMed]
- Turi, M.; Anilkumar Sithara, A.; Hofmanová, L.; Žihala, D.; Radhakrishnan, D.; Vdovin, A.; Knápková, S.; Ševčíková, T.; Chyra, Z.; Jelínek, T.; et al. Transcriptome Analysis of Diffuse Large B-Cell Lymphoma Cells Inducibly Expressing MyD88 L265P Mutation Identifies Upregulated CD44, LGALS3, NFKBIZ, and BATF as Downstream Targets of Oncogenic NF-κB Signaling. Int. J. Mol. Sci. 2023, 24, 5623. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.A.; Bren, G.D.; Pennington, K.N.; Trushin, S.A.; Asin, S.; Paya, C.V. Serine 32 and serine 36 of IkappaBalpha are directly phosphorylated by protein kinase CKII in vitro. J. Mol. Biol. 1999, 290, 839–850. [Google Scholar] [CrossRef] [PubMed]
- El Yaagoubi, O.M.; Oularbi, L.; Bouyahya, A.; Samaki, H.; El Antri, S.; Aboudkhil, S. The role of the ubiquitin-proteasome pathway in skin cancer development: 26S proteasome-activated NF-κB signal transduction. Cancer Biol. Ther. 2021, 22, 479–492. [Google Scholar] [CrossRef]
- Kent, L.N.; Li, Y.; Wakle-Prabagaran, M.; Naqvi, M.Z.; Weil, S.G.; England, S.K. Blocking the BKCa channel induces NF-κB nuclear translocation by increasing nuclear calcium concentration. Biol. Reprod. 2022, 106, 441–448. [Google Scholar] [CrossRef]
- Zhong, X.; Zhang, Z.; Shen, H.; Xiong, Y.; Shah, Y.M.; Liu, Y.; Fan, X.G.; Rui, L. Hepatic NF-κB-Inducing Kinase and Inhibitor of NF-κB Kinase Subunit α Promote Liver Oxidative Stress, Ferroptosis, and Liver Injury. Hepatol. Commun. 2021, 5, 1704–1720. [Google Scholar] [CrossRef]
- Dejardin, E.; Droin, N.M.; Delhase, M.; Haas, E.; Cao, Y.; Makris, C.; Li, Z.W.; Karin, M.; Ware, C.F.; Green, D.R. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity 2002, 17, 525–535. [Google Scholar] [CrossRef] [Green Version]
- Song, P.; Zhou, Y.; Coughlan, K.A.; Dai, X.; Xu, H.; Viollet, B.; Zou, M.H. Adenosine monophosphate-activated protein kinase-α2 deficiency promotes vascular smooth muscle cell migration via S-phase kinase-associated protein 2 upregulation and E-cadherin downregulation. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 2800–2809. [Google Scholar] [CrossRef] [Green Version]
- Saxon, J.A.; Cheng, D.S.; Han, W.; Polosukhin, V.V.; McLoed, A.G.; Richmond, B.W.; Gleaves, L.A.; Tanjore, H.; Sherrill, T.P.; Barham, W.; et al. p52 Overexpression Increases Epithelial Apoptosis, Enhances Lung Injury, and Reduces Survival after Lipopolysaccharide Treatment. J. Immunol. 2016, 196, 1891–1899. [Google Scholar] [CrossRef] [Green Version]
- Budke, B.; Zhong, A.; Sullivan, K.; Park, C.; Gittin, D.I.; Kountz, T.S.; Connell, P.P. Noncanonical NF-κB factor p100/p52 regulates homologous recombination and modulates sensitivity to DNA-damaging therapy. Nucleic Acids Res. 2022, 50, 6251–6263. [Google Scholar] [CrossRef]
- Vallabhapurapu, S.; Karin, M. Regulation and function of NF-kappaB transcription factors in the immune system. Ann. Rev. Immunol. 2009, 27, 693–733. [Google Scholar] [CrossRef]
- Bacher, S.; Meier-Soelch, J.; Kracht, M.; Schmitz, M.L. Regulation of Transcription Factor NF-κB in Its Natural Habitat: The Nucleus. Cells 2021, 10, 753. [Google Scholar] [CrossRef]
- Yang, Y. Research on NF-κB Target Gene Profile and Its Regulatory Network. Ph.D. Thesis, Southeast University, Nanjing, China, 2016. [Google Scholar] [CrossRef]
- Shishodia, S.; Aggarwal, B.B. Nuclear factor-kappaB activation mediates cellular transformation, proliferation, invasion angiogenesis and metastasis of cancer. Cancer Treat. Res. 2004, 119, 139–173. [Google Scholar]
- Li, W.; Cai, H.; Ren, L.; Yang, Y.; Yang, H.; Liu, J.; Li, S.; Zhang, Y.; Zheng, X.; Tan, W.; et al. Sphingosine kinase 1 promotes growth of glioblastoma by increasing inflammation mediated by the NF-κB /IL-6/STAT3 and JNK/PTX3 pathways. Acta Pharm. Sin. B 2022, 12, 4390–4406. [Google Scholar] [CrossRef] [PubMed]
- Chai, R.C.; Chang, Y.Z.; Chang, X.; Pang, B.; An, S.Y.; Zhang, K.N.; Chang, Y.H.; Jiang, T.; Wang, Y.Z. YTHDF2 facilitates UBXN1 mRNA decay by recognizing METTL3-mediated m(6)A modification to activate NF-κB and promote the malignant progression of glioma. J. Hematol. Oncol. 2021, 14, 109. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Kong, T.; Shao, Z.; Liu, M.; Zhang, R.; Zhang, S.; Kong, Q.; Chen, J.; Cheng, B.; Wang, C. Orexin-A alleviates astrocytic apoptosis and inflammation via inhibiting OX1R-mediated NF-κB and MAPK signaling pathways in cerebral ischemia/reperfusion injury. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166230. [Google Scholar] [CrossRef] [PubMed]
