Endoreplication—Why Are We Not Using Its Full Application Potential?
Abstract
:1. Introduction
1.1. Basic Differences between the Cell Cycle and the Endocycle
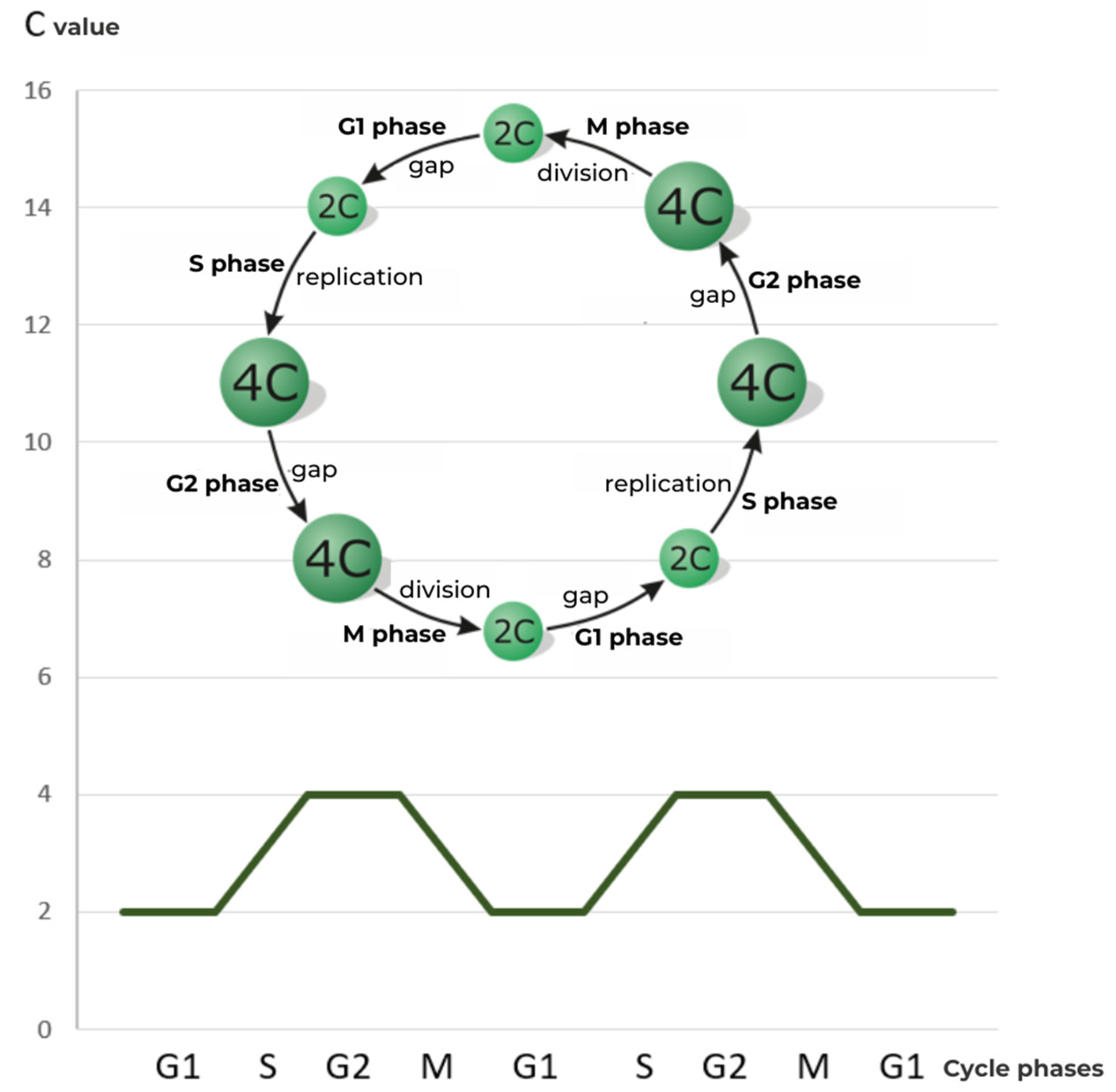
1.1.1. Classic Cell Cycle
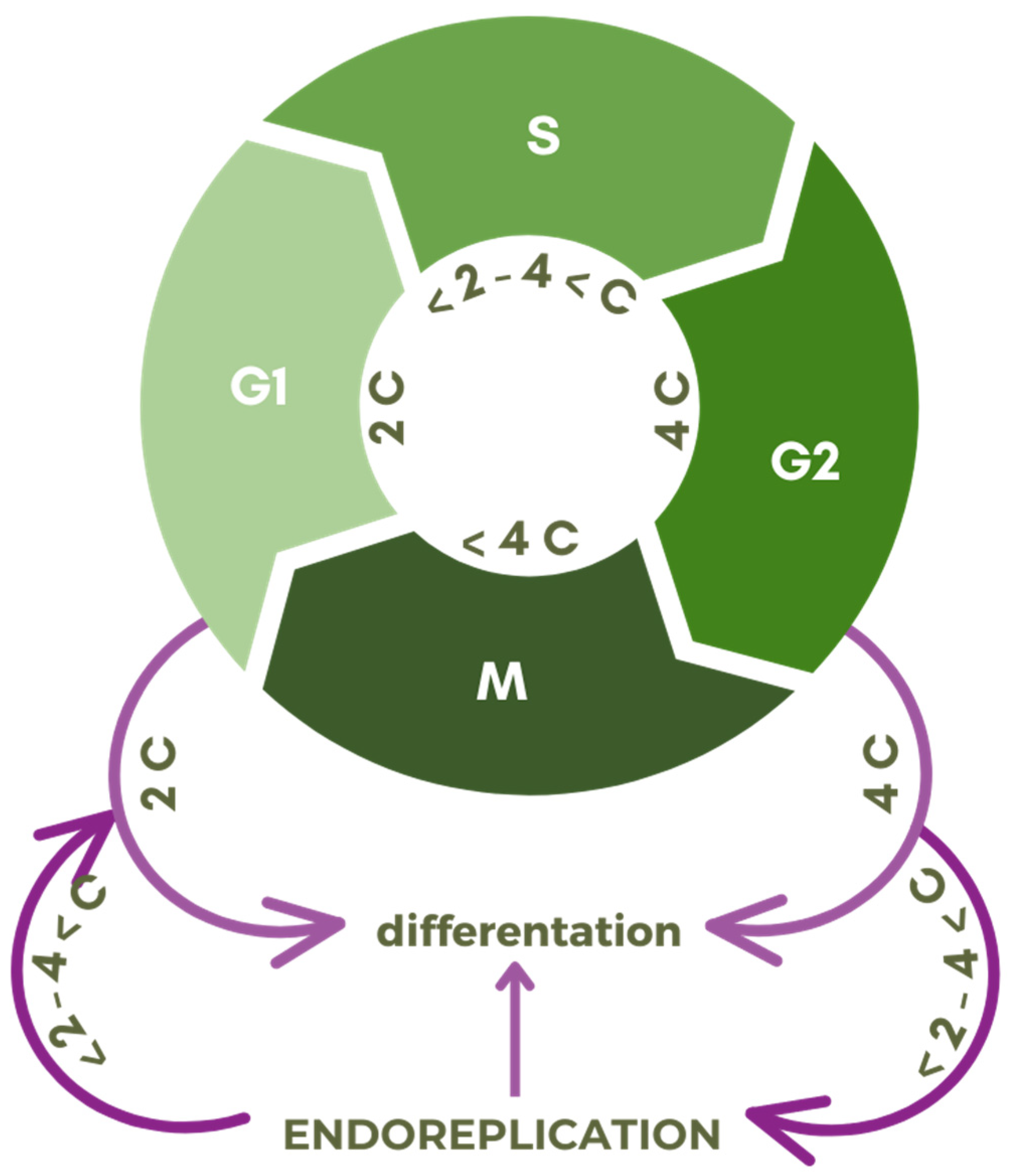
1.1.2. Foundations of Endoreplication
1.1.3. Endocycle Types
Endoreduplication
Endomitosis
Incomplete Replication
Amplification
2. Endoreplication in a Molecular Shortcut
3. What Do Plants Need Endocycles for?
4. Evolutionary Role of Endoreplication
5. The Universality of Endocycles
6. Endocycle Limitations
7. Difficulties in Exploiting Endoreplication by Humans
8. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, H.O.; Davidson, J.M.; Duronio, R.J. Endoreplication: Polyploidy with purpose. Genes Dev. 2009, 23, 2461–2477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagl, W. DNA endoreduplication and polyteny understood as evolutionary strategies. Nature 1976, 261, 614–615. [Google Scholar] [CrossRef]
- Gregory, T.R. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol. Rev. 2001, 76, 65–101. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J. Genetic and Epigenetic Mechanisms for Gene Expression and Phenotypic Variation in Plant Polyploids. Annu. Rev. Plant Biol. 2007, 58, 377–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boudolf, V.; Vlieghe, K.; Beemster, G.T.; Magyar, Z.; Acosta, J.A.T.; Maes, S.; Van Der Schueren, E.; Inzé, D.; De Veylder, L. The Plant-Specific Cyclin-Dependent Kinase CDKB1;1 and Transcription Factor E2Fa-DPa Control the Balance of Mitotically Dividing and Endoreduplicating Cells in Arabidopsis. Plant Cell 2004, 16, 2683–2692. [Google Scholar] [CrossRef] [Green Version]
- De Storme, N.; Copenhaver, G.P.; Geelen, D. Production of Diploid Male Gametes in Arabidopsis by Cold-Induced Destabi-lization of Postmeiotic Radial Microtubule Arrays. Plant Physiol. 2012, 160, 1808–1826. [Google Scholar] [CrossRef] [Green Version]
- Kitagawa, M.; Jackson, D. Control of Meristem Size. Annu. Rev. Plant Biol. 2019, 70, 269–291. [Google Scholar] [CrossRef]
- Fox, D.T.; Soltis, D.E.; Soltis, P.S.; Ashman, T.-L.; Van de Peer, Y. Polyploidy: A Biological Force From Cells to Ecosystems. Trends Cell Biol. 2020, 30, 688–694. [Google Scholar] [CrossRef]
- Frawley, L.E.; Orr-Weaver, T.L. Polyploidy. Curr. Biol. 2015, 25, R353–R358. [Google Scholar] [CrossRef] [Green Version]
- Tate, J.A.; Soltis, D.E.; Soltis, P.S.; Gregory, T.R. The Evolution of the Genome; Academic Press: Cambridge, MA, USA, 2005; Volume 2005, pp. 371–426. [Google Scholar]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef]
- Bardil, A.; Tayalé, A.; Parisod, C. Evolutionary dynamics of retrotransposons following autopolyploidy in the Buckler Mustard species complex. Plant J. 2015, 82, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.P.; Dutta, A. DNA Replication in Eukaryotic Cells. Annu. Rev. Biochem. 2002, 71, 333–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cebolla, A.; Vinardell, J.M.; Kiss, E.; Oláh, B.; Roudier, F.; Kondorosi, A.; Kondorosi, E. The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 1999, 18, 4476–4484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jovtchev, G.; Schubert, V.; Meister, A.; Barow, M.; Schubert, I. Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet. Genome Res. 2006, 114, 77–82. [Google Scholar] [CrossRef]
