Mechanisms of NURR1 Regulation: Consequences for Its Biological Activity and Involvement in Pathology
Abstract
:1. Introduction
2. NURR1 in Pathology
3. Regulation of NURR1 Activity by Protein-Protein Interactions
4. Regulation of NURR1 by Phosphorylation
5. Regulation of NURR1 by SUMOylation
6. Regulation of the Nuclear-Cytoplasmic Distribution of NURR1
7. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| α-SYN | α-Synuclein |
| AKT | Protein kinase B (PKB) |
| BAX | Bcl-2-associated X protein |
| CoREST | REST corepressor 1 |
| CRM1 | Chromosomal region maintenance 1 |
| DBD | DNA-binding domain |
| DJ-1 | Protein deglycase |
| DR5 | Retinoic Acid Receptor Response Element |
| ERK | Extracellular signal-regulated kinases |
| FOXP3 | Forkhead box P3 |
| GR | Glucocorticoid receptor |
| GSK3 | Glycogen synthase kinase-3 |
| HDAC | Histone deacetylase |
| MAPK | Mitogen-activated protein kinase |
| MSK | Mitogen and stress-activated protein kinase |
| NBRE | Nerve growth factor-induced clone B (NGFIB) response element |
| NES | Nuclear export signal |
| NLS | Nuclear localization signal |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NOR-1 | Neuron-derived orphan receptor 1 |
| NPC | Nuclear pore complex |
| NUR77 | Nuclear receptor subfamily 4 group A member 1 |
| NURR1 | Nuclear receptor-related 1 protein |
| LBD | Ligand-binding domain |
| p57Kip2 | Cyclin-dependent kinase inhibitor 1C (CDKN1C) |
| PARK7 | Parkinson’s disease protein 7 |
| PD | Parkison’s disease |
| PFF | Pre-formed fibrils |
| PIASγ | Protein inhibitor of activated STAT isoform γ |
| PITX3 | Paired Like Homeodomain 3 |
| PTMs | Post-translational modifications |
| RUNX1 | Runt-related transcription factor 1 |
| RSK | Ribosomal s6 kinase |
| RXR | Retinoid X receptor |
| SMRT | Silencing mediator of retinoic acid and thyroid hormone receptor |
| SUMO | Small Ubiquitin-like Modifier |
| UPS | Ubiquitin-Proteasome System |
References
- Jankovic, J.; Chen, S.; Le, W.D. The role of Nurr1 in the development of dopaminergic neurons and Parkinson’s disease. Prog. Neurobiol. 2005, 77, 128–138. [Google Scholar] [CrossRef]
- Liu, N.; Baker, H. Activity-dependent Nurr1 and NGFI-B gene expression in adult mouse olfactory bulb. Neuroreport 1999, 10, 747–751. [Google Scholar] [CrossRef]
- Moon, M.; Jeong, I.; Kim, C.H.; Kim, J.; Lee, P.K.; Mook-Jung, I.; Leblanc, P.; Kim, K.S. Correlation between orphan nuclear receptor Nurr1 expression and amyloid deposition in 5XFAD mice, an animal model of Alzheimer’s disease. J. Neurochem. 2015, 132, 254–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Bruemmer, D. NR4A Orphan Nuclear Receptors in Cardiovascular Biology. Drug Discov. Today Dis. Mech. 2009, 6, e43–e48. [Google Scholar] [CrossRef] [Green Version]
- Saucedo-Cardenas, O.; Kardon, R.; Ediger, T.R.; Lydon, J.P.; Conneely, O.M. Cloning and structural organization of the gene encoding the murine nuclear receptor transcription factor, NURR1. Gene 1997, 187, 135–139. [Google Scholar] [CrossRef]
