Phospho-RNA-Seq Highlights Specific Small RNA Profiles in Plasma Extracellular Vesicles
Abstract
:1. Introduction
2. Results
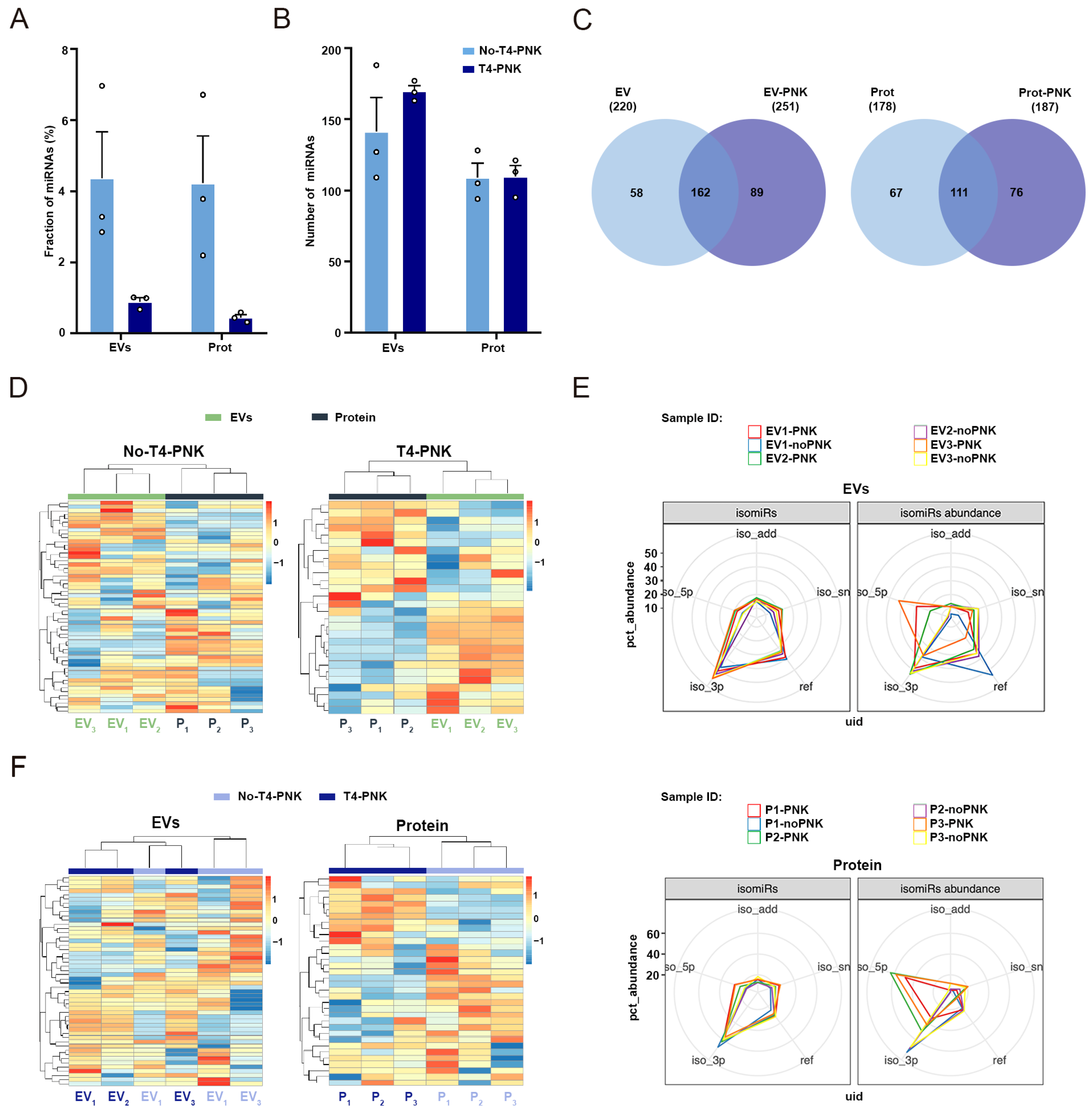
2.1. Study Design and General Characterization of sRNA Profiles
2.2. Phospho-sRNA Seq Stresses Specific sRNA Profiles in the EV and Non-EV Plasma Compartments
2.3. T4-PNK Highlights Different isomiRs Profiles in the EV and Non-EV Plasma Compartments
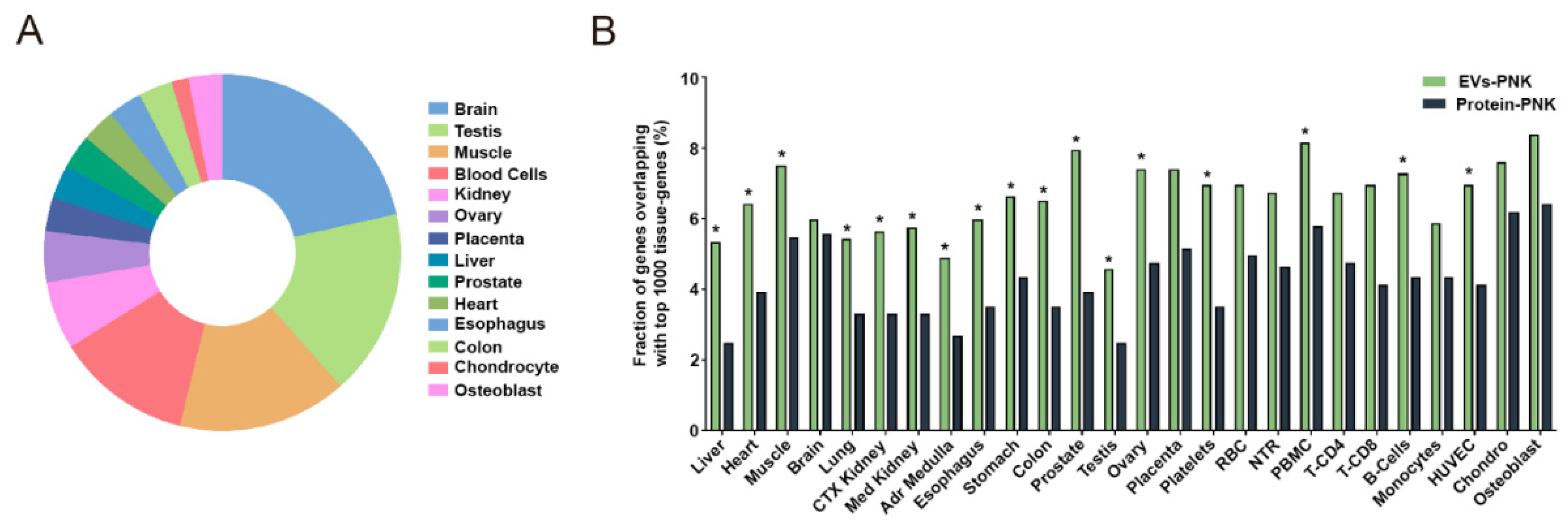
2.4. T4-PNK Increases the Proportion of Tissue-Expressed Gene Fragments in EVs
3. Discussion
4. Materials and Methods
4.1. Blood Collection and Sample Processing
4.2. Extracellular Vesicles Isolation
4.3. Quantification and Morphological Characterization of the EVs
4.4. Western Blot Analysis
4.5. Library Preparation and Sequencing
4.6. Bioinformatic Analysis for sRNA Characterization
4.7. Data Analysis and Statistical Testing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quinn, J.F.; Patel, T.; Wong, D.; Das, S.; Freedman, J.E.; Laurent, L.; Carter, B.S.; Hochberg, F.H.; Van Keuren-Jensen, K.; Huentelman, M.J.; et al. Extracellular RNAs: Development as biomarkers of human disease. J. Extracell. Vesicles 2015, 4, 27495. [Google Scholar] [CrossRef] [PubMed]
- Fritz, J.V.; Heintz-Buschart, A.; Ghosal, A.; Wampach, L.; Etheridge, A.; Galas, D.; Wilmes, P. Sources and Functions of Extracellular Small RNAs in Human Circulation. Annu. Rev. Nutr. 2016, 36, 301–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Max, K.E.A.; Bertram, K.; Akat, K.M.; Bogardus, K.A.; Li, J.; Morozov, P.; Ben-Dov, I.Z.; Li, X.; Weiss, Z.R.; Azizian, A.; et al. Human plasma and serum extracellular small RNA reference profiles and their clinical utility. Proc. Natl. Acad. Sci. USA 2018, 115, E5334–E5343. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Chen, P.; Yu, C.; Shi, Q.; Wei, S.; Li, Y.; Qi, H.; Cao, Q.; Guo, C.; Wu, X.; et al. Hypoxic bone marrow mesenchymal stromal cells-derived exosomal miR-182-5p promotes liver regeneration via FOXO1 -mediated macrophage polarization. FASEB J. 2022, 36, 334. [Google Scholar] [CrossRef]
- Sánchez, G.B.; Bunn, K.E.; Pua, H.H.; Rafat, M. Extracellular vesicles: Mediators of intercellular communication in tissue injury and disease. Cell Commun. Signal. 2021, 19, 106. [Google Scholar] [CrossRef]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef]
- Maia, J.; Caja, S.; Moraes, M.C.S.; Couto, N.; Costa-Silva, B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef] [Green Version]
- Hill, A.F.; Pegtel, D.M.; Lambertz, U.; Leonardi, T.; O’Driscoll, L.; Pluchino, S.; Ter-Ovanesyan, D.; Hoen, E.N.N’. ISEV position paper: Extracellular vesicle RNA analysis and bioinformatics. J. Extracell. Vesicles 2013, 2, 2. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [Green Version]