- Beaulieu, J.M.; Leitch, I.J.; Patel, S.; Pendharkar, A.; Knight, C.A. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytol. 2008, 179, 975–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butterfass, T. A nucleotypic control of chloroplast reproduction. Protoplasma 1983, 118, 71–74. [Google Scholar] [CrossRef]
- Umeda, M.; Ikeuchi, M.; Ishikawa, M.; Ito, T.; Nishihama, R.; Kyozuka, J.; Torii, K.U.; Satake, A.; Goshima, G.; Sakakibara, H. Plant stem cell research is uncovering the secrets of longevity and persistent growth. Plant J. 2021, 106, 326–335. [Google Scholar] [CrossRef]
- Sabelli, P.A. Replicate and die for your own good: Endoreduplication and cell death in the cereal endosperm. J. Cereal Sci. 2012, 56, 9–20. [Google Scholar] [CrossRef]
- Bourdon, M.; Pirrello, J.; Cheniclet, C.; Coriton, O.; Bourge, M.; Brown, S.; Moïse, A.; Peypelut, M.; Rouyère, V.; Renaudin, J.-P.; et al. Evidence for karyoplasmic homeostasis during endoreduplication and a ploidy-dependent increase in gene transcription during tomato fruit growth. Development 2012, 139, 3817–3826. [Google Scholar] [CrossRef] [Green Version]
- Chao, D.-Y.; Dilkes, B.; Luo, H.; Douglas, A.; Yakubova, E.; Lahner, B.; Salt, D.E. Polyploids Exhibit Higher Potassium Uptake and Salinity Tolerance in Arabidopsis. Science 2013, 341, 658–659. [Google Scholar] [CrossRef] [Green Version]
- Hannweg, K.; Steyn, W.; Bertling, I. In vitro-induced tetraploids of Plectranthus esculentus are nematode-tolerant and have enhanced nutritional value. Euphytica 2016, 207, 343–351. [Google Scholar] [CrossRef]
- Schubert, V.; Klatte, M.; Pecinka, A.; Meister, A.; Jasencakova, Z.; Schubert, I. Sister Chromatids Are Often Incompletely Aligned in Meristematic and Endopolyploid Interphase Nuclei of Arabidopsis thaliana. Genetics 2006, 172, 467–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biskup, A.; Izmailow, R. Endosperm Development in Seeds of Echium vulgare L. [Boraginaceae] from Polluted Sites. Acta Biol. Cracoviensia Ser. Bot. 2004, 46, 39–44. [Google Scholar]
- Oksala, T.; Therman, E. Endomitosis in Tapetal Cells of Eremurus (Liliaceae). Am. J. Bot. 1977, 64, 866–872. [Google Scholar] [CrossRef]
- D’amato, F. Role of Polyploidy in Reproductive Organs and Tissues. In Embryology of Angiosperms; Springer: Berlin/Heidelberg, Germany, 1984; pp. 519–566. [Google Scholar] [CrossRef]
- Nagl, W. Gene Amplification and Related Events. In Somaclonal Variation in Crop Improvement I.; Bajaj, Y.P.S., Ed.; Bio-Technology in Agriculture and Forestry; Springer: Berlin/Heidelberg, Germany, 1990; pp. 153–201. [Google Scholar] [CrossRef]
- Cook, G.S.; Grønlund, A.L.; Siciliano, I.; Spadafora, N.; Amini, M.; Herbert, R.J.; Bitonti, M.B.; Graumann, K.; Francis, D.; Rogers, H.J. Plant WEE1 kinase is cell cycle regulated and removed at mitosis via the 26S proteasome machinery. J. Exp. Bot. 2013, 64, 2093–2106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Veylder, L.; Beeckman, T.; Beemster, G.T.; Engler, J.d.A.; Ormenese, S.; Maes, S.; Naudts, M.; Van Der Schueren, E.; Jacqmard, A.; Engler, G.; et al. Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 2002, 21, 1360–1368. [Google Scholar] [CrossRef] [Green Version]