- Wang, Z.; Benoit, G.; Liu, J.; Prasad, S.; Aarnisalo, P.; Liu, X.; Xu, H.; Walker, N.P.; Perlmann, T. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 2003, 423, 555–560. [Google Scholar] [CrossRef]
- Baker, K.D.; Shewchuk, L.M.; Kozlova, T.; Makishima, M.; Hassell, A.; Wisely, B.; Caravella, J.A.; Lambert, M.H.; Reinking, J.L.; Krause, H.; et al. The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell 2003, 113, 731–742. [Google Scholar] [CrossRef] [Green Version]
- Maxwell, M.A.; Muscat, G.E. The NR4A subgroup: Immediate early response genes with pleiotropic physiological roles. Nucl. Recept. Signal 2006, 4, e002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakaria, M.; Haque, M.E.; Cho, D.Y.; Azam, S.; Kim, I.S.; Choi, D.K. Molecular Insights into NR4A2(Nurr1): An Emerging Target for Neuroprotective Therapy Against Neuroinflammation and Neuronal Cell Death. Mol. Neurobiol. 2019, 56, 5799–5814. [Google Scholar] [CrossRef]
- Bushue, N.; Wan, Y.J. Retinoid pathway and cancer therapeutics. Adv. Drug Deliv. Rev. 2010, 62, 1285–1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacchetti, P.; Mitchell, T.R.; Granneman, J.G.; Bannon, M.J. Nurr1 enhances transcription of the human dopamine transporter gene through a novel mechanism. J. Neurochem. 2001, 76, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Hermanson, E.; Joseph, B.; Castro, D.; Lindqvist, E.; Aarnisalo, P.; Wallen, A.; Benoit, G.; Hengerer, B.; Olson, L.; Perlmann, T. Nurr1 regulates dopamine synthesis and storage in MN9D dopamine cells. Exp. Cell Res. 2003, 288, 324–334. [Google Scholar] [CrossRef]
- Hermanson, E.; Borgius, L.; Bergsland, M.; Joodmardi, E.; Perlmann, T. Neuropilin1 is a direct downstream target of Nurr1 in the developing brain stem. J. Neurochem. 2006, 97, 1403–1411. [Google Scholar] [CrossRef]
- Wallen, A.A.; Castro, D.S.; Zetterstrom, R.H.; Karlen, M.; Olson, L.; Ericson, J.; Perlmann, T. Orphan nuclear receptor Nurr1 is essential for Ret expression in midbrain dopamine neurons and in the brain stem. Mol. Cell Neurosci. 2001, 18, 649–663. [Google Scholar] [CrossRef]
- Jo, A.Y.; Kim, M.Y.; Lee, H.S.; Rhee, Y.H.; Lee, J.E.; Baek, K.H.; Park, C.H.; Koh, H.C.; Shin, I.; Lee, Y.S.; et al. Generation of dopamine neurons with improved cell survival and phenotype maintenance using a degradation-resistant nurr1 mutant. Stem Cells 2009, 27, 2238–2246. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Yague, A.J.; Lastres-Becker, I.; Stefanis, L.; Vassilatis, D.K.; Cuadrado, A. alpha-Synuclein Induces the GSK-3-Mediated Phosphorylation and Degradation of NURR1 and Loss of Dopaminergic Hallmarks. Mol. Neurobiol. 2021, 58, 6697–6711. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Castelao, B.; Losada, F.; Ahicart, P.; Castano, J.G. The N-terminal region of Nurr1 (a.a 1-31) is essential for its efficient degradation by the ubiquitin proteasome pathway. PLoS ONE 2013, 8, e55999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Yague, A.J.; Rada, P.; Rojo, A.I.; Lastres-Becker, I.; Cuadrado, A. Nuclear import and export signals control the subcellular localization of Nurr1 protein in response to oxidative stress. J. Biol. Chem. 2013, 288, 5506–5517. [Google Scholar] [CrossRef] [Green Version]