- Graner, M.W.; Alzate, O.; Dechkovskaia, A.M.; Keene, J.D.; Sampson, J.H.; Mitchell, D.A.; Bigner, D.D. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2008, 23, 1541–1557. [Google Scholar] [CrossRef] [Green Version]
- Sung, S.; Kim, J.; Jung, Y. Liver-Derived Exosomes and Their Implications in Liver Pathobiology. Int. J. Mol. Sci. 2019, 19, 3715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabbiano, F.; Corsi, J.; Gurrieri, E.; Trevisan, C.; Notarangelo, M.; D’Agostino, V.G. RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? J. Extracell. Vesicles 2020, 10, e12043. [Google Scholar] [CrossRef] [PubMed]
- Schageman, J.; Zeringer, E.; Li, M.; Barta, T.; Lea, K.; Gu, J.; Magdaleno, S.; Setterquist, R.; Vlassov, A.V. The Complete Exosome Workflow Solution: From Isolation to Characterization of RNA Cargo. BioMed Res. Int. 2013, 2013, 253957. [Google Scholar] [CrossRef]
- Lässer, C.; Shelke, G.V.; Yeri, A.; Kim, D.-K.; Crescitelli, R.; Raimondo, S.; Sjöstrand, M.; Gho, Y.S.; Van Keuren Jensen, K.; Lötvall, J. Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next-generation sequencing. RNA Biol. 2017, 14, 58–72. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasan, S.; Yeri, A.; Cheah, P.S.; Chung, A.; Danielson, K.; De Hoff, P.; Filant, J.; Laurent, C.D.; Laurent, L.D.; Magee, R.; et al. Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation. Cell 2019, 177, 446–462.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albanese, M.; Chen, Y.-F.A.; Hüls, C.; Gärtner, K.; Tagawa, T.; Mejias-Perez, E.; Keppler, O.T.; Göbel, C.; Zeidler, R.; Shein, M.; et al. MicroRNAs are minor constituents of extracellular vesicles that are rarely delivered to target cells. PLOS Genet. 2021, 17, e1009951. [Google Scholar] [CrossRef]
- Goodarzi, H.; Liu, X.; Nguyen, H.C.; Zhang, S.; Fish, L.; Tavazoie, S.F. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015, 161, 790–802. [Google Scholar] [CrossRef] [Green Version]
- Balatti, V.; Nigita, G.; Veneziano, D.; Drusco, A.; Stein, G.S.; Messier, T.L.; Farina, N.H.; Lian, J.B.; Tomasello, L.; Liu, C.-G.; et al. tsRNA signatures in cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 8071–8076. [Google Scholar] [CrossRef]
- Martinez, B.V.; Dhahbi, J.M.; Nunez Lopez, Y.O.; Lamperska, K.; Golusinski, P.; Luczewski, L.; Kolenda, T.; Atamna, H.; Spindler, S.R.; Golusiński, W.; et al. Circulating small non coding RNA signature in head and neck squamous cell carcinoma. Oncotarget 2015, 6, 19246–19263. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Vella, L.J.; Barnham, K.J.; McLean, C.; Masters, C.L.; Hill, A.F. Small RNA fingerprinting of Alzheimer’s disease frontal cortex extracellular vesicles and their comparison with peripheral extracellular vesicles. J. Extracell. Vesicles 2020, 9, 1766822. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Tosar, J.P.; Witwer, K.; Cayota, A. Revisiting Extracellular RNA Release, Processing, and Function. Trends Biochem. Sci. 2021, 46, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Freedman, J.E.; Gerstein, M.; Mick, E.; Rozowsky, J.; Levy, D.; Kitchen, R.; Das, S.; Shah, R.; Danielson, K.; Beaulieu, L.; et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat. Commun. 2016, 7, 11106. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Zhou, T.; Chen, Q. Exploring the expanding universe of small RNAs. Nature 2022, 24, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [Green Version]
- Akat, K.M.; Lee, Y.A.; Hurley, A.; Morozov, P.; Max, K.E.; Brown, M.; Bogardus, K.; Sopeyin, A.; Hildner, K.; Diacovo, T.G.; et al. Detection of circulating extracellular mRNAs by modified small-RNA-sequencing analysis. J. Clin. Investig. 2019, 4, e127317. [Google Scholar] [CrossRef] [Green Version]
- Giraldez, M.D.; Spengler, R.M.; Etheridge, A.; Goicochea, A.J.; Tuck, M.; Choi, S.W.; Galas, D.J.; Tewari, M. Phospho-RNA-seq: A modified small RNA-seq method that reveals circulating mRNA and lncRNA fragments as potential biomarkers in human plasma. EMBO J. 2019, 38, e101695. [Google Scholar] [CrossRef]
- Mizenko, R.R.; Brostoff, T.; Rojalin, T.; Koster, H.J.; Swindell, H.S.; Leiserowitz, G.S.; Wang, A.; Carney, R.P. Tetraspanins are unevenly distributed across single extracellular vesicles and bias sensitivity to multiplexed cancer biomarkers. J. Nanobiotechnol. 2021, 19, 250. [Google Scholar] [CrossRef]
- Jung, K.O.; Jo, H.; Yu, J.H.; Gambhir, S.S.; Pratx, G. Development and MPI tracking of novel hypoxia-targeted theranostic exosomes. Biomaterials 2018, 177, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Gámez-Valero, A.; Monguió-Tortajada, M.; Carreras-Planella, L.; La Franquesa, M.; Beyer, K.; Borràs, F.E. Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles’ characteristics compared to precipitating agents. Sci. Rep. 2016, 6, 33641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2012, 1820, 940–948. [Google Scholar] [CrossRef]
- Monguió-Tortajada, M.; Roura, S.; Gálvez-Montón, C.; Pujal, J.M.; Aran, G.; Sanjurjo, L.; Franquesa, M.; Sarrias, M.-R.; Bayes-Genis, A.; Borràs, F.E. Nanosized UCMSC-derived extracellular vesicles but not conditioned medium exclusively inhibit the inflammatory response of stimulated T cells: Implications for nanomedicine. Theranostics 2017, 7, 270–284. [Google Scholar] [CrossRef]