- Ge, M.X.; Shi, Y.K.; Liu, D. Tripartite motif-containing 25 facilitates immunosuppression and inhibits apoptosis of glioma via activating NF-κB. Exp. Biol. Med. 2022, 247, 1529–1541. [Google Scholar] [CrossRef]
- Obad, S.; Brunnström, H.; Vallon-Christersson, J.; Borg, A.; Drott, K.; Gullberg, U. Staf50 is a novel p53 target gene conferring reduced clonogenic growth of leukemic U-937 cells. Oncogene 2004, 23, 4050–4059. [Google Scholar] [CrossRef] [Green Version]
- Eldin, P.; Papon, L.; Oteiza, A.; Brocchi, E.; Lawson, T.G.; Mechti, N. TRIM22 E3 ubiquitin ligase activity is required to mediate antiviral activity against encephalomyocarditis virus. J. Gen. Virol. 2009, 90, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Ding, K.; Luo, T.; Zhang, X.; Chen, A.; Zhang, D.; Li, G.; Thorsen, F.; Huang, B.; Li, X.; et al. TRIM22 activates NF-κB signaling in glioblastoma by accelerating the degradation of IκBα. Cell Death Differ. 2021, 28, 367–381. [Google Scholar] [CrossRef]
- Fei, X.; Wu, X.; Dou, Y.N.; Sun, K.; Guo, Q.; Zhang, L.; Li, S.; Wei, J.; Huan, Y.; He, X.; et al. TRIM22 orchestrates the proliferation of GBMs and the benefits of TMZ by coordinating the modification and degradation of RIG-I. Mol. Ther. Oncolytics 2022, 26, 413–428. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Li, B.; Zhao, R.; Pan, Z.; Zhang, S.; Qiu, W.; Guo, Q.; Qi, Y.; Gao, Z.; Fan, Y.; et al. Hypoxia-induced circADAMTS6 in a TDP43-dependent manner accelerates glioblastoma progression via ANXA2/NF-κB pathway. Oncogene 2023, 42, 138–153. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.Z.; Chai, R.C.; Pang, B.; Chang, X.; An, S.Y.; Zhang, K.N.; Jiang, T.; Wang, Y.Z. METTL3 enhances the stability of MALAT1 with the assistance of HuR via m6A modification and activates NF-κB to promote the malignant progression of IDH-wildtype glioma. Cancer Lett. 2021, 511, 36–46. [Google Scholar] [CrossRef]
- Zhao, T.; Zeng, J.; Xu, Y.; Su, Z.; Chong, Y.; Ling, T.; Xu, H.; Shi, H.; Zhu, M.; Mo, Q.; et al. Chitinase-3 like-protein-1 promotes glioma progression via the NF-κB signaling pathway and tumor microenvironment reprogramming. Theranostics 2022, 12, 6989–7008. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bao, G.; Zhang, M.; Xiang, J.; Zhou, H.; Wahafu, A.; Wu, W.; Ma, X.; Huo, L.; Bai, X.; et al. CRB2 enhances malignancy of glioblastoma via activation of the NF-κB pathway. Exp. Cell Res. 2022, 414, 113077. [Google Scholar] [CrossRef]
- Xiang, J.; Alafate, W.; Wu, W.; Wang, Y.; Li, X.; Xie, W.; Bai, X.; Li, R.; Wang, M.; Wang, J. NEK2 enhances malignancies of glioblastoma via NIK/NF-κB pathway. Cell Death Dis. 2022, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wahafu, A.; Wu, W.; Xiang, J.; Huo, L.; Ma, X.; Wang, N.; Liu, H.; Bai, X.; Xu, D.; et al. FABP5 enhances malignancies of lower-grade gliomas via canonical activation of NF-κB signaling. J. Cell. Mol. Med. 2021, 25, 4487–4500. [Google Scholar] [CrossRef]
- Wang, H.; Xu, Q.; Dong, X.; Guan, Z.; Wang, Z.; Hao, Y.; Lu, R.; Chen, L. Gold nanoparticles enhances radiosensitivity in glioma cells by inhibiting TRAF6/NF-κB induced CCL2 expression. Heliyon 2023, 9, e14362. [Google Scholar] [CrossRef]
- Xie, K.; Zhou, D.; Fang, C.; Pu, R.; Zhu, Z. Inhibition of NF-κB activation by BAY 11-7821 suppresses the proliferation and inflammation of glioma cells through inducing autophagy. Transl. Cancer Res. 2022, 11, 403–413. [Google Scholar] [CrossRef]
- Scuderi, S.A.; Lanza, M.; Casili, G.; Esposito, F.; Colarossi, C.; Giuffrida, D.; Irene, P.; Cuzzocrea, S.; Esposito, E.; Campolo, M. TBK1 Inhibitor Exerts Antiproliferative Effect on Glioblastoma Multiforme Cells. Oncol. Res. 2021, 28, 779–790. [Google Scholar] [CrossRef]
- Li, S.; He, Y.; Chen, K.; Sun, J.; Zhang, L.; He, Y.; Yu, H.; Li, Q. RSL3 Drives Ferroptosis through NF-κB Pathway Activation and GPX4 Depletion in Glioblastoma. Oxidative Med. Cell. Longev. 2021, 2021, 2915019. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, B.; Lu, P.; Wang, J.; Chen, C.; Yin, Y.; Wan, Q.; Wang, J.; Jiao, J.; Fang, X.; et al. The positive regulatory loop of TCF4N/p65 promotes glioblastoma tumourigenesis and chemosensitivity. Clin. Transl. Med. 2022, 12, e1042. [Google Scholar] [CrossRef] [PubMed]