- Carpentier, R.; Sacchetti, P.; Segard, P.; Staels, B.; Lefebvre, P. The glucocorticoid receptor is a co-regulator of the orphan nuclear receptor Nurr1. J. Neurochem. 2008, 104, 777–789. [Google Scholar] [CrossRef]
- Wallen-Mackenzie, A.; Mata de Urquiza, A.; Petersson, S.; Rodriguez, F.J.; Friling, S.; Wagner, J.; Ordentlich, P.; Lengqvist, J.; Heyman, R.A.; Arenas, E.; et al. Nurr1-RXR heterodimers mediate RXR ligand-induced signaling in neuronal cells. Genes Dev. 2003, 17, 3036–3047. [Google Scholar] [CrossRef] [Green Version]
- Sacchetti, P.; Dwornik, H.; Formstecher, P.; Rachez, C.; Lefebvre, P. Requirements for heterodimerization between the orphan nuclear receptor Nurr1 and retinoid X receptors. J. Biol. Chem. 2002, 277, 35088–35096. [Google Scholar] [CrossRef] [Green Version]
- Arredondo, C.; Orellana, M.; Vecchiola, A.; Pereira, L.A.; Galdames, L.; Andres, M.E. PIASgamma enhanced SUMO-2 modification of Nurr1 activation-function-1 domain limits Nurr1 transcriptional synergy. PLoS ONE 2013, 8, e55035. [Google Scholar] [CrossRef] [Green Version]
- Galleguillos, D.; Vecchiola, A.; Fuentealba, J.A.; Ojeda, V.; Alvarez, K.; Gomez, A.; Andres, M.E. PIASgamma represses the transcriptional activation induced by the nuclear receptor Nurr1. J. Biol. Chem. 2004, 279, 2005–2011. [Google Scholar] [CrossRef] [Green Version]
- Amm, I.; Sommer, T.; Wolf, D.H. Protein quality control and elimination of protein waste: The role of the ubiquitin-proteasome system. Biochim. Biophys. Acta 2014, 1843, 182–196. [Google Scholar] [CrossRef] [Green Version]
- Chu, Y.; Le, W.; Kompoliti, K.; Jankovic, J.; Mufson, E.J.; Kordower, J.H. Nurr1 in Parkinson’s disease and related disorders. J. Comp. Neurol. 2006, 494, 495–514. [Google Scholar] [CrossRef] [Green Version]
- Le, W.D.; Xu, P.; Jankovic, J.; Jiang, H.; Appel, S.H.; Smith, R.G.; Vassilatis, D.K. Mutations in NR4A2 associated with familial Parkinson disease. Nat. Genet. 2003, 33, 85–89. [Google Scholar] [CrossRef]
- Kambey, P.A.; Liu, W.Y.; Wu, J.; Bosco, B.; Nadeem, I.; Kanwore, K.; Gao, D.S. Single-nuclei RNA sequencing uncovers heterogenous transcriptional signatures in Parkinson’s disease associated with nuclear receptor-related factor 1 defect. Neural Regen. Res. 2023, 18, 2037–2046. [Google Scholar]
- Zetterstrom, R.H.; Solomin, L.; Jansson, L.; Hoffer, B.J.; Olson, L.; Perlmann, T. Dopamine neuron agenesis in Nurr1-deficient mice. Science 1997, 276, 248–250. [Google Scholar] [CrossRef]
- Jeon, S.G.; Yoo, A.; Chun, D.W.; Hong, S.B.; Chung, H.; Kim, J.I.; Moon, M. The Critical Role of Nurr1 as a Mediator and Therapeutic Target in Alzheimer’s Disease-related Pathogenesis. Aging Dis. 2020, 11, 705–724. [Google Scholar] [CrossRef]
- Moon, M.; Jung, E.S.; Jeon, S.G.; Cha, M.Y.; Jang, Y.; Kim, W.; Lopes, C.; Mook-Jung, I.; Kim, K.S. Nurr1 (NR4A2) regulates Alzheimer’s disease-related pathogenesis and cognitive function in the 5XFAD mouse model. Aging Cell 2019, 18, e12866. [Google Scholar] [CrossRef] [Green Version]