- De Veylder, L.; Larkin, J.C.; Schnittger, A. Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 2011, 16, 624–634. [Google Scholar] [CrossRef]
- Gómez, M.S.; Sheridan, M.L.; Casati, P. E2Fb and E2Fa transcription factors independently regulate the DNA damage re-sponse after ultraviolet B exposure in Arabidopsis. Plant J. 2022, 109, 1098–1115. [Google Scholar] [CrossRef]
- Grafi, G.; Larkins, B.A. Endoreduplication in Maize Endosperm: Involvement of M Phase—Promoting Factor Inhibition and Induction of S Phase—Related Kinases. Science 1995, 269, 1262–1264. [Google Scholar] [CrossRef]
- Kołodziejczyk, I.; Kaźmierczak, A.; Posmyk, M.M. Melatonin Application Modifies Antioxidant Defense and Induces Endoreplication in Maize Seeds Exposed to Chilling Stress. Int. J. Mol. Sci. 2021, 22, 8628. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, G.; Gao, F.; Xu, R.; Li, N.; Zhang, Y.; Li, Y. Transcriptional repression of the APC/C activator genes CCS52A1/A2 by the mediator complex subunit MED16 controls endoreduplication and cell growth in Arabidopsis. Plant Cell 2019, 31, 1899–1912. [Google Scholar] [CrossRef]
- Mahapatra, K.; Roy, S. SOG1 transcription factor promotes the onset of endoreduplication under salinity stress in Arabidopsis. Sci. Rep. 2021, 11, 11659. [Google Scholar] [CrossRef]
- Nowack, M.K.; Harashima, H.; Dissmeyer, N.; Zhao, X.; Bouyer, D.; Weimer, A.K.; De Winter, F.; Yang, F.; Schnittger, A. Genetic Framework of Cyclin-Dependent Kinase Function in Arabidopsis. Dev. Cell 2012, 22, 1030–1040. [Google Scholar] [CrossRef] [Green Version]
- Sozzani, R.; Cui, H.; Moreno-Risueno, M.A.; Busch, W.; Van Norman, J.M.; Vernoux, T.; Brady, S.M.; Dewitte, W.; Murray, J.A.H.; Benfey, P.N. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 2010, 466, 128–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Z.; Ji, Y.; Sun, L.; Xu, X.; Fan, D.; Zhong, H.; Liang, Z.; Gunther, F. Changes in production potentials of rapeseed in the Yangtze River Basin of China under climate change: A multi-model ensemble approach. J. Geogr. Sci. 2018, 28, 1700–1714. [Google Scholar] [CrossRef] [Green Version]
- Vlad, D.; Kierzkowski, D.; Rast, M.I.; Vuolo, F.; Ioio, R.D.; Galinha, C.; Gan, X.; Hajheidari, M.; Hay, A.; Smith, R.S.; et al. Leaf Shape Evolution Through Duplication, Regulatory Diversification, and Loss of a Homeobox Gene. Science 2014, 343, 780–783. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Konate, M.; Sai, N.; Tesfamicael, K.G.; Cavagnaro, T.; Gilliham, M.; Breen, J.; Metcalfe, A.; Stephen, J.R.; De Bei, R.; et al. Global DNA Methylation Patterns Can Play a Role in Defining Terroir in Grapevine (Vitis vinifera cv. Shiraz). Front. Plant Sci. 2017, 8, 1860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshiyama, K.O. SOG1: A master regulator of the DNA damage response in plants. Genes Genet. Syst. 2015, 90, 209–216. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Gu, M.; Xia, X.; Shi, K.; Zhou, Y.; Yu, J. Effects of Phenylcarboxylic Acids on Mitosis, Endoreduplication and Expression of Cell Cycle-Related Genes in Roots of Cucumber (Cucumis sativus L.). J. Chem. Ecol. 2009, 35, 679–688. [Google Scholar] [CrossRef]
