Genome-Wide Identification of WRKY Gene Family and Functional Characterization of CcWRKY25 in Capsicum chinense
Abstract
:1. Introduction
2. Results
2.1. Identification and Sequence Analysis of Genome-Wide WRKYs in C. chinense
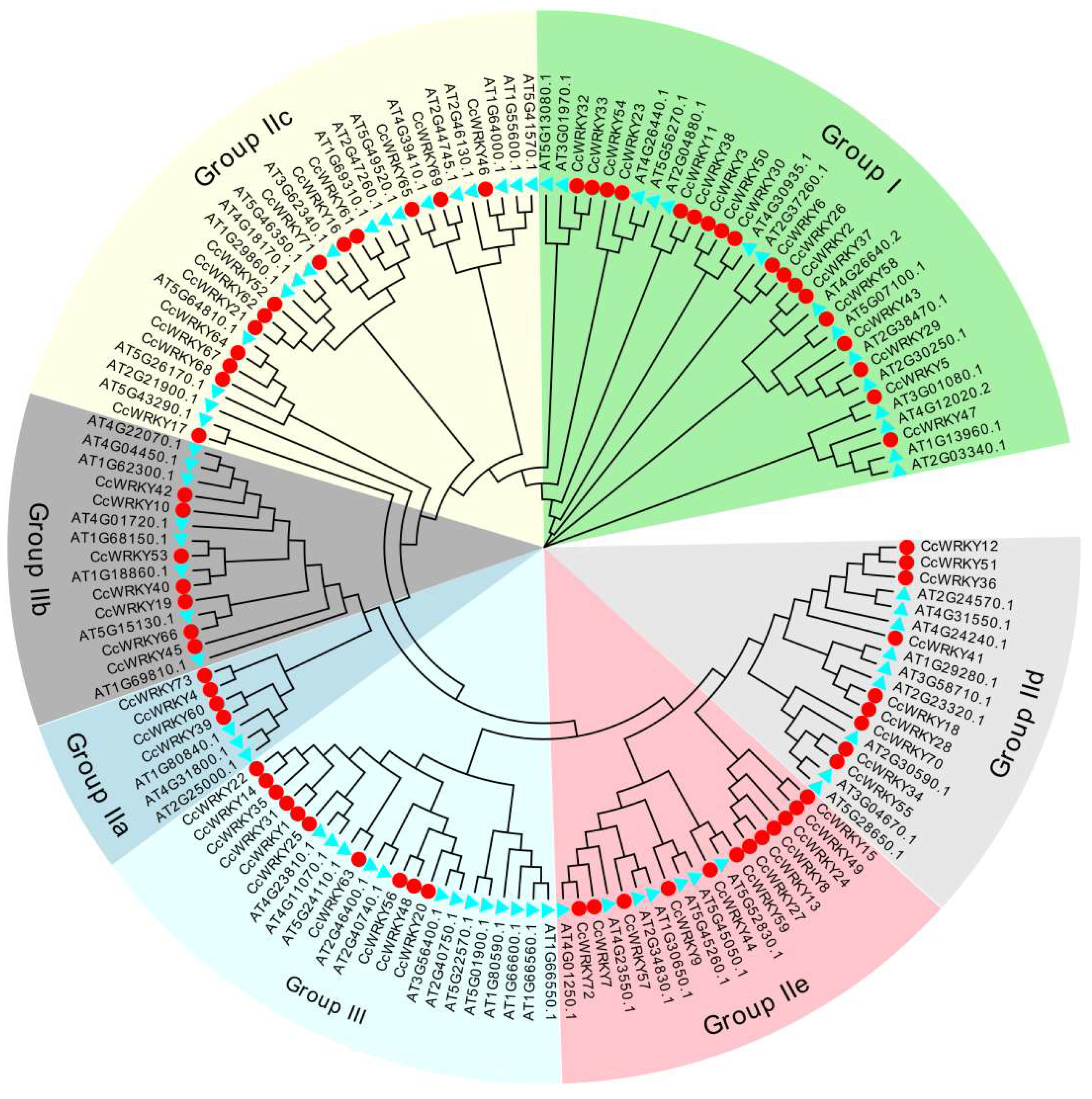
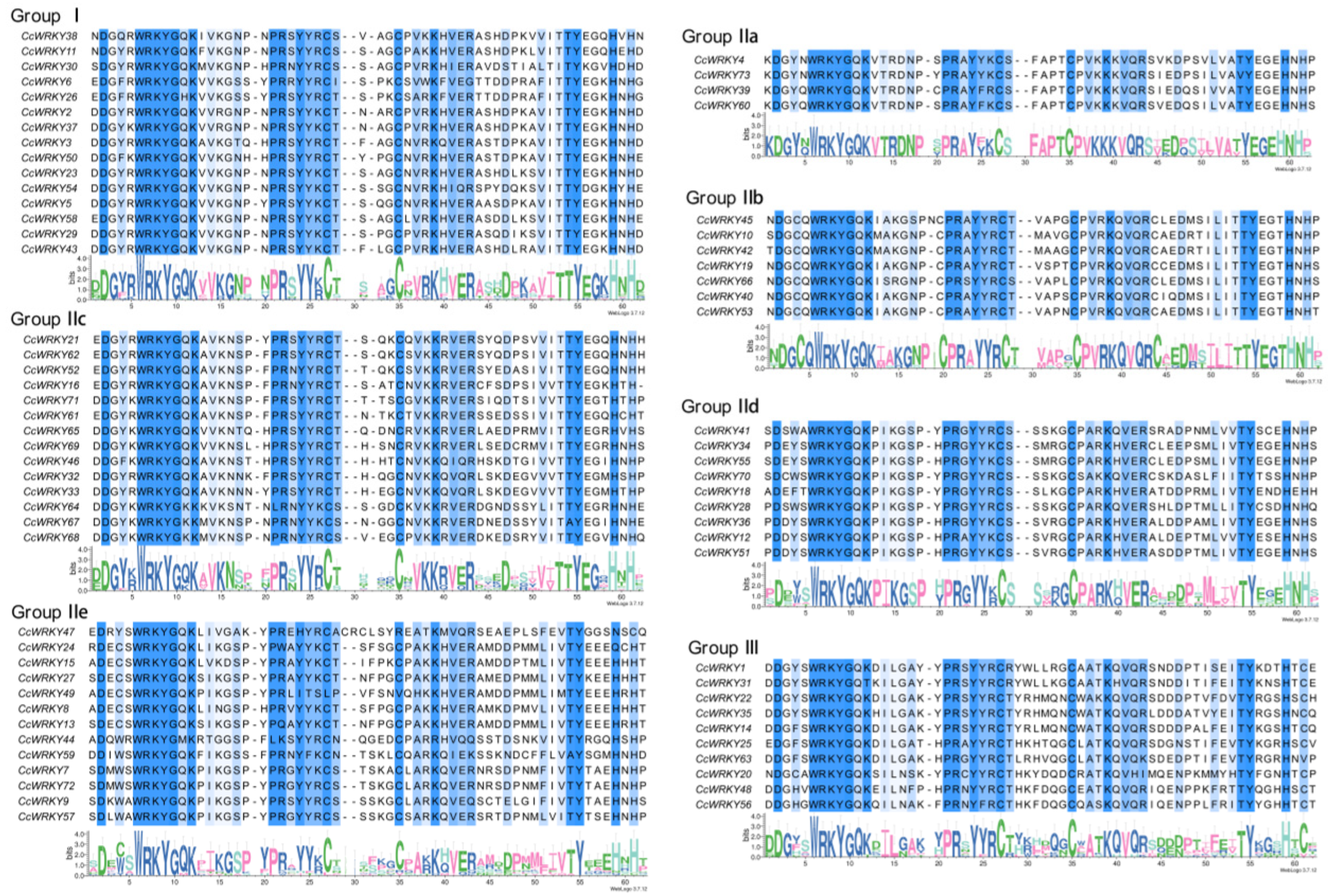
2.2. Multiple Sequence Alignment and Phylogenetic Analysis of CcWRKY Gene Family
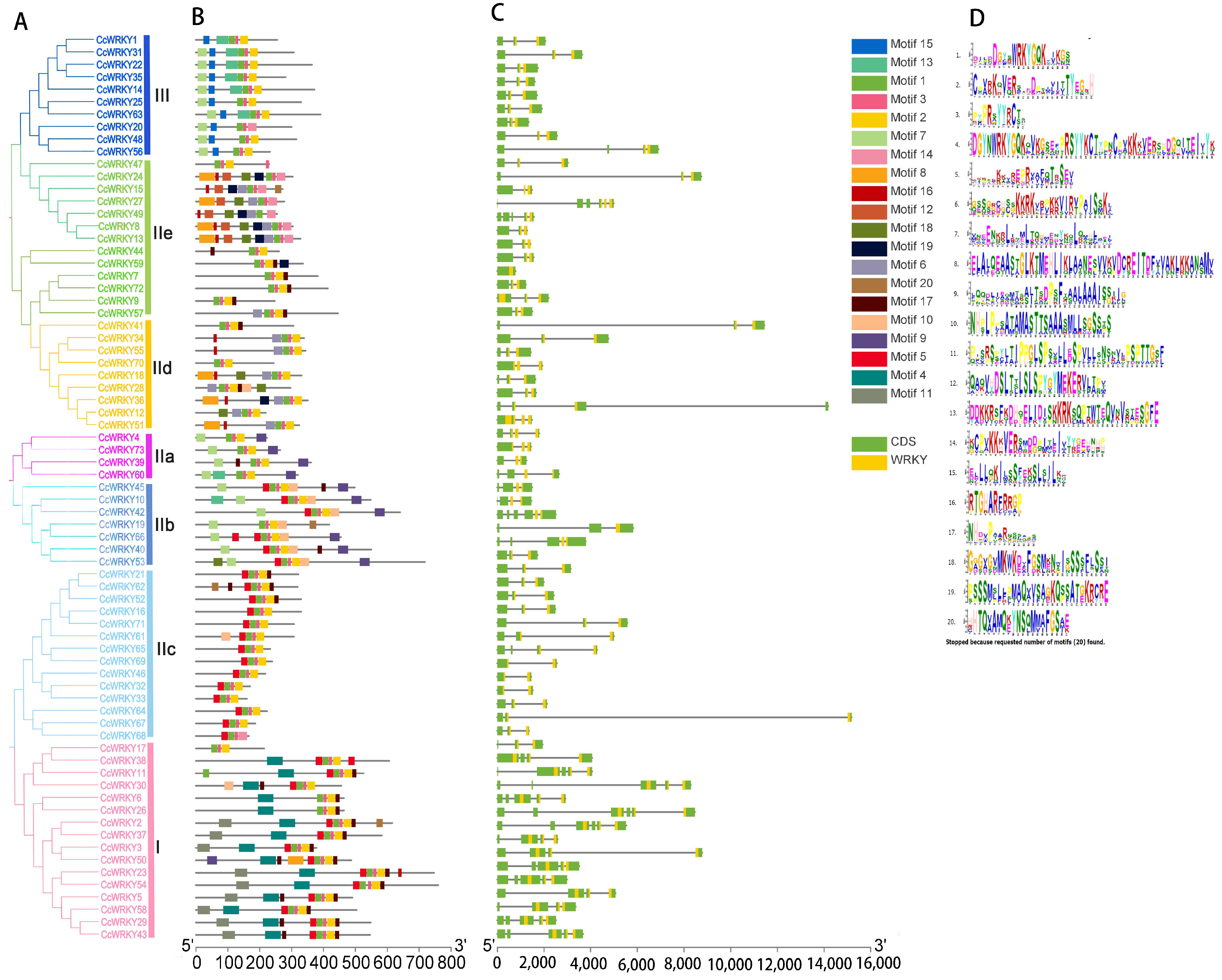
2.3. Analysis of Gene Structure, Conserved Motifs, and Structural Domains of the CcWRKY Gene Family
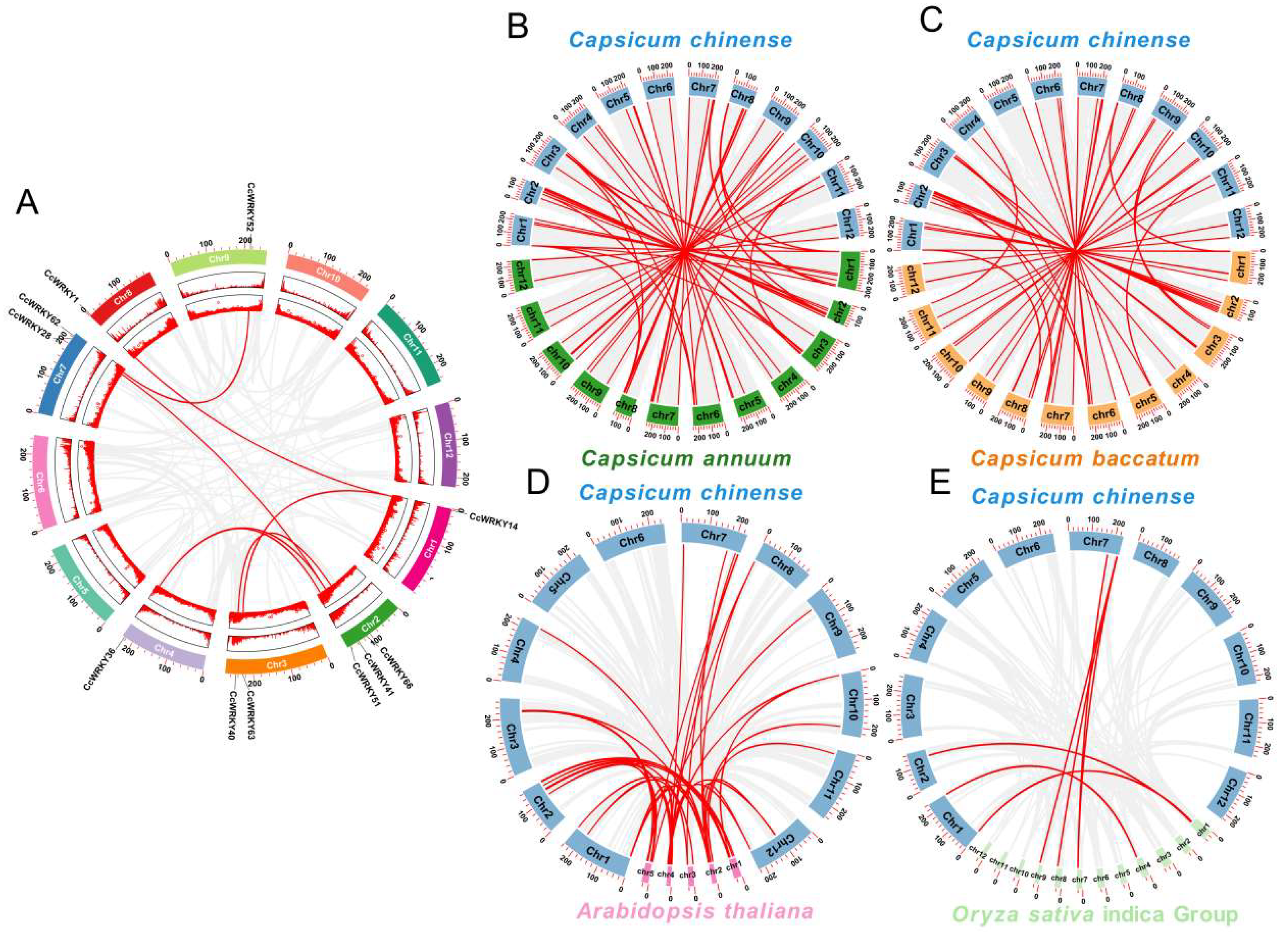
2.4. Collinearity Analysis of WRKY Gene Family in C. chinense
2.5. Screening for Spiciness-Associated CcWRKY Transcription Factors
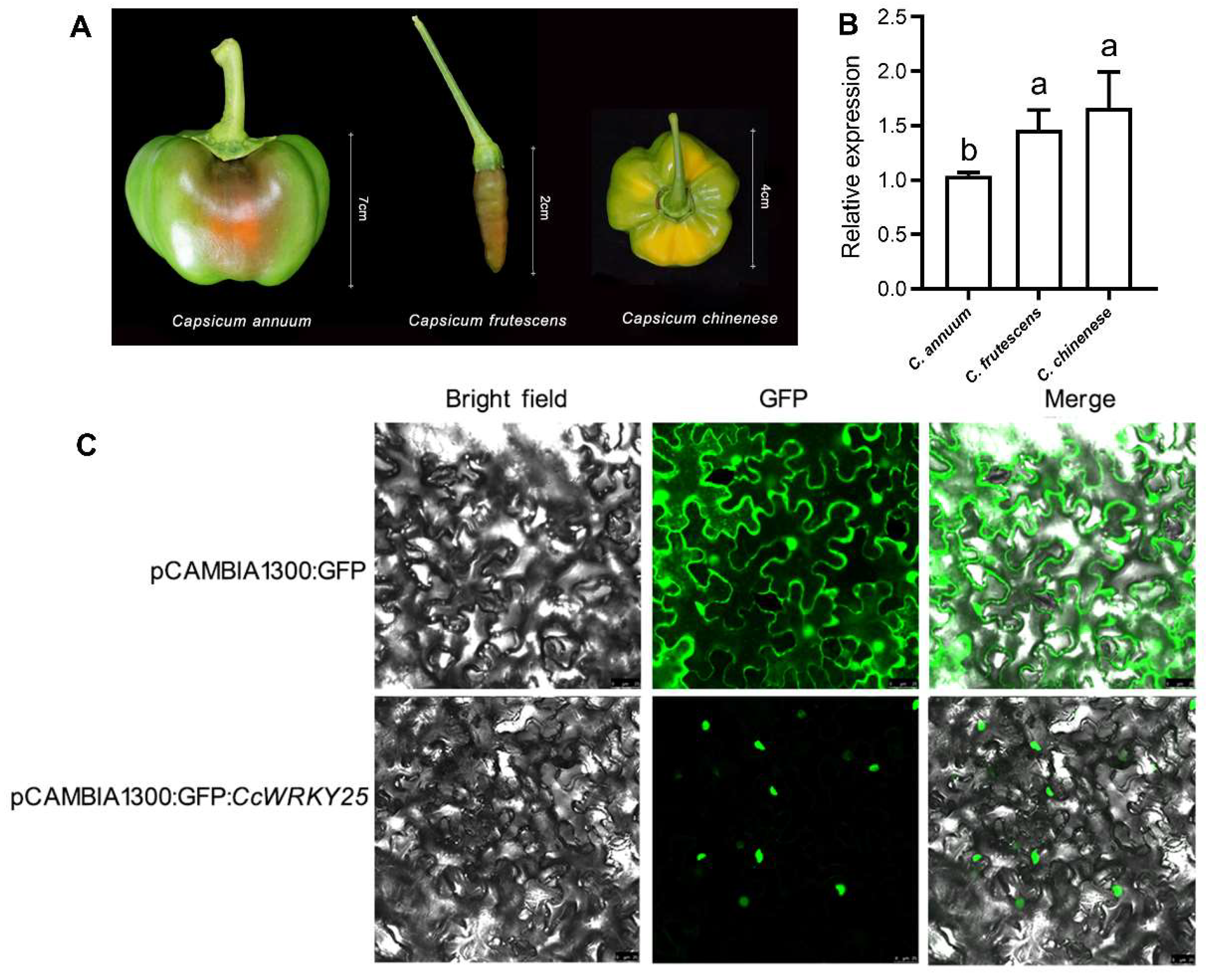
2.6. Cloning and Subcellular Localization of CcWRKY25
2.7. Heterologous Expression of CcWRKY25
2.8. Effects of Silencing CcWRKY25 on the Biosynthesis of Capsaicinoids and Lignin
3. Discussion
3.1. WRKY Transcription Factors in C. chinense
3.2. Functional Characterization of CcWRKY25
4. Materials and Methods
4.1. Plant Materials
4.2. Bioinformatic Analysis of CcWRKY Gene Family
4.3. Expression Patterns of CcWRKYs
4.4. RNA Extraction and RT-qPCR Analysis
4.5. Cloning and Subcellular Localization Assay of CcWRKY25
4.6. CcWRKY25 Transformation in Arabidopsis
4.7. Silencing the CcWRKY25 Gene in Huangdenglong Pepper
4.8. Measurement of Lignin, Flavonoids, and Capsaicinoid Content