- Montarolo, F.; Perga, S.; Martire, S.; Navone, D.N.; Marchet, A.; Leotta, D.; Bertolotto, A. Altered NR4A Subfamily Gene Expression Level in Peripheral Blood of Parkinson’s and Alzheimer’s Disease Patients. Neurotox. Res. 2016, 30, 338–344. [Google Scholar] [CrossRef]
- Rojas, P.; Joodmardi, E.; Hong, Y.; Perlmann, T.; Ogren, S.O. Adult mice with reduced Nurr1 expression: An animal model for schizophrenia. Mol. Psychiatry 2007, 12, 756–766. [Google Scholar] [CrossRef] [Green Version]
- Hawk, J.D.; Abel, T. The role of NR4A transcription factors in memory formation. Brain Res. Bull. 2011, 85, 21–29. [Google Scholar] [CrossRef] [Green Version]
- Bridi, M.S.; Abel, T. The NR4A orphan nuclear receptors mediate transcription-dependent hippocampal synaptic plasticity. Neurobiol. Learn. Mem. 2013, 105, 151–158. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.H.; Han, B.S.; Moon, J.; Kim, D.J.; Shin, J.; Rajan, S.; Nguyen, Q.T.; Sohn, M.; Kim, W.G.; Han, M.; et al. Nuclear receptor Nurr1 agonists enhance its dual functions and improve behavioral deficits in an animal model of Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2015, 112, 8756–8761. [Google Scholar] [CrossRef]
- Barneda-Zahonero, B.; Servitja, J.M.; Badiola, N.; Minano-Molina, A.J.; Fado, R.; Saura, C.A.; Rodriguez-Alvarez, J. Nurr1 protein is required for N-methyl-D-aspartic acid (NMDA) receptor-mediated neuronal survival. J. Biol. Chem. 2012, 287, 11351–11362. [Google Scholar] [CrossRef] [Green Version]
- Espana, J.; Valero, J.; Minano-Molina, A.J.; Masgrau, R.; Martin, E.; Guardia-Laguarta, C.; Lleo, A.; Gimenez-Llort, L.; Rodriguez-Alvarez, J.; Saura, C.A. beta-Amyloid disrupts activity-dependent gene transcription required for memory through the CREB coactivator CRTC1. J. Neurosci. 2010, 30, 9402–9410. [Google Scholar] [CrossRef] [Green Version]
- Terzioglu-Usak, S.; Negis, Y.; Karabulut, D.S.; Zaim, M.; Isik, S. Cellular Model of Alzheimer’s Disease: Abeta1-42 Peptide Induces Amyloid Deposition and a Decrease in Topo Isomerase IIbeta and Nurr1 Expression. Curr. Alzheimer Res. 2017, 14, 636–644. [Google Scholar] [CrossRef]
- Hara, K.; Matsukawa, N.; Yasuhara, T.; Xu, L.; Yu, G.; Maki, M.; Kawase, T.; Hess, D.C.; Kim, S.U.; Borlongan, C.V. Transplantation of post-mitotic human neuroteratocarcinoma-overexpressing Nurr1 cells provides therapeutic benefits in experimental stroke: In vitro evidence of expedited neuronal differentiation and GDNF secretion. J. Neurosci. Res. 2007, 85, 1240–1251. [Google Scholar] [CrossRef]
- Xie, X.; Peng, L.; Zhu, J.; Zhou, Y.; Li, L.; Chen, Y.; Yu, S.; Zhao, Y. miR-145-5p/Nurr1/TNF-alpha Signaling-Induced Microglia Activation Regulates Neuron Injury of Acute Cerebral Ischemic/Reperfusion in Rats. Front. Mol. Neurosci. 2017, 10, 383. [Google Scholar] [CrossRef] [Green Version]