- Markopoulos, G.S. G against Glioma: G protein inhibitory α subunit 2 (Gαi2) as a novel glioma target. Int. J. Biol. Sci. 2023, 19, 1007–1008. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Ding, W.; Xiang, Y.; Wang, X.; Yang, J. Calponin 3 Acts as a Potential Diagnostic and Prognostic Marker and Promotes Glioma Cell Proliferation, Migration, and Invasion. World Neurosurg. 2022, 165, e721–e731. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Voshart, D.; Paridaen, J.; Oosterhof, N.; Liang, D.; Thiruvalluvan, A.; Zuhorn, I.S.; den Dunnen, W.F.A.; Zhang, G.; Lin, H.; et al. CD146 increases stemness and aggressiveness in glioblastoma and activates YAP signaling. Cell. Mol. Life Sci. CMLS 2022, 79, 398. [Google Scholar] [CrossRef]
- Wu, J.; Shen, S.; Liu, T.; Ren, X.; Zhu, C.; Liang, Q.; Cui, X.; Chen, L.; Cheng, P.; Cheng, W.; et al. Chemerin enhances mesenchymal features of glioblastoma by establishing autocrine and paracrine networks in a CMKLR1-dependent manner. Oncogene 2022, 41, 3024–3036. [Google Scholar] [CrossRef]
- Wu, W.; Wang, Y.; Niu, C.; Wahafu, A.; Huo, L.; Guo, X.; Xiang, J.; Li, X.; Xie, W.; Bai, X.; et al. Retinol binding protein 1-dependent activation of NF- κB signaling enhances the malignancy of non-glioblastomatous diffuse gliomas. Cancer Sci. 2022, 113, 517–528. [Google Scholar] [CrossRef]
- Guo, L.; Chen, Y.; Hu, S.; Gao, L.; Tang, N.; Liu, R.; Qin, Y.; Ren, C.; Du, S. GDF15 expression in glioma is associated with malignant progression, immune microenvironment, and serves as a prognostic factor. CNS Neurosci. Ther. 2022, 28, 158–171. [Google Scholar] [CrossRef]
- Liu, H.; Xing, R.; Ou, Z.; Zhao, J.; Hong, G.; Zhao, T.J.; Han, Y.; Chen, Y. G-protein-coupled receptor GPR17 inhibits glioma development by increasing polycomb repressive complex 1-mediated ROS production. Cell Death Dis. 2021, 12, 610. [Google Scholar] [CrossRef]
- Shen, D.; Tian, L.; Yang, F.; Li, J.; Li, X.; Yao, Y.; Lam, E.W.; Gao, P.; Jin, B.; Wang, R. ADO/hypotaurine: A novel metabolic pathway contributing to glioblastoma development. Cell Death Discov. 2021, 7, 21. [Google Scholar] [CrossRef]
- Ahmad, S.; Abbas, M.; Ullah, M.F.; Aziz, M.H.; Beylerli, O.; Alam, M.A.; Syed, M.A.; Uddin, S.; Ahmad, A. Long non-coding RNAs regulated NF-κB signaling in cancer metastasis: Micromanaging by not so small non-coding RNAs. Semin. Cancer Biol. 2022, 85, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Ashrafizadeh, M.; Zarrabi, A.; Mostafavi, E.; Aref, A.R.; Sethi, G.; Wang, L.; Tergaonkar, V. Non-coding RNA-based regulation of inflammation. Semin. Immunol. 2022, 59, 101606. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, X.; Cui, X.; Tan, Y.; Wang, Q.; Wang, Y.; Xu, C.; Fang, C.; Kang, C. LncRNA PRADX-mediated recruitment of PRC2/DDX5 complex suppresses UBXN1 expression and activates NF-κB activity, promoting tumorigenesis. Theranostics 2021, 11, 4516–4530. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Xu, R.; Ding, K.; Bao, G.; Zhang, X.; Huang, B.; Wang, X.; Martinez, A.; Wang, X.; Li, G.; et al. Long Noncoding RNA SChLAP1 Forms a Growth-Promoting Complex with HNRNPL in Human Glioblastoma through Stabilization of ACTN4 and Activation of NF-κB Signaling. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 6868–6881. [Google Scholar] [CrossRef] [Green Version]
- Tang, G.; Luo, L.; Zhang, J.; Zhai, D.; Huang, D.; Yin, J.; Zhou, Q.; Zhang, Q.; Zheng, G. lncRNA LINC01057 promotes mesenchymal differentiation by activating NF-κB signaling in glioblastoma. Cancer Lett. 2021, 498, 152–164. [Google Scholar] [CrossRef]
- Pflug, K.; Lee, D.; McFadden, K.; Herrera, L.; Sitcheran, R. Transcriptional Induction of NF-kB-Inducing Kinase by E2F4/5 Facilitates Collective Invasion of Glioma Cells. Res. Sq. 2023. preprint. [Google Scholar] [CrossRef]
- Connolly, N.P.; Galisteo, R.; Xu, S.; Bar, E.E.; Peng, S.; Tran, N.L.; Ames, H.M.; Kim, A.J.; Woodworth, G.F.; Winkles, J.A. Elevated fibroblast growth factor-inducible 14 expression transforms proneural-like gliomas into more aggressive and lethal brain cancer. Glia 2021, 69, 2199–2214. [Google Scholar] [CrossRef]
- Yang, Z.; Ye, X.; Zhang, Y.; Huang, Y.; Chen, J.; Zeng, Y.; Chen, J. ECRG4 acts as a tumor suppressor in nasopharyngeal carcinoma by suppressing the AKT/GSK3β/β-catenin signaling pathway. Cytotechnology 2022, 74, 231–243. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, L.; Feng, Y.; Wang, L.; Liu, X.; Li, Y. UBR5 promotes migration and invasion of glioma cells by regulating the ECRG4/NF-ҡB pathway. J. Biosci. 2022, 47, 45. [Google Scholar] [CrossRef]