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thul, S.T.; Darokar, M.P.; Shasany, A.K.; Khanuja, S.P. Molecular profiling for genetic variability in Capsicum species based on ISSR and RAPD markers. Mol. Biotechnol. 2012, 51, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-H.; Lu, W.C.; Wang, C.W.; Chan, Y.C.; Chen, M.K. Capsaicin induces cell cycle arrest and apoptosis in human KB cancer cells. BMC Complement. Altern. Med. 2013, 13, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, B.; Zhou, X.; Chen, C.; Chen, C.; Chen, K.; Chen, M.; Liu, S.; Chen, G.; Cao, B.; Cao, F. Coexpression network analysis reveals an MYB transcriptional activator involved in capsaicinoid biosynthesis in hot peppers. Hortic. Res. 2020, 7, 162. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Lovett, J.L.; Shedden, K.; Strassmann, B.I.; Vincenz, C. Targeted RNA-seq improves efficiency, resolution, and accuracy of allele specific expression for human term placentas. G3 2021, 11, jkab176. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.; Lee, D.G.; Back, S.; Hong, J.P.; Jang, S.; Han, K.; Kang, B.-C. Genetic mapping revealed that the Pun2 gene in Capsicum chacoense encodes a putative aminotransferase. Front. Plant. Sci. 2022, 13, 1039393. [Google Scholar] [CrossRef] [PubMed]
- Aza-González, C.; Núñez-Palenius, H.G.; Ochoa-Alejo, N. Molecular biology of capsaicinoid biosynthesis in chili pepper (Capsicum spp.). Plant Cell Rep. 2011, 30, 695–706. [Google Scholar] [CrossRef]
- Tanaka, Y.; Nakashima, F.; Kirii, E.; Goto, T.; Yoshida, Y.; Yasuba, K.I. Difference in capsaicinoid biosynthesis gene expression in the pericarp reveals elevation of capsai-cinoid contents in chili peppers (Capsicum chinense). Plant Cell Rep. 2017, 36, 267–279. [Google Scholar] [CrossRef]