- Turchinovich, A.; Surowy, H.; Serva, A.; Zapatka, M.; Lichter, P.; Burwinkel, B. Capture and Amplification by Tailing and Switching (CATS). RNA Biol. 2014, 11, 817–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozowsky, J.; Kitchen, R.R.; Park, J.J.; Galeev, T.R.; Diao, J.; Warrell, J.; Thistlethwaite, W.; Subramanian, S.L.; Milosavljevic, A.; Gerstein, M. exceRpt: A Comprehensive Analytic Platform for Extracellular RNA Profiling. Cell Syst. 2019, 8, 352–357.e3. [Google Scholar] [CrossRef] [Green Version]
- Pantano, L.; Estivill, X.; Martí, E. A non-biased framework for the annotation and classification of the non-miRNA small RNA transcriptome. Bioinformatics 2011, 27, 3202–3203. [Google Scholar] [CrossRef] [Green Version]
- Pantano, L.; Pantano, F.; Marti, E.; Sui, S.H. Visualization of the small RNA transcriptome using seqclusterViz. F1000Research 2019, 8, 232. [Google Scholar] [CrossRef]
- Pantano, L.; Estivill, X.; Martí, E. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res. 2010, 38, e34. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Zhang, Y.; Tan, D.; Zhang, X.; Yan, M.; Zhang, Y.; Franklin, R.; Shahbazi, M.; Mackinlay, K.; Liu, S.; et al. PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nature 2021, 23, 424–436. [Google Scholar] [CrossRef]
- Liu, X.; Yu, X.; Zack, D.J.; Zhu, H.; Qian, J. TiGER: A database for tissue-specific gene expression and regulation. BMC Bioinform. 2008, 9, 271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Cherlin, T.; Magee, R.; Jing, Y.; Pliatsika, V.; Loher, P.; Rigoutsos, I. Ribosomal RNA fragmentation into short RNAs (rRFs) is modulated in a sex- and population of origin-specific manner. BMC Biol. 2020, 18, 38. [Google Scholar] [CrossRef] [PubMed]
- Tosar, J.P.; Cayota, A. Extracellular tRNAs and tRNA-derived fragments. RNA Biol. 2020, 17, 1149–1167. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Yang, S.; Huang, J.; Zheng, H.; He, Y.; Wang, L. Distinct Extracellular RNA Profiles in Different Plasma Components. Front. Genet. 2021, 12, 564780. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Muller, H.; Marzi, M.; Nicassio, F. IsomiRage: From Functional Classification to Differential Expression of miRNA Isoforms. Front. Bioeng. Biotechnol. 2014, 2, 38. [Google Scholar] [CrossRef] [Green Version]
- Tomasello, L.; Distefano, R.; Nigita, G.; Croce, C.M. The MicroRNA Family Gets Wider: The IsomiRs Classification and Role. Front. Cell Dev. Biol. 2021, 9, 668648. [Google Scholar] [CrossRef]
- Vickers, K.C.; Sethupathy, P.; Baran-Gale, J.; Remaley, A.T. Complexity of microRNA function and the role of isomiRs in lipid homeostasis. J. Lipid Res. 2013, 54, 1182–1191. [Google Scholar] [CrossRef] [Green Version]