- Che, M.; Lan, Q. RIT1 Promotes Glioma Proliferation and Invasion via the AKT/ERK/NF-ĸB Signaling Pathway. J. Mol. Neurosci. MN 2022, 72, 1547–1556. [Google Scholar] [CrossRef]
- Yan, T.; Tan, Y.; Deng, G.; Sun, Z.; Liu, B.; Wang, Y.; Yuan, F.; Sun, Q.; Hu, P.; Gao, L.; et al. TGF-β induces GBM mesenchymal transition through upregulation of CLDN4 and nuclear translocation to activate TNF-α/NF-κB signal pathway. Cell Death Dis. 2022, 13, 339. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, S.; Li, H.L.; Luo, H.; Wu, X.; Lu, J.; Wang, H.W.; Chen, Y.; Chen, D.; Wu, W.T.; et al. FOSL1 promotes proneural-to-mesenchymal transition of glioblastoma stem cells via UBC9/CYLD/NF-κB axis. Mol. Ther. J. Am. Soc. Gene Ther. 2022, 30, 2568–2583. [Google Scholar] [CrossRef] [PubMed]
- Babu, D.; Chintal, R.; Panigrahi, M.; Phanithi, P.B. Distinct expression and function of breast cancer metastasis suppressor 1 in mutant P53 glioblastoma. Cell. Oncol. 2022, 45, 1451–1465. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Gong, K.; Beckley, N.; Zhang, Y.; Yang, X.; Chkheidze, R.; Hatanpaa, K.J.; Garzon-Muvdi, T.; Koduru, P.; Nayab, A.; et al. EGFR ligand shifts the role of EGFR from oncogene to tumour suppressor in EGFR-amplified glioblastoma by suppressing invasion through BIN3 upregulation. Nat. Cell Biol. 2022, 24, 1291–1305. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, L.; Song, Y.; Jiang, Y.; Zhang, D.; Wang, R.; Hu, T.; Han, S. MCM8 is regulated by EGFR signaling and promotes the growth of glioma stem cells through its interaction with DNA-replication-initiating factors. Oncogene 2021, 40, 4615–4624. [Google Scholar] [CrossRef]
- Li, S.; Jiang, X.; Guan, M.; Zhang, Y.; Cao, Y.; Zhang, L. The overexpression of GPX8 is correlated with poor prognosis in GBM patients. Front. Genet. 2022, 13, 898204. [Google Scholar] [CrossRef]
- Chen, L.; Zhou, J.; Li, L.; Zhao, J.; Li, H.; Zheng, W.; Xu, J.; Jing, Z. SLC39A7 promotes malignant behaviors in glioma via the TNF-α-mediated NF-κB signaling pathway. J. Cancer 2021, 12, 4530–4541. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, L.; Qi, Y.; Li, H.; Pan, S. GLIS family zinc finger 3 promoting cell malignant behaviors and NF-κB signaling in glioma. Brain Res. 2021, 1770, 147623. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, H.; Zhang, Z.; Xu, J.; Qi, Y.; Xue, H.; Gao, Z.; Zhao, R.; Wang, S.; Zhang, S.; et al. Cell surface GRP78 regulates BACE2 via lysosome-dependent manner to maintain mesenchymal phenotype of glioma stem cells. J. Exp. Clin. Cancer Res. CR 2021, 40, 20. [Google Scholar] [CrossRef]
- Wang, Z.; Shi, Y.; Ying, C.; Jiang, Y.; Hu, J. Hypoxia-induced PLOD1 overexpression contributes to the malignant phenotype of glioblastoma via NF-κB signaling. Oncogene 2021, 40, 1458–1475. [Google Scholar] [CrossRef]
- Wei, L.; Li, L.; Liu, L.; Yu, R.; Li, X.; Luo, Z. Knockdown of Annexin-A1 Inhibits Growth, Migration and Invasion of Glioma Cells by Suppressing the PI3K/Akt Signaling Pathway. ASN Neuro 2021, 13, 17590914211001218. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhao, J.; Liu, Y.; Hu, J.; Gao, L.; Wang, H.; Cui, D. CircKPNB1 mediates a positive feedback loop and promotes the malignant phenotypes of GSCs via TNF-α/NF-κB signaling. Cell Death Dis. 2022, 13, 697. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Hu, J.; Li, S.; Zhou, C.; Feng, M.; Li, L.; Gao, Y.; Chen, X.; Wu, X.; Cao, Y.; et al. LINC01393, a Novel Long Non-Coding RNA, Promotes the Cell Proliferation, Migration and Invasion through MiR-128-3p/NUSAP1 Axis in Glioblastoma. Int. J. Mol. Sci. 2023, 24, 5878. [Google Scholar] [CrossRef] [PubMed]
- Bonafé, G.A.; Dos Santos, J.S.; Fernandes, A.; Ziegler, J.V.; Marson, F.A.L.; Rocha, T.; Carvalho, P.O.; Ortega, M.M. Anti-Migratory Effect of Dipotassium Glycyrrhizinate on Glioblastoma Cell Lines: Microarray Data for the Identification of Key MicroRNA Signatures. Front. Oncol. 2022, 12, 819599. [Google Scholar] [CrossRef]