- Mazourek, M.; Pujar, A.; Borovsky, Y.; Paran, I.; Mueller, L.; Jahn, M.M. A dynamic interface for capsaicinoid systems biology. Plant Physiol. 2009, 150, 1806–1821. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Park, M.; Yeom, S.I.; Kim, Y.M.; Lee, J.M.; Lee, H.A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.T. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef]
- Keyhaninejad, N.; Curry, J.; Romero, J.; O’Connell, M.A. Fruit specific variability in capsaicinoid accumulation and transcription of structural and regulatory genes in Capsicum fruit. Plant Sci. 2014, 215, 59–68. [Google Scholar] [CrossRef] [Green Version]
- Arce-Rodríguez, M.L.; Ochoa-Alejo, N. An R2R3-MYB transcription factor regulates capsaicinoid biosynthesis. Plant Physiol. 2017, 174, 1359–1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, B.; Chen, C.; Song, J.; Zheng, P.; Wang, J.; Wei, J.; Cai, W.; Chen, S.; Cai, Y.; Yuan, Y. The Capsicum MYB31 regulates capsaicinoid biosynthesis in the pepper pericarp. Plant Physiol. Biochem. 2022, 176, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Sun, B.; Cai, W.; Zhou, X.; Mao, Y.; Chen, C.; Wei, J.; Cao, B.; Chen, C.; Chen, G.; et al. Natural variations in the MYB transcription factor MYB31 determine the evolution of extremely pungent peppers. New Phytol. 2019, 223, 922–938. [Google Scholar] [CrossRef]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY gene family in rice (Oryza sativa). J. Integr. Plant Biol. 2007, 49, 827–842. [Google Scholar] [CrossRef]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef]