- Soltys, D.; Rudzińska-Langwald, A.; Kurek, W.; Szajko, K.; Sliwinska, E.; Bogatek, R.; Gniazdowska, A. Phytotoxic cyanamide affects maize (Zea mays) root growth and root tip function: From structure to gene expression. J. Plant Physiol. 2014, 171, 565–575. [Google Scholar] [CrossRef]
- Deng, B.; Du, W.; Liu, C.; Sun, W.; Tian, S.; Dong, H. Antioxidant Response to Drought, Cold and Nutrient Stress in Two Ploidy Levels of Tobacco Plants: Low Resource Requirement Confers Polytolerance in Polyploids? Plant Growth Regul. 2012, 66, 37–47. [Google Scholar] [CrossRef]
- Godfree, R.C.; Marshall, D.J.; Young, A.G.; Miller, C.H.; Mathews, S. Empirical evidence of fixed and homeostatic patterns of polyploid advantage in a keystone grass exposed to drought and heat stress. R. Soc. Open Sci. 2017, 4, 170934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forrester, N.J.; Ashman, T. Autopolyploidy alters nodule-level interactions in the legume—Rhizobium mutualism. Am. J. Bot. 2020, 107, 179–185. [Google Scholar] [CrossRef] [Green Version]
- Hias, N.; Svara, A.; Keulemans, J.W. Effect of Polyploidisation on the Response of Apple (Malus domestica Borkh.) to Venturia Inaequalis Infection. Eur. J. Plant Pathol. 2018, 151, 515–526. [Google Scholar] [CrossRef]
- Collett, C.E.; Harberd, N.P.; Leyser, O. Hormonal Interactions in the Control of Arabidopsis Hypocotyl Elongation. Plant Physiol. 2000, 124, 553–562. [Google Scholar] [CrossRef] [Green Version]
- Nagl, W. Endopolyploidy and Polyteny in Differentiation and Evolution; Sole distributors for the U.S.A. and Canada; Elsevier: Amsterdam, The Netherlands, 1978. [Google Scholar]
- Nowicka, A.; Kovacik, M.; Tokarz, B.; Vrána, J.; Zhang, Y.; Weigt, D.; Doležel, J.; Pecinka, A. Dynamics of endoreduplication in developing barley seeds. J. Exp. Bot. 2021, 72, 268–282. [Google Scholar] [CrossRef] [PubMed]
- Su’udi, M.; Cha, J.-Y.; Ahn, I.-P.; Kwak, Y.-S.; Woo, Y.-M.; Son, D. Functional characterization of a B-type cell cycle switch 52 in rice (OsCCS52B). Plant Cell Tissue Organ Cult. (PCTOC) 2012, 111, 101–111. [Google Scholar] [CrossRef]
- Chevalier, C.; Nafati, M.; Mathieu-Rivet, E.; Bourdon, M.; Frangne, N.; Cheniclet, C.; Renaudin, J.-P.; Gévaudant, F.; Hernould, M. Elucidating the functional role of endoreduplication in tomato fruit development. Ann. Bot. 2011, 107, 1159–1169. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira, P.N.; da Silva, L.F.C.; Eloy, N.B. The role of APC/C in cell cycle dynamics, growth and development in cereal crops. Front. Plant Sci. 2022, 13, 987919. [Google Scholar] [CrossRef]
- Bhosale, R.; Boudolf, V.; Cuevas, F.; Lu, R.; Eekhout, T.; Hu, Z.; Van Isterdael, G.; Lambert, G.M.; Xu, F.; Nowack, M.K.; et al. A Spatiotemporal DNA Endoploidy Map of the Arabidopsis Root Reveals Roles for the Endocycle in Root Development and Stress Adaptation. Plant Cell 2018, 30, 2330–2351. [Google Scholar] [CrossRef] [Green Version]