- Nan, Y.; Guo, L.; Zhen, Y.; Wang, L.; Ren, B.; Chen, X.; Lu, Y.; Yu, K.; Zhong, Y.; Huang, Q. miRNA-451 regulates the NF-κB signaling pathway by targeting IKKβ to inhibit glioma cell growth. Cell Cycle 2021, 20, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hao, Y.; Zhang, A.; Yang, W.; Wei, W.; Wang, G.; Jia, Z. miR-19a/b promote EMT and proliferation in glioma cells via SEPT7-AKT-NF-κB pathway. Mol. Ther. Oncolytics 2021, 20, 290–305. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Wang, H.; Chen, E.; Bian, E.; Xu, Y.; Ji, X.; Yang, Z.; Hua, X.; Zhang, Y.; Zhao, B. LncRNA-ATB promotes TGF-β-induced glioma cells invasion through NF-κB and P38/MAPK pathway. J. Cell. Physiol. 2019, 234, 23302–23314. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Xu, C.; Li, Y.; Tang, B.; Xie, S.; Hong, T.; Zeng, E. Long non-coding RNA LINC00526 represses glioma progression via forming a double negative feedback loop with AXL. J. Cell. Mol. Med. 2019, 23, 5518–5531. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.S.; Peng, S.F.; Wu, R.S.; Chueh, F.S.; Huang, W.W.; Chen, P.Y.; Kuo, C.L.; Huang, A.C.; Liao, C.L.; Hsia, T.C. Bisdemethoxycurcumin suppresses human osteosarcoma U-2 OS cell migration and invasion via affecting the PI3K/Akt/NF-κB, PI3K/Akt/GSK3β and MAPK signaling pathways in vitro. Oncol. Rep. 2022, 48, 210. [Google Scholar] [CrossRef]
- Bi, F.; Wang, J.; Zheng, X.; Xiao, J.; Zhi, C.; Gu, J.; Zhang, Y.; Li, J.; Miao, Z.; Wang, Y.; et al. HSP60 participates in the anti-glioma effects of curcumin. Exp. Ther. Med. 2021, 21, 204. [Google Scholar] [CrossRef]
- Lv, F.; Du, Q.; Li, L.; Xi, X.; Liu, Q.; Li, W.; Liu, S. Eriodictyol inhibits glioblastoma migration and invasion by reversing EMT via downregulation of the P38 MAPK/GSK-3β/ZEB1 pathway. Eur. J. Pharmacol. 2021, 900, 174069. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Liu, L.; Li, X.; Chen, W.; Lu, Z. Tat-NTS Suppresses the Proliferation, Migration and Invasion of Glioblastoma Cells by Inhibiting Annexin-A1 Nuclear Translocation. Cell. Mol. Neurobiol. 2022, 42, 2715–2725. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.Z.; Xu, S.Y. Fentanyl inhibits cell invasion and migration by modulating NF-κB activation in glioma. Brain Res. 2023, 1809, 148356. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.P.; Carmody, R.J. NF-κB and the Transcriptional Control of Inflammation. Int. Rev. Cell Mol. Biol. 2018, 335, 41–84. [Google Scholar] [PubMed]
- Zhang, H.; Song, Y.; Yang, H.; Liu, Z.; Gao, L.; Liang, X.; Ma, C. Tumor cell-intrinsic Tim-3 promotes liver cancer via NF-κB/IL-6/STAT3 axis. Oncogene 2018, 37, 2456–2468. [Google Scholar] [CrossRef]
- Guo, Q.; Shen, S.; Guan, G.; Zhu, C.; Zou, C.; Cao, J.; Cheng, W.; Xu, X.; Yu, J.; Lin, Z.; et al. Cancer cell intrinsic TIM-3 induces glioblastoma progression. iScience 2022, 25, 105329. [Google Scholar] [CrossRef]
- Chen, R.; Chen, C.; Han, N.; Guo, W.; Deng, H.; Wang, Y.; Ding, Y.; Zhang, M. Annexin-1 is an oncogene in glioblastoma and causes tumour immune escape through the indirect upregulation of interleukin-8. J. Cell. Mol. Med. 2022, 26, 4343–4356. [Google Scholar] [CrossRef]
- Yi, K.; Cui, X.; Liu, X.; Wang, Y.; Zhao, J.; Yang, S.; Xu, C.; Yang, E.; Xiao, M.; Hong, B.; et al. PTRF/Cavin-1 as a Novel RNA-Binding Protein Expedites the NF-κB/PD-L1 Axis by Stabilizing lncRNA NEAT1, Contributing to Tumorigenesis and Immune Evasion in Glioblastoma. Front. Immunol. 2021, 12, 802795. [Google Scholar] [CrossRef]
- Icard, P.; Shulman, S.; Farhat, D.; Steyaert, J.M.; Alifano, M.; Lincet, H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer. Chemother. 2018, 38, 1–11. [Google Scholar] [CrossRef]
- Chen, Z.; Bao, H.; Long, J.; Zhao, P.; Hu, X.; Wang, H.; Zhang, Y.; Yang, J.; Zhuge, Q.; Xia, L. GBE1 Promotes Glioma Progression by Enhancing Aerobic Glycolysis through Inhibition of FBP1. Cancers 2023, 15, 1594. [Google Scholar] [CrossRef]
- Kamradt, M.L.; Jung, J.U.; Pflug, K.M.; Lee, D.W.; Fanniel, V.; Sitcheran, R. NIK promotes metabolic adaptation of glioblastoma cells to bioenergetic stress. Cell Death Dis. 2021, 12, 271. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Tong, Y.; Jiang, X.; Meng, Y.; Jiang, H.; Du, L.; Wu, Q.; Li, S.; Luo, S.; Li, M.; et al. Aerobic glycolysis promotes tumor immune evasion by hexokinase2-mediated phosphorylation of IκBα. Cell Metab. 2022, 34, 1312–1324.e1316. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, R.; Qu, X.; Yu, H.; Chu, H.; Zhang, Y.; Zhu, W.; Wu, X.; Gao, H.; Tao, B.; et al. α-Ketoglutarate-Activated NF-κB Signaling Promotes Compensatory Glucose Uptake and Brain Tumor Development. Mol. Cell 2019, 76, 148–162.e147. [Google Scholar] [CrossRef] [PubMed]
- Batash, R.; Asna, N.; Schaffer, P.; Francis, N.; Schaffer, M. Glioblastoma Multiforme, Diagnosis and Treatment; Recent Literature Review. Curr. Med. Chem. 2017, 24, 3002–3009. [Google Scholar] [CrossRef]