- Xiang, X.H.; Wu, X.R.; Hao, J.T.; Yang, M.L.; Yang, F.; Chen, G.; Liu, G.S.; Wang, Y.Y. Genome-wide identification and expression analysis of the WRKY gene family in common tobacco (Nicotiana tabacum L.). Hereditas 2016, 38, 840–856. [Google Scholar] [CrossRef]
- Mishra, P.; Tripathi, A.; Kashyap, S.P.; Aamir, M.; Tiwari, K.N.; Singh, V.; Tiwari, S.K. In silico mining of WRKY TFs through Solanum melongena L. and Solanum incanum L. transcriptomes and identification of SiWRKY53 as a source of resistance to bacterial wilt. Plant Gene 2021, 26, 100278. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X.; Chen, Y.; Liu, Y.; Wu, Y.; Ren, S.; Li, L. Identification, evolution and expression analysis of WRKY gene family in Eucommia ulmoides. Genomics 2021, 113, 3294–3309. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Scarpeci, T.E.; Anor, M.I.; Mueller-Roeber, B.; Valle, E.M. Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol. Biol. 2013, 83, 265–277. [Google Scholar] [CrossRef]
- Yu, Y.; Liu, Z.; Wang, L.; Kim, S.G.; Seo, P.J.; Qiao, M.; Wang, N.; Li, S.; Cao, X.; Park, C.M. WRKY 71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J. 2016, 85, 96–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markulin, L.; Corbin, C.; Renouard, S.; Drouet, S.; Durpoix, C.; Mathieu, C.; Lopez, T.; Auguin, D.; Hano, C.; Lainé, É. Characterization of LuWRKY36, a flax transcription factor promoting secoisolariciresinol biosynthesis in response to Fusarium oxysporum elicitors in Linum usitatissimum L. hairy roots. Planta 2019, 250, 347–366. [Google Scholar] [CrossRef]