- Kondorosi, E.; Roudier, F.; Gendreau, E. Plant Cell-Size Control: Growing by Ploidy? Curr. Opin. Plant Biol. 2000, 3, 488–492. [Google Scholar] [CrossRef]
- Weber, J.; Georgiev, V.; Pavlov, A.; Bley, T. Flow cytometric investigations of diploid and tetraploid plants and in vitro cultures of Datura stramonium and Hyoscyamus niger. Cytometry A. 2008, 73, 931–939. [Google Scholar] [CrossRef]
- Galbraith, D.W.; Harkins, K.R.; Knapp, S. Systemic Endopolyploidy in Arabidopsis thaliana. Plant Physiol. 1991, 96, 985–989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, H.D.; Westoby, M. The Relationship Between Nuclear DNA Content and Leaf Strategy in Seed Plants. Ann. Bot. 2005, 96, 1321–1330. [Google Scholar] [CrossRef] [PubMed]
- Tenaillon, M.I.; Manicacci, D.; Nicolas, S.D.; Tardieu, F.; Welcker, C. Testing the link between genome size and growth rate in maize. PeerJ 2016, 4, e2408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krahulcová, A.; Trávníček, P.; Krahulec, F.; Rejmánek, M. Small genomes and large seeds: Chromosome numbers, genome size and seed mass in diploid Aesculus species (Sapindaceae). Ann. Bot. 2017, 119, 957–964. [Google Scholar]
- Bilinski, P.; Albert, P.S.; Berg, J.J.; Birchler, J.A.; Grote, M.N.; Lorant, A.; Quezada, J.; Swarts, K.; Yang, J.; Ross-Ibarra, J. Parallel altitudinal clines reveal trends in adaptive evolution of genome size in Zea mays. PLOS Genet. 2018, 14, e1007162. [Google Scholar] [CrossRef] [Green Version]
- Puttick, M.N.; Clark, J.; Donoghue, P.C. Size is not everything: Rates of genome size evolution, not C-value, correlate with speciation in angiosperms. Proc. R. Soc. B Biol. Sci. 2015, 282, 20152289. [Google Scholar] [CrossRef]
- Igea, J.; Miller, E.F.; Papadopulos, A.S.; Tanentzap, A.J. Seed size and its rate of evolution correlate with species diversification across angiosperms. PLoS Biol. 2017, 15, e2002792. [Google Scholar] [CrossRef] [Green Version]
- Chumová, Z.; Záveská, E.; Ponert, J.; Schmidt, P.A.; Trávníček, P. Partial endoreplication stimulates diversification in the species-richest lineage of orchids. bioRxiv 2020. [Google Scholar] [CrossRef]
- Pyšek, P.; Skálová, H.; Čuda, J.; Guo, W.Y.; Suda, J.; Doležal, J.; Kauzál, O.; Lambertini, C.; Lučanová, M.; Mandáková, T.; et al. Small genome separates native and invasive populations in an ecologically important cosmopolitan grass. Ecology 2018, 99, 79–90. [Google Scholar] [CrossRef] [Green Version]
- Pellicer, J.; Hidalgo, O.; Dodsworth, S.; Leitch, I.J. Genome Size Diversity and Its Impact on the Evolution of Land Plants. Genes 2018, 9, 88. [Google Scholar] [CrossRef] [Green Version]
- Simonin, K.A.; Roddy, A.B. Genome downsizing, physiological novelty, and the global dominance of flowering plants. PLOS Biol. 2018, 16, e2003706. [Google Scholar] [CrossRef] [Green Version]
- Smarda, P.; Bureš, P.; Horová, L.; Leitch, I.J.; Mucina, L.; Pacini, E.; Tichý, L.; Grulich, V.; Rotreklová, O. Ecological and evo-lutionary significance of genomic GC content diversity in monocots. Proc. Natl. Acad. Sci. USA 2014, 111, E4096–E4102. [Google Scholar] [CrossRef]
- Veleba, A.; Šmarda, P.; Zedek, F.; Horová, L.; Šmerda, J.; Bureš, P. Evolution of genome size and genomic GC content in carnivorous holokinetics (Droseraceae). Ann. Bot. 2017, 119, 409–416. [Google Scholar] [CrossRef] [Green Version]