- Ghosh, D.; Nandi, S.; Bhattacharjee, S. Combination therapy to checkmate Glioblastoma: Clinical challenges and advances. Clin. Transl. Med. 2018, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Werlenius, K.; Stragliotto, G.; Strandeus, M.; Blomstrand, M.; Carén, H.; Jakola, A.S.; Rydenhag, B.; Dyregaard, D.; Dzhandzhugazyan, K.N.; Kirkin, A.F.; et al. A randomized phase II trial of efficacy and safety of the immunotherapy ALECSAT as an adjunct to radiotherapy and temozolomide for newly diagnosed glioblastoma. Neuro-Oncol. Adv. 2021, 3, vdab156. [Google Scholar] [CrossRef]
- Raghava Kurup, R.; Oakes, E.K.; Vadlamani, P.; Nwosu, O.; Danthi, P.; Hundley, H.A. ADAR3 activates NF-κB signaling and promotes glioblastoma cell resistance to temozolomide. Sci. Rep. 2022, 12, 13362. [Google Scholar] [CrossRef]
- Liao, X.; Li, Z.; Zheng, H.; Qian, W.; Zhang, S.; Chen, S.; Li, X.; Tang, M.; Xu, Y.; Yu, R.; et al. Downregulation of BASP1 promotes temozolomide resistance in gliomas via epigenetic activation of the FBXO32/NF-κB/MGMT axis. Mol. Cancer Res. MCR 2023, OF1–OF16. [Google Scholar] [CrossRef]
- Zhang, J.; Guo, Z.; Xie, Q.; Zhong, C.; Gao, X.; Yang, Q. Tryptophan hydroxylase 1 drives glioma progression by modulating the serotonin/L1CAM/NF-κB signaling pathway. BMC Cancer 2022, 22, 457. [Google Scholar] [CrossRef]
- Richard, S.A.; Eugene, K.D. The Pivotal Immunomodulatory and Anti-Inflammatory Effect of Histone-Lysine N-Methyltransferase in the Glioma Microenvironment: Its Biomarker and Therapy Potentials. Anal. Cell. Pathol. 2021, 2021, 4907167. [Google Scholar] [CrossRef]
- Stillger, M.N.; Chen, C.Y.; Lai, Z.W.; Li, M.; Schäfer, A.; Pagenstecher, A.; Nimsky, C.; Bartsch, J.W.; Schilling, O. Changes in calpain-2 expression during glioblastoma progression predisposes tumor cells to temozolomide resistance by minimizing DNA damage and p53-dependent apoptosis. Cancer Cell Int. 2023, 23, 49. [Google Scholar] [CrossRef] [PubMed]
- Tong, F.; Zhao, J.X.; Fang, Z.Y.; Cui, X.T.; Su, D.Y.; Liu, X.; Zhou, J.H.; Wang, G.X.; Qiu, Z.J.; Liu, S.Z.; et al. MUC1 promotes glioblastoma progression and TMZ resistance by stabilizing EGFRvIII. Pharmacol. Res. 2023, 187, 106606. [Google Scholar] [CrossRef]
- Shi, Z.F.; Li, G.Z.; Zhai, Y.; Pan, C.Q.; Wang, D.; Yu, M.C.; Liu, C.; Zhang, W.; Yu, X.G. EGFRvIII Promotes the Proneural-Mesenchymal Transition of Glioblastoma Multiforme and Reduces Its Sensitivity to Temozolomide by Regulating the NF-κB/ALDH1A3 Axis. Genes 2023, 14, 651. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Jiao, B.H.; Wu, J.L.; Yang, J.K.; Hu, Y.H.; Cui, K. Mechanism of RIP2 enhancing stemness of glioma cells induces temozolomide resistance. CNS Neurosci. Ther. 2022, 28, 2319–2330. [Google Scholar] [CrossRef] [PubMed]
- Witte, K.E.; Slotta, C.; Lütkemeyer, M.; Kitke, A.; Coras, R.; Simon, M.; Kaltschmidt, C.; Kaltschmidt, B. PLEKHG5 regulates autophagy, survival and MGMT expression in U251-MG glioblastoma cells. Sci. Rep. 2020, 10, 21858. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.H.; Jiao, B.H.; Wang, C.Y.; Wu, J.L. Regulation of temozolomide resistance in glioma cells via the RIP2/NF-κB/MGMT pathway. CNS Neurosci. Ther. 2021, 27, 552–563. [Google Scholar] [CrossRef]
- Han, D.; Zhang, N.; Zhao, S.; Liu, H.; Wang, X.; Yang, M.; Wang, S.; Li, Y.; Liu, Z.; Teng, L. AKIP1 promotes glioblastoma viability, mobility and chemoradiation resistance via regulating CXCL1 and CXCL8 mediated NF-κB and AKT pathways. Am. J. Cancer Res. 2021, 11, 1185–1205. [Google Scholar]
- Yu, J.; Shi, J.; Yuan, F.; Yin, W.; Zeng, H.; Ge, L.; Li, H.; Wang, X. Kavain ablates the radio-resistance of IDH-wildtype glioblastoma by targeting LITAF/NF-κB pathway. Cell. Oncol. 2023, 46, 179–193. [Google Scholar] [CrossRef]
- Gao, Z.; Xu, J.; Fan, Y.; Zhang, Z.; Wang, H.; Qian, M.; Zhang, P.; Deng, L.; Shen, J.; Xue, H.; et al. ARPC1B promotes mesenchymal phenotype maintenance and radiotherapy resistance by blocking TRIM21-mediated degradation of IFI16 and HuR in glioma stem cells. J. Exp. Clin. Cancer Res. CR 2022, 41, 323. [Google Scholar] [CrossRef]
- McLaughlin, M.; Patin, E.C.; Pedersen, M.; Wilkins, A.; Dillon, M.T.; Melcher, A.A.; Harrington, K.J. Inflammatory microenvironment remodelling by tumour cells after radiotherapy. Nat. Rev. Cancer 2020, 20, 203–217. [Google Scholar] [CrossRef]