- Vuillermot, S.; Joodmardi, E.; Perlmann, T.; Ove Ogren, S.; Feldon, J.; Meyer, U. Schizophrenia-relevant behaviors in a genetic mouse model of constitutive Nurr1 deficiency. Genes Brain Behav. 2011, 10, 589–603. [Google Scholar] [CrossRef]
- Weinstein, J.J.; Chohan, M.O.; Slifstein, M.; Kegeles, L.S.; Moore, H.; Abi-Dargham, A. Pathway-Specific Dopamine Abnormalities in Schizophrenia. Biol. Psychiatry 2017, 81, 31–42. [Google Scholar] [CrossRef] [Green Version]
- Xing, G.; Zhang, L.; Russell, S.; Post, R. Reduction of dopamine-related transcription factors Nurr1 and NGFI-B in the prefrontal cortex in schizophrenia and bipolar disorders. Schizophr. Res. 2006, 84, 36–56. [Google Scholar] [CrossRef]
- Arredondo, C.; Gonzalez, M.; Andres, M.E.; Gysling, K. Opposite effects of acute and chronic amphetamine on Nurr1 and NF-kappaB p65 in the rat ventral tegmental area. Brain Res. 2016, 1652, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Luo, C.; Tu, G.; Li, C.; Liu, Y.; Liu, W.; Lam Yung, K.K.; Mo, Z. Rhynchophylline Downregulates Phosphorylated cAMP Response Element Binding Protein, Nuclear Receptor-related-1, and Brain-derived Neurotrophic Factor Expression in the Hippocampus of Ketamine-induced Conditioned Place Preference Rats. Pharmacogn. Mag. 2018, 14, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Volkow, N.D.; Wang, G.J.; Kollins, S.H.; Wigal, T.L.; Newcorn, J.H.; Telang, F.; Fowler, J.S.; Zhu, W.; Logan, J.; Ma, Y.; et al. Evaluating dopamine reward pathway in ADHD: Clinical implications. JAMA 2009, 302, 1084–1091. [Google Scholar] [CrossRef]
- Vuillermot, S.; Joodmardi, E.; Perlmann, T.; Ogren, S.O.; Feldon, J.; Meyer, U. Prenatal immune activation interacts with genetic Nurr1 deficiency in the development of attentional impairments. J. Neurosci. 2012, 32, 436–451. [Google Scholar] [CrossRef] [Green Version]
- Wan, P.K.; Siu, M.K.; Leung, T.H.; Mo, X.T.; Chan, K.K.; Ngan, H.Y. Role of Nurr1 in Carcinogenesis and Tumor Immunology: A State of the Art Review. Cancers 2020, 12, 3044. [Google Scholar] [CrossRef] [PubMed]
- Safe, S.; Karki, K. The Paradoxical Roles of Orphan Nuclear Receptor 4A (NR4A) in Cancer. Mol. Cancer Res. 2021, 19, 180–191. [Google Scholar] [CrossRef] [PubMed]
- Zarei, M.; Shrestha, R.; Johnson, S.; Yu, Z.; Karki, K.; Vaziri-Gohar, A.; Epps, J.; Du, H.; Suva, L.; Zarei, M.; et al. Nuclear Receptor 4A2 (NR4A2/NURR1) Regulates Autophagy and Chemoresistance in Pancreatic Ductal Adenocarcinoma. Cancer Res. Commun. 2021, 1, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Hamers, A.A.; Hanna, R.N.; Nowyhed, H.; Hedrick, C.C.; de Vries, C.J. NR4A nuclear receptors in immunity and atherosclerosis. Curr. Opin. Lipidol. 2013, 24, 381–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mix, K.S.; McMahon, K.; McMorrow, J.P.; Walkenhorst, D.E.; Smyth, A.M.; Petrella, B.L.; Gogarty, M.; Fearon, U.; Veale, D.; Attur, M.G.; et al. Orphan nuclear receptor NR4A2 induces synoviocyte proliferation, invasion, and matrix metalloproteinase 13 transcription. Arthritis Rheum. 2012, 64, 2126–2136. [Google Scholar] [CrossRef] [PubMed]