- Singh, A.K.; Kumar, S.R.; Dwivedi, V.; Rai, A.; Pal, S.; Shasany, A.K.; Nagegowda, D.A. A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol. 2017, 215, 1115–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Song, G.; Zhang, F.; Shu, X.; Cheng, G.; Zhuang, W.; Wang, T.; Li, Y.; Wang, Z. Characterization of the WRKY gene family related to anthocyanin biosynthesis and the regulation mechanism under drought stress and methyl jasmonate treatment in Lycoris radiata. Int. J. Mol. Sci. 2023, 24, 2423. [Google Scholar] [CrossRef] [PubMed]
- Antonious, G.F.; Berke, T.; Jarret, R.L. Pungency in Capsicum chinense: Variation among countries of origin. J. Environ. Sci. Health B 2009, 44, 179–184. [Google Scholar] [CrossRef]
- Kim, S.; Park, J.; Yeom, S.I.; Kim, Y.M.; Seo, E.; Kim, K.T.; Kim, M.S.; Lee, J.M.; Cheong, K.; Shin, H.S. New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 2017, 18, 210. [Google Scholar] [CrossRef] [Green Version]
- Meng, D.; Li, Y.; Bai, Y.; Li, M.; Cheng, L. Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress. Plant Physiol. Biochem. 2016, 103, 71–83. [Google Scholar] [CrossRef]
- Dou, L.; Zhang, X.; Pang, C.; Song, M.; Wei, H.; Fan, S.; Yu, S. Genome-wide analysis of the WRKY gene family in cotton. Mol. Genet. Genom. 2014, 289, 1103–1121. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, M.; Zhang, X.; Hao, B.; Kaushik, S.; Pan, Y. WRKY gene family evolution in Arabidopsis thaliana. Genetica 2011, 139, 973–983. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahebi, M.; Hanafi, M.M.; Rafii, M.; Mahmud, T.; Azizi, P.; Osman, M.; Abiri, R.; Taheri, S.; Kalhori, N.; Shabanimofrad, M. Improvement of drought tolerance in rice (Oryza sativa L.): Genetics, genomic tools, and the WRKY gene family. Biomed. Res. Int. 2018, 2018, 3158474. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.L.; Guo, Z.-J.; Wang, H.H.; Li, J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005, 12, 9–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Verk, M.C.; Pappaioannou, D.; Neeleman, L.; Bol, J.F.; Linthorst, H.J. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 2008, 146, 1983–1995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Wang, W.; Wang, D.; Hu, S.; Zhang, Q.; Wang, Z.; Cui, L. Genome-wide identification and characterization of the WRKY gene family in Scutellaria baicalensis georgi under diverse abiotic stress. Int. J. Mol. Sci. 2022, 23, 4225. [Google Scholar] [CrossRef]
- Liu, K.; Yang, Q.; Yang, T.; Yang, F.; Wang, R.; Cong, J.; Li, G. Transcriptome-based identification and expression profiling of AP2/ERF members in Caragana intermedia and functional analysis of CiDREB3. Mol. Biol. Rep. 2021, 48, 7953–7965. [Google Scholar] [CrossRef]
- Govindarajan, V.S. Capsicum--production, technology, chemistry, and quality. Part III. Chemistry of the color, aroma, and pungency stimuli. Crit. Rev. Food Sci. Nutr. 1986, 24, 245–355. [Google Scholar] [CrossRef]
- Iwai, K.; Suzuki, T.; Fujiwake, H. Formation and accumulation of pungent principle of hot pepper fruits, capsaicin and its analogues, in Capsicum annuun var. Annuun cv. karayatsubusa at different growth stages after flowering. J. AGR Food Chem. 1979, 43, 2493–2498. [Google Scholar] [CrossRef] [Green Version]
- Mattus-Araya, E.; Guajardo, J.; Herrera, R.; Moya-León, M.A. ABA speeds up the progress of color in developing F. chiloensis fruit through the activation of PAL, CHS and ANS, key genes of the phenylpropanoid/flavonoid and anthocyanin pathways. Int. J. Mol. Sci. 2022, 23, 3854. [Google Scholar] [CrossRef]