- Hayashi, H.; Yonesaka, K.; Nakamura, A.; Fujimoto, D.; Azuma, K.; Sakata, S.; Tachihara, M.; Ikeda, S.; Yokoyama, T.; Hataji, O.; et al. Alternating therapy with osimertinib and afatinib for treatment-naive patients with EGFR-mutated advanced non-small cell lung cancer: A single-group, open-label phase 2 trial (WJOG10818L). Lung Cancer 2022, 168, 38–45. [Google Scholar] [CrossRef]
- Song, Y.; Wu, Y.L.; Cao, L.J.; Chen, J.H.; Ma, Z.Y.; Cui, J.W.; Wang, J.; Liu, H.B.; Ding, J.Y.; Hu, M. Efficacy and Safety of Gefitinib as Third-line Treatment in NSCLC Patients With Activating EGFR Mutations Treated With First-line Gefitinib Followed by Second-line Chemotherapy: A Single-Arm, Prospective, Multicenter Phase II Study (RE-CHALLENGE, CTONG1304). Am. J. Clin. Oncol. 2019, 42, 432–439. [Google Scholar]
- Ye, T.; Chen, R.; Zhou, Y.; Zhang, J.; Zhang, Z.; Wei, H.; Xu, Y.; Wang, Y.; Zhang, Y. Salvianolic acid A (Sal A) suppresses malignant progression of glioma and enhances temozolomide (TMZ) sensitivity via repressing transgelin-2 (TAGLN2) mediated phosphatidylinositol-3-kinase (PI3K)/protein kinase B (Akt) pathway. Bioengineered 2022, 13, 11646–11655. [Google Scholar] [CrossRef] [PubMed]
- Shishodia, S.; Azu, N.; Rosenzweig, J.A.; Jackson, D.A. Guggulsterone for Chemoprevention of Cancer. Curr. Pharm. Des. 2016, 22, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.B.; Chen, X.Z.; Yu, Z.L.; Xue, F. Guggulsterone from Commiphora mukul potentiates anti-glioblastoma efficacy of temozolomide in vitro and in vivo via down-regulating EGFR/PI3K/Akt signaling and NF-κB activation. J. Ethnopharmacol. 2023, 301, 115855. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Mao, L.; Huang, H.; Tang, L.; Jiang, H.; Zhang, Y.; Mu, Q. Hesperetin ameliorates glioblastoma by inhibiting proliferation, inducing apoptosis, and suppressing metastasis. Transl. Cancer Res. 2022, 11, 1781–1794. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.Y.; Lee, S.C.; Liu, H.C.; Peng, S.F.; Chueh, F.S.; Lu, T.J.; Lee, H.T.; Chou, Y.C. Phenethyl Isothiocyanate Suppresses the Proinflammatory Cytokines in Human Glioblastoma Cells through the PI3K/Akt/NF-κB Signaling Pathway In Vitro. Oxidative Med. Cell. Longev. 2022, 2022, 2108289. [Google Scholar] [CrossRef]
- Jia, C.; Zhao, Y.; Huang, H.; Fan, K.; Xie, T.; Xie, M. Apigenin sensitizes radiotherapy of mouse subcutaneous glioma through attenuations of cell stemness and DNA damage repair by inhibiting NF-κB/HIF-1α-mediated glycolysis. J. Nutr. Biochem. 2022, 107, 109038. [Google Scholar] [CrossRef]
- Rotondo, R.; Oliva, M.A.; Arcella, A. The Sesquiterpene Lactone Cynaropicrin Manifests Strong Cytotoxicity in Glioblastoma Cells U-87 MG by Induction of Oxidative Stress. Biomedicines 2022, 10, 1583. [Google Scholar] [CrossRef]
- Su, J.; Yin, W.; Huo, M.; Yao, Q.; Ding, L. Induction of apoptosis in glioma cells by lycorine via reactive oxygen species generation and regulation of NF-κB pathways. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2023, 396, 1247–1255. [Google Scholar] [CrossRef]
- Tang, Q.; Cao, H.; Tong, N.; Liu, Y.; Wang, W.; Zou, Y.; Xu, L.; Zeng, Z.; Xu, W.; Yin, Z.; et al. Tubeimoside-I sensitizes temozolomide-resistant glioblastoma cells to chemotherapy by reducing MGMT expression and suppressing EGFR induced PI3K/Akt/mTOR/NF-κB-mediated signaling pathway. Phytomedicine Int. J. Phytother. Phytopharm. 2022, 99, 154016. [Google Scholar] [CrossRef]
- Chiang, I.T.; Liu, Y.C.; Liu, H.S.; Ali, A.A.A.; Chou, S.Y.; Hsu, T.I.; Hsu, F.T. Regorafenib Reverses Temozolomide-Induced CXCL12/CXCR4 Signaling and Triggers Apoptosis Mechanism in Glioblastoma. Neurother. J. Am. Soc. Exp. NeuroTherapeutics 2022, 19, 616–634. [Google Scholar] [CrossRef]
- Babu, D.; Mudiraj, A.; Yadav, N.; Chandrashekhar, Y.B.V.K.; Panigrahi, M.; Prakash Babu, P. Rabeprazole has efficacy per se and reduces resistance to temozolomide in glioma via EMT inhibition. Cell. Oncol. 2021, 44, 889–905. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Qian, W.; Li, X.; Wei, W. NF-κB Inhibitor Myrislignan Induces Ferroptosis of Glioblastoma Cells via Regulating Epithelial-Mesenchymal Transformation in a Slug-Dependent Manner. Oxidative Med. Cell. Longev. 2023, 2023, 7098313. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Li, M.; Huang, Q.; Xie, L.; Huang, Z. Monacolin K Induces Apoptosis of Human Glioma U251 Cells by Triggering ROS-Mediated Oxidative Damage and Regulating MAPKs and NF-κB Pathways. ACS Chem. Neurosci. 2023, 14, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.R.; Cheng, C.C.; Lee, A.L.; Shieh, J.C.; Wu, H.J.; Huang, A.P.; Hsu, Y.H.; Young, T.H. Poly(allylguanidine)-Coated Surfaces Regulate TGF-β in Glioblastoma Cells to Induce Apoptosis via NF-κB Pathway Activation. ACS Appl. Mater. Interfaces 2021, 13, 59400–59410. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ye, Y.; Zhu, X.; Jin, T.; Yi, W.; Gu, L.; Xiong, X. Meisoindigo inhibits cellular proliferation via down-regulation of the PI3K/Akt pathway and induces cellular apoptosis in glioblastoma U87 cells. Acta Biochim. Pol. 2021, 68, 309–315. [Google Scholar]