- Engelen-Eigles, G.; Jones, R.J.; Phillips, R.L. DNA endoreduplication in maize endosperm cells: The effect of exposure to short-term high temperature. Plant Cell Environ. 2000, 23, 657–663. [Google Scholar] [CrossRef]
- Coate, J.E.; Bar, H.; Doyle, J.J. Extensive Translational Regulation of Gene Expression in an Allopolyploid (Glycine dolichocarpa). Plant Cell 2014, 26, 136–150. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Escobar, O.A.; Chomicki, G.; Condamine, F.L.; Karremans, A.P.; Bogarín, D.; Matzke, N.J.; Silvestro, D.; Antonelli, A. Recent origin and rapid speciation of Neotropical orchids in the world’s richest plant biodiversity hotspot. New Phytol. 2017, 215, 891–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chumová, Z.; Záveská, E.; Hloušková, P.; Ponert, J.; Schmidt, P.A.; Čertner, M.; Mandáková, T.; Trávníček, P. Repeat prolif-eration and partial endoreplication jointly shape the patterns of genome size evolution in orchids. Plant J. 2021, 107, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Knight, C.A.; Clancy, R.B.; Götzenberger, L.; Dann, L.; Beaulieu, J.M. On the Relationship between Pollen Size and Genome Size. J. Bot. 2010, 2010, 612017. [Google Scholar] [CrossRef] [Green Version]
- Francis, D.; Davies, M.S.; Barlow, P.W. A Strong Nucleotypic Effect on the Cell Cycle Regardless of Ploidy Level. Ann. Bot. 2008, 101, 747–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šímová, I.; Herben, T. Geometrical constraints in the scaling relationships between genome size, cell size and cell cycle length in herbaceous plants. Proc. R. Soc. B Boil. Sci. 2011, 279, 867–875. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.R. Quantitative Changes in Nuclear DNA Accompanying Postgermination Embryonic Development in Vanda (Orchidaceae). Am. J. Bot. 1968, 55, 1036–1041. [Google Scholar] [CrossRef]
- del Pozo, J.C.; Ramirez-Parra, E. Deciphering the Molecular Bases for Drought Tolerance in Arabidopsis Autotetraploids. Plant Cell Environ. 2014, 37, 2722–2737. [Google Scholar] [CrossRef] [PubMed]
- Biradar, D.P.; Rayburn, A.L.; Bullock, D.G. Endopolyploidy in Diploid and Tetraploid Maize (Zea mays L.). Ann. Bot. 1993, 71, 417–421. [Google Scholar] [CrossRef]
- Trávníček, P.; Čertner, M.; Ponert, J.; Chumová, Z.; Jersáková, J.; Suda, J. Diversity in genome size and GC content shows adaptive potential in orchids and is closely linked to partial endoreplication, plant life-history traits and climatic conditions. New Phytol. 2019, 224, 1642–1656. [Google Scholar] [CrossRef]
- Trávníček, P.; Ponert, J.; Urfus, T.; Jersáková, J.; Vrána, J.; Hřibová, E.; Doležel, J.; Suda, J. Challenges of flow-cytometric estimation of nuclear genome size in orchids, a plant group with both whole-genome and progressively partial endoreplication. Cytom. Part A 2015, 87, 958–966. [Google Scholar] [CrossRef]
- Kinoshita, I.; Sanbe, A.; Yokomura, E.-I. Increases in Nuclear DNA Content without Mitosis in Benzyladenine-treated Primary Leaves of Intact and Decapitated Bean Plants. J. Exp. Bot. 1991, 42, 667–672. [Google Scholar] [CrossRef]