- Ma, D.; Constabel, C.P. MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef]
- Shi, J.; Yan, X.; Sun, T.; Shen, Y.; Shi, Q.; Wang, W.; Bao, M.; Luo, H.; Nian, F.; Ning, G. Homeostatic regulation of flavonoid and lignin biosynthesis in phenylpropanoid pathway of transgenic tobacco. Gene 2022, 809, 146017. [Google Scholar] [CrossRef]
- Egan, A.N.; Moore, S.; Stellari, G.M.; Kang, B.C.; Jahn, M.M. Tandem gene duplication and recombination at the AT3 locus in the Solanaceae, a gene essential for capsaicinoid biosynthesis in Capsicum. PLoS ONE 2019, 14, e0210510. [Google Scholar] [CrossRef]
- Zhang, Z.X.; Zhao, S.N.; Liu, G.F.; Huang, Z.M.; Cao, Z.M.; Cheng, S.H.; Lin, S.S. Discovery of putative capsaicin biosynthetic genes by RNA-Seq and digital gene expression analysis of pepper. Sci. Rep. 2016, 6, 34121. [Google Scholar] [CrossRef]
- Fornalé, S.; Capellades, M.; Encina, A.; Wang, K.; Irar, S.; Lapierre, C.; Ruel, K.; Joseleau, J.P.; Berenguer, J.; Puigdomènech, P. Altered lignin biosynthesis improves cellulosic bioethanol production in transgenic maize plants down-regulated for cinnamyl alcohol dehydrogenase. Mol. Plant 2012, 5, 817–830. [Google Scholar] [CrossRef] [Green Version]
- Dwivedi, U.N.; Campbell, W.H.; Yu, J.; Datla, R.S.; Bugos, R.C.; Chiang, V.L.; Podila, G.K. Modification of lignin biosynthesis in transgenic Nicotiana through expression of an antisense O-methyltransferase gene from Populus. Plant Mol. Biol. 1994, 26, 61–71. [Google Scholar] [CrossRef]