- Volmar, M.N.M.; Cheng, J.; Alenezi, H.; Richter, S.; Haug, A.; Hassan, Z.; Goldberg, M.; Li, Y.; Hou, M.; Herold-Mende, C.; et al. Cannabidiol converts NF-κB into a tumor suppressor in glioblastoma with defined antioxidative properties. Neuro-Oncology 2021, 23, 1898–1910. [Google Scholar] [CrossRef]
- Rotondo, R.; Oliva, M.A.; Staffieri, S.; Castaldo, S.; Giangaspero, F.; Arcella, A. Implication of Lactucopicrin in Autophagy, Cell Cycle Arrest and Oxidative Stress to Inhibit U87Mg Glioblastoma Cell Growth. Molecules 2020, 25, 5843. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, X.; Tang, H.; Pan, Y.; Hu, B.; Huang, G. Rosmarinic acid inhibits cell proliferation, migration, and invasion and induces apoptosis in human glioma cells. Int. J. Mol. Med. 2021, 47, 67. [Google Scholar] [CrossRef]
- Saha, S.; Zhang, Y.; Wilson, B.; Abounader, R.; Dutta, A. The tumor-suppressive long noncoding RNA DRAIC inhibits protein translation and induces autophagy by activating AMPK. J. Cell Sci. 2021, 134, jcs259306. [Google Scholar] [CrossRef]
- Guo, A.; Fang, G.; Lin, Z.; Zheng, S.; Zhuang, Z.; Lin, R.; Lin, Y. Overexpression of lncRNA IRAIN restrains the progression and Temozolomide resistance of glioma via repressing IGF-1R-PI3K-NF-κB signaling pathway. Histol. Histopathol. 2022, 37, 543–554. [Google Scholar]
| Drugs | Source | Testing Index | Efficacy |
|---|---|---|---|
| Guggulsterone [136] | Myrrh | EGFR; PI3K; AKT; p-p65 | Inhibiting drug resistance |
| Hesperetin [137] | Citrus fruits | PI3K; AKT; p-AKT; p65 | Inhibiting proliferation; Inducing apoptosis; suppressing metastasis |
| Phenethyl isothiocyanate [138] | Cruciferous vegetables | PI3K; p-AKT;p-p65; p-IKKα/β; IKKα/β | Inhibiting drug resistance |
| Apigenin [139] | Celery | p65 | Improving sensitivity to radiotherapy |
| Cynaropicrin [140] | Cynara scolymus | ERK; p65 | Promoting the pharmacodynamics of TMZ |
| Lycorine [141] | Lycoris | p-p65 | Inhibiting proliferation; reducing drug resistance |
| Tubeimoside-I [142] | Bolbostemma paniculatum | EGFR; p-PI3K; p-AKT; p-mTOR; p-p65 | Promoting the pharmacodynamics of TMZ |
| Regorafenib [143] | Compound | p-ERK; p-p65 | Reducing drug resistance |
| Rabeprazole [144] | Aciphex tablets | p-p65; p- IκBα | Reducing drug resistance |
| Myrislignan [145] | Myristica fragrans | p-p65 | Inducing ferroptosis |
| Monacolin K [146] | Compound | JNK; ERK; p65; IκBα | Inducing apoptosis |
| Allylamine hydrochloride [147] | Biomaterials | p-65 | Inducing apoptosis |
| Meisoindigo [148] | Compound | PI3K; AKT; p-AKT; p65; p-p65 | Inducing apoptosis |
| Cannabidiol [149] | Cannabis sativa plants | p65; p-p65 | Antioxidative; Inducing apoptosis |
| Lactucopicrin [150] | Lactucavirosa | p65 | Promoting autophagy; inhibiting proliferation; oxidative stress |
| Rosmarinic acid [151] | Rosemary | PI3K; AKT; p-AKT; p65 | Inhibiting proliferation; suppressing migration and invasion; inducing apoptosis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, P.; Xu, J.; Cui, H. The Recent Research Progress of NF-κB Signaling on the Proliferation, Migration, Invasion, Immune Escape and Drug Resistance of Glioblastoma. Int. J. Mol. Sci. 2023, 24, 10337. https://doi.org/10.3390/ijms241210337
Shi P, Xu J, Cui H. The Recent Research Progress of NF-κB Signaling on the Proliferation, Migration, Invasion, Immune Escape and Drug Resistance of Glioblastoma. International Journal of Molecular Sciences. 2023; 24(12):10337. https://doi.org/10.3390/ijms241210337
Chicago/Turabian StyleShi, Pengfei, Jie Xu, and Hongjuan Cui. 2023. "The Recent Research Progress of NF-κB Signaling on the Proliferation, Migration, Invasion, Immune Escape and Drug Resistance of Glioblastoma" International Journal of Molecular Sciences 24, no. 12: 10337. https://doi.org/10.3390/ijms241210337