- Paige, K.N. Overcompensation, environmental stress, and the role of endoreduplication. Am. J. Bot. 2018, 105, 1105–1108. [Google Scholar] [CrossRef]
- Castellano, M.M.; del Pozo, J.C.; Ramirez-Parra, E.; Brown, S.; Gutierrez, C. Expression and Stability of Arabidopsis CDC6 Are Associated with Endoreplication. Plant Cell 2001, 13, 2671–2686. [Google Scholar] [CrossRef] [Green Version]
- Scholes, D.R.; Paige, K.N. Plasticity in ploidy: A generalized response to stress. Trends Plant Sci. 2015, 20, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Larkins, B.A.; Dilkes, B.P.; Dante, R.A.; Coelho, C.M.; Woo, Y.; Liu, Y. Investigating the hows and whys of DNA endoreduplication. J. Exp. Bot. 2001, 52, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Koprivý, L.; Fráková, V.; Kolarčik, V.; Mártonfiová, L.; Dudáš, M.; Mártonfi, P. Genome size and endoreplication in two pairs of cytogenetically contrasting species of Pulmonaria (Boraginaceae) in Central Europe. AoB Plants 2022, 14, plac036. [Google Scholar] [CrossRef]
- Dosier, L.W.; Riopel, J.L. Origin, Development, and Growth of Differentiating Trichoblasts in Elodea Canadensis. Am. J. Bot. 1978, 65, 813–822. [Google Scholar] [CrossRef]
- Nagl, W. The Phaseolus suspensor and its polytene chromosomes. Z. Für Pflanzenphysiol. 1974, 73, 1–44. [Google Scholar] [CrossRef]
- Kausch, A.P.; Horner, H.T. Increased nuclear DNA content in raphide crystal idioblasts during development in Vanilla planifolia L. (Orchidaceae). Eur. J. Cell Biol. 1984, 33, 7–12. [Google Scholar]
- Vitousek, P.M.; Porder, S.; Houlton, B.Z.; Chadwick, O.A. Terrestrial phosphorus limitation: Mechanisms, implications, and nitrogen–phosphorus interactions. Ecol. Appl. 2010, 20, 5–15. [Google Scholar] [CrossRef] [Green Version]
- Šmarda, P.; Hejcman, M.; Březinová, A.; Horová, L.; Steigerová, H.; Zedek, F.; Bureš, P.; Hejcmanová, P.; Schellberg, J. Effect of phosphorus availability on the selection of species with different ploidy levels and genome sizes in a long-term grassland fertilization experiment. New Phytol. 2013, 200, 911–921. [Google Scholar] [CrossRef]
- Melaragno, J.E.; Mehrotra, B.; Coleman, A.W. Relationship between Endopolyploidy and Cell Size in Epidermal Tissue of Arabidopsis. Plant Cell 1993, 5, 1661–1668. [Google Scholar] [CrossRef]
- Cookson, S.J.; Radziejwoski, A.; Granier, C. Cell and Leaf Size Plasticity in Arabidopsis: What Is the Role of Endoreduplication? Plant Cell Environ. 2006, 29, 1273–1283. [Google Scholar] [CrossRef]
- Dudits, D.; Cserháti, M.; Miskolczi, P.; Horváth, G.V. 1 The Growing Family of Plant Cyclin-Dependent Kinases with Multiple Functions in Cellular and Developmental Regulation. In Annual Plant Reviews, Cell Cycle Control and Plant Development; Wiley: Hoboken, NJ, USA, 2008; p. 1. [Google Scholar]
- Bainard, J.D.; Bainard, L.D.; Henry, T.A.; Fazekas, A.J.; Newmaster, S.G. A multivariate analysis of variation in genome size and endoreduplication in angiosperms reveals strong phylogenetic signal and association with phenotypic traits. New Phytol. 2012, 196, 1240–1250. [Google Scholar] [CrossRef] [PubMed]