- Ali, I.; Sher, H.; Ali, A.; Hussain, S.; Ullah, Z. Simplified floral dip transformation method of Arabidopsis thaliana. J. Microbiol. Methods 2022, 197, 106492. [Google Scholar] [CrossRef]
- Kim, J.; Park, M.; Jeong, E.S.; Lee, J.M.; Choi, D. Harnessing anthocyanin-rich fruit: A visible reporter for tracing virus-induced gene silencing in pepper fruit. Plant Methods 2017, 13, 3. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, Y.; Hosokawa, M.; Miwa, T.; Watanabe, T.; Yazawa, S. Newly mutated pu-tative-aminotransferase in nonpungent pepper (Capsicum annuum) results in biosynthesis of capsinoids, capsaicinoid analogues. J. Agric. Food Chem. 2010, 58, 1761–1767. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Wu, D.; Zhang, W.; Shu, H.; Sun, P.; Huang, C.; Deng, Q.; Wang, Z.; Cheng, S. Genome-Wide Identification of WRKY Gene Family and Functional Characterization of CcWRKY25 in Capsicum chinense. Int. J. Mol. Sci. 2023, 24, 11389. https://doi.org/10.3390/ijms241411389
Zhang L, Wu D, Zhang W, Shu H, Sun P, Huang C, Deng Q, Wang Z, Cheng S. Genome-Wide Identification of WRKY Gene Family and Functional Characterization of CcWRKY25 in Capsicum chinense. International Journal of Molecular Sciences. 2023; 24(14):11389. https://doi.org/10.3390/ijms241411389
Chicago/Turabian StyleZhang, Liping, Dan Wu, Wei Zhang, Huangying Shu, Peixia Sun, Chuang Huang, Qin Deng, Zhiwei Wang, and Shanhan Cheng. 2023. "Genome-Wide Identification of WRKY Gene Family and Functional Characterization of CcWRKY25 in Capsicum chinense" International Journal of Molecular Sciences 24, no. 14: 11389. https://doi.org/10.3390/ijms241411389