- Lan, C.; Peng, H.; McGowan, E.M.; Hutvagner, G.; Li, J. An isomiR expression panel based novel breast cancer classification approach using improved mutual information. BMC Med. Genom. 2018, 11, 118. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Zheng, Z.; Chen, P.; Wu, M. Tumor classification and biomarker discovery based on the 5′isomiR expression level. BMC Cancer 2019, 19, 127. [Google Scholar] [CrossRef] [PubMed]
- Zelli, V.; Compagnoni, C.; Capelli, R.; Corrente, A.; Cornice, J.; Vecchiotti, D.; Di Padova, M.; Zazzeroni, F.; Alesse, E.; Tessitore, A. Emerging Role of isomiRs in Cancer: State of the Art and Recent Advances. Genes 2021, 12, 1447. [Google Scholar] [CrossRef] [PubMed]
- Glogovitis, I.; Yahubyan, G.; Würdinger, T.; Koppers-Lalic, D.; Baev, V. isomiRs–Hidden Soldiers in the miRNA Regulatory Army, and How to Find Them? Biomolecules 2020, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Alsop, E.; Meechoovet, B.; Kitchen, R.; Sweeney, T.; Beach, T.G.; Serrano, G.E.; Hutchins, E.; Ghiran, I.; Reiman, R.; Syring, M.; et al. A Novel Tissue Atlas and Online Tool for the Interrogation of Small RNA Expression in Human Tissues and Biofluids. Front. Cell Dev. Biol. 2022, 10, 804164. [Google Scholar] [CrossRef] [PubMed]
- Gerstberger, S.; Hafner, M.; Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 829–845. [Google Scholar] [CrossRef]
- Munir, J.; Yoon, J.K.; Ryu, S. Therapeutic miRNA-Enriched Extracellular Vesicles: Current Approaches and Future Prospects. Cells 2020, 9, 2271. [Google Scholar] [CrossRef] [PubMed]
- Trotta, C.R.; Miao, F.; Arn, E.A.; Stevens, S.W.; Ho, C.K.; Rauhut, R.; Abelson, J.N. The Yeast tRNA Splicing Endonuclease: A Tetrameric Enzyme with Two Active Site Subunits Homologous to the Archaeal tRNA Endonucleases. Cell 1997, 89, 849–858. [Google Scholar] [CrossRef] [Green Version]
- Luhtala, N.; Parker, R. T2 Family ribonucleases: Ancient enzymes with diverse roles. Trends Biochem. Sci. 2010, 35, 253–259. [Google Scholar] [CrossRef] [Green Version]
- Brennan-Laun, S.E.; Ezelle, H.J.; Li, X.-L.; Hassel, B.A. RNase-L Control of Cellular mRNAs: Roles in Biologic Functions and Mechanisms of Substrate Targeting. J. Interf. Cytokine Res. 2014, 34, 275–288. [Google Scholar] [CrossRef] [Green Version]
- Upton, J.-P.; Wang, L.; Han, D.; Wang, E.S.; Huskey, N.E.; Lim, L.; Truitt, M.; McManus, M.T.; Ruggero, D.; Goga, A.; et al. IRE1α Cleaves Select microRNAs During ER Stress to Derepress Translation of Proapoptotic Caspase-2. Science 2012, 338, 818–822. [Google Scholar] [CrossRef] [Green Version]
- Rath, S.; Donovan, J.; Whitney, G.; Chitrakar, A.; Wang, W.; Korennykh, A. Human RNase L tunes gene expression by selectively destabilizing the microRNA-regulated transcriptome. Proc. Natl. Acad. Sci. USA 2015, 112, 15916–15921. [Google Scholar] [CrossRef] [PubMed]