- Lang, L.; Schnittger, A. Endoreplication—A means to an end in cell growth and stress response. Curr. Opin. Plant Biol. 2020, 54, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Bekaert, M.; Edger, P.P.; Pires, J.C.; Conant, G.C. Two-Phase Resolution of Polyploidy in the Arabidopsis Metabolic Network Gives Rise to Relative and Absolute Dosage Constraints. Plant Cell 2011, 23, 1719–1728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Rocher, E.J.; Harkins, K.R.; Galbraith, D.W.; Bohnert, H.J. Developmentally Regulated Systemic Endopolyploidy in Suc-culents with Small Genomes. Science 1990, 250, 99–101. [Google Scholar] [CrossRef]
- Ahrens, C.W.; James, E.A.; Miller, A.D.; Scott, F.; Aitken, N.C.; Jones, A.W.; Lu-Irving, P.; Borevitz, J.O.; Cantrill, D.J.; Rymer, P.D. Spatial, climate and ploidy factors drive genomic diversity and resilience in the widespread grass Themeda triandra. Mol. Ecol. 2020, 29, 3872–3888. [Google Scholar] [CrossRef]
- Kobayashi, H. Variations of endoreduplication and its potential contribution to endosperm development in rice (Oryza sativa L.). Plant Prod. Sci. 2019, 22, 227–241. [Google Scholar] [CrossRef] [Green Version]
- Piet, Q.; Droc, G.; Marande, W.; Sarah, G.; Bocs, S.; Klopp, C.; Bourge, M.; Siljak-Yakovlev, S.; Bouchez, O.; Lopez-Roques, C.; et al. A chromosome-level, haplotype-phased Vanilla planifolia genome highlights the challenge of partial endoreplication for accurate whole-genome assembly. Plant Commun. 2022, 3, 100330. [Google Scholar] [CrossRef]
- Meng, X.; Dang, H.Q.; Kapler, G.M. Developmentally Programmed Switches in DNA Replication: Gene Amplification and Genome-Wide Endoreplication in Tetrahymena. Microorganisms 2023, 11, 491. [Google Scholar] [CrossRef]
- Brasil, J.N.; Costa, C.N.M.; Cabral, L.M.; Ferreira, P.C.G.; Hemerly, A.S. The plant cell cycle: Pre-Replication complex formation and controls. Genet. Mol. Biol. 2017, 40, 276–291. [Google Scholar] [CrossRef] [Green Version]
- Takatsuka, H.; Umeda, M. Hormonal control of cell division and elongation along differentiation trajectories in roots. J. Exp. Bot. 2014, 65, 2633–2643. [Google Scholar] [CrossRef] [Green Version]
- Bhosale, R.; Maere, S.; De Veylder, L. Endoreplication as a potential driver of cell wall modifications. Curr. Opin. Plant Biol. 2019, 51, 58–65. [Google Scholar] [CrossRef] [PubMed]


| Primary Inhibitors of CDK/Cyc | Secondary | |
|---|---|---|
| Inhibitors | Activators | |
| TOP6B | ||
| APC/CCS52 | MED16 | |
| WEE1 | LIM1 | |
| SMRs | SOG1 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kołodziejczyk, I.; Tomczyk, P.; Kaźmierczak, A. Endoreplication—Why Are We Not Using Its Full Application Potential? Int. J. Mol. Sci. 2023, 24, 11859. https://doi.org/10.3390/ijms241411859
Kołodziejczyk I, Tomczyk P, Kaźmierczak A. Endoreplication—Why Are We Not Using Its Full Application Potential? International Journal of Molecular Sciences. 2023; 24(14):11859. https://doi.org/10.3390/ijms241411859
Chicago/Turabian StyleKołodziejczyk, Izabela, Przemysław Tomczyk, and Andrzej Kaźmierczak. 2023. "Endoreplication—Why Are We Not Using Its Full Application Potential?" International Journal of Molecular Sciences 24, no. 14: 11859. https://doi.org/10.3390/ijms241411859