- Greulich, W.; Wagner, M.; Gaidt, M.M.; Stafford, C.; Cheng, Y.; Linder, A.; Carell, T.; Hornung, V. TLR8 Is a Sensor of RNase T2 Degradation Products. Cell 2019, 179, 1264–1275.e13. [Google Scholar] [CrossRef] [PubMed]
- Rashad, S.; Niizuma, K.; Tominaga, T. tRNA cleavage: A new insight. Neural Regen. Res. 2020, 15, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Salzman, D.W.; Nakamura, K.; Nallur, S.; Dookwah, M.T.; Metheetrairut, C.; Slack, F.J.; Weidhaas, J.B. miR-34 activity is modulated through 5′-end phosphorylation in response to DNA damage. Nat. Commun. 2016, 7, 10954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czerniak, T.; Saenz, J.P. Lipid membranes modulate the activity of RNA through sequence-dependent interactions. Proc. Natl. Acad. Sci. USA 2022, 119, e2119235119. [Google Scholar] [CrossRef]
- Pandey, K.K.; Madhry, D.; Kumar, Y.R.; Malvankar, S.; Sapra, L.; Srivastava, R.K.; Bhattacharyya, S.; Verma, B. Regulatory roles of tRNA-derived RNA fragments in human pathophysiology. Mol. Ther.—Nucleic Acids 2021, 26, 161–173. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Stahl, P.D.; Raposo, G. Extracellular Vesicles: Exosomes and Microvesicles, Integrators of Homeostasis. Physiology 2019, 34, 169–177. [Google Scholar] [CrossRef]
- Pua, H.H.; Happ, H.C.; Gray, C.J.; Mar, D.J.; Chiou, N.-T.; Hesse, L.E.; Ansel, K.M. Increased Hematopoietic Extracellular RNAs and Vesicles in the Lung during Allergic Airway Responses. Cell Rep. 2019, 26, 933–944.e4. [Google Scholar] [CrossRef] [Green Version]
- Konstantinidou, A.; Mougios, V.; Sidossis, L.S. Acute Exercise Alters the Levels of Human Saliva miRNAs Involved in Lipid Metabolism. Int. J. Sport. Med. 2016, 37, 584–588. [Google Scholar] [CrossRef]
- Shah, R.; Yeri, A.; Das, A.; Courtright-Lim, A.; Ziegler, O.; Gervino, E.; Ocel, J.; Pinzon, P.Q.; Wooster, L.; Bailey, C.S.; et al. Small RNA-seq during acute maximal exercise reveal RNAs involved in vascular inflammation and cardiometabolic health: Brief report. Am. J. Physiol. Circ. Physiol. 2017, 313, H1162–H1167. [Google Scholar] [CrossRef] [PubMed]
- Murillo, O.D.; Thistlethwaite, W.; Rozowsky, J.; Subramanian, S.L.; Lucero, R.; Shah, N.; Jackson, A.R.; Srinivasan, S.; Chung, A.; Laurent, C.D.; et al. exRNA Atlas Analysis Reveals Distinct Extracellular RNA Cargo Types and Their Carriers Present across Human Biofluids. Cell 2019, 177, 463–477.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shurtleff, M.J.; Yao, J.; Qin, Y.; Nottingham, R.M.; Temoche-Diaz, M.M.; Schekman, R.; Lambowitz, A.M. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. USA 2017, 114, E8987–E8995. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Vermesh, O.; Mani, V.; Ge, T.J.; Madsen, S.J.; Sabour, A.; Hsu, E.-C.; Gowrishankar, G.; Kanada, M.; Jokerst, J.V.; et al. The Exosome Total Isolation Chip. ACS Nano 2017, 11, 10712–10723. [Google Scholar] [CrossRef]
- Böing, A.N.; van der Pol, E.; Grootemaat, A.E.; Coumans, F.A.W.; Sturk, A.; Nieuwland, R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 2014, 3, 23430. [Google Scholar] [CrossRef] [PubMed]
- Sáenz-Cuesta, M.; Arbelaiz, A.; Oregi, A.; Irizar, H.; Osorio-Querejeta, I.; Muñoz-Culla, M.; Banales, J.; Falcón-Pérez, J.M.; Olascoaga, J.; Otaegui, D. Methods for extracellular vesicles isolation in a hospital setting. Front. Immunol. 2015, 6, 50. [Google Scholar] [CrossRef]
- Gámez-Valero, A.; Campdelacreu, J.; Vilas, D.; Ispierto, L.; Reñé, R.; Álvarez, R.; Armengol, M.P.; Borràs, F.E.; Beyer, K. Exploratory study on microRNA profiles from plasma-derived extracellular vesicles in Alzheimer’s disease and dementia with Lewy bodies. Transl. Neurodegener. 2019, 8, 31. [Google Scholar] [CrossRef] [Green Version]
- Monguió-Tortajada, M.; Morón-Font, M.; Gámez-Valero, A.; Carreras-Planella, L.; Borràs, F.E.; Franquesa, M. Extracellular-Vesicle Isolation from Different Biological Fluids by Size-Exclusion Chromatography. Curr. Protoc. Stem Cell Biol. 2019, 49, e82. [Google Scholar] [CrossRef]
- Lozano-Ramos, I.; Bancu, I.; Oliveira-Tercero, A.; Armengol, M.P.; Menezes-Neto, A.; Del Portillo, H.A.; Lauzurica-Valdemoros, R.; Borràs, F.E. Size-exclusion chromatography-based enrichment of extracellular vesicles from urine samples. J. Extracell. Vesicles 2015, 4, 27369. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solaguren-Beascoa, M.; Gámez-Valero, A.; Escaramís, G.; Herrero-Lorenzo, M.; Ortiz, A.M.; Minguet, C.; Gonzalo, R.; Bravo, M.I.; Costa, M.; Martí, E. Phospho-RNA-Seq Highlights Specific Small RNA Profiles in Plasma Extracellular Vesicles. Int. J. Mol. Sci. 2023, 24, 11653. https://doi.org/10.3390/ijms241411653
Solaguren-Beascoa M, Gámez-Valero A, Escaramís G, Herrero-Lorenzo M, Ortiz AM, Minguet C, Gonzalo R, Bravo MI, Costa M, Martí E. Phospho-RNA-Seq Highlights Specific Small RNA Profiles in Plasma Extracellular Vesicles. International Journal of Molecular Sciences. 2023; 24(14):11653. https://doi.org/10.3390/ijms241411653
Chicago/Turabian StyleSolaguren-Beascoa, Maria, Ana Gámez-Valero, Georgia Escaramís, Marina Herrero-Lorenzo, Ana M. Ortiz, Carla Minguet, Ricardo Gonzalo, Maria Isabel Bravo, Montserrat Costa, and Eulàlia Martí. 2023. "Phospho-RNA-Seq Highlights Specific Small RNA Profiles in Plasma Extracellular Vesicles" International Journal of Molecular Sciences 24, no. 14: 11653. https://doi.org/10.3390/ijms241411653