- Zagani, R.; Hamzaoui, N.; Cacheux, W.; de Reynies, A.; Terris, B.; Chaussade, S.; Romagnolo, B.; Perret, C.; Lamarque, D. Cyclooxygenase-2 inhibitors down-regulate osteopontin and Nr4A2-new therapeutic targets for colorectal cancers. Gastroenterology 2009, 137, 1358–1366.e3. [Google Scholar] [CrossRef] [PubMed]
- Ke, N.; Claassen, G.; Yu, D.H.; Albers, A.; Fan, W.; Tan, P.; Grifman, M.; Hu, X.; Defife, K.; Nguy, V.; et al. Nuclear hormone receptor NR4A2 is involved in cell transformation and apoptosis. Cancer Res. 2004, 64, 8208–8212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Yang, J.; Zou, Y.; Huang, G.L.; He, Z.W. Orphan nuclear receptor nurr1 as a potential novel marker for progression in human prostate cancer. Asian Pac. J. Cancer Prev. 2013, 14, 2023–2028. [Google Scholar] [CrossRef] [Green Version]
- Ji, L.; Gong, C.; Ge, L.; Song, L.; Chen, F.; Jin, C.; Zhu, H.; Zhou, G. Orphan nuclear receptor Nurr1 as a potential novel marker for progression in human pancreatic ductal adenocarcinoma. Exp. Ther. Med. 2017, 13, 551–559. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Cai, H.; Ma, L.; Ding, Y.; Tan, X.; Chang, W.; Guan, W.; Liu, Y.; Shen, Q.; Yu, Y.; et al. Expression of orphan nuclear receptor NR4A2 in gastric cancer cells confers chemoresistance and predicts an unfavorable postoperative survival of gastric cancer patients with chemotherapy. Cancer 2013, 119, 3436–3445. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Cai, H.; Ma, L.; Ding, Y.; Tan, X.; Liu, Y.; Su, T.; Yu, Y.; Chang, W.; Zhang, H.; et al. Nuclear orphan receptor NR4A2 confers chemoresistance and predicts unfavorable prognosis of colorectal carcinoma patients who received postoperative chemotherapy. Eur. J. Cancer 2013, 49, 3420–3430. [Google Scholar] [CrossRef]
- Perlmann, T.; Jansson, L. A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes. Dev. 1995, 9, 769–782. [Google Scholar] [CrossRef] [Green Version]
- Aarnisalo, P.; Kim, C.H.; Lee, J.W.; Perlmann, T. Defining requirements for heterodimerization between the retinoid X receptor and the orphan nuclear receptor Nurr1. J. Biol. Chem. 2002, 277, 35118–35123. [Google Scholar] [CrossRef] [Green Version]
- Nishihara, E.; O’Malley, B.W.; Xu, J. Nuclear receptor coregulators are new players in nervous system development and function. Mol. Neurobiol. 2004, 30, 307–325. [Google Scholar] [CrossRef]
- Yu, X.; Shang, J.; Kojetin, D.J. Molecular basis of ligand-dependent Nurr1-RXRalpha activation. Elife 2023, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Forman, B.M.; Umesono, K.; Chen, J.; Evans, R.M. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell 1995, 81, 541–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajan, S.; Jang, Y.; Kim, C.H.; Kim, W.; Toh, H.T.; Jeon, J.; Song, B.; Serra, A.; Lescar, J.; Yoo, J.Y.; et al. PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function. Nat. Chem. Biol. 2020, 16, 876–886. [Google Scholar] [CrossRef] [PubMed]
- Martens, C.; Bilodeau, S.; Maira, M.; Gauthier, Y.; Drouin, J. Protein-protein interactions and transcriptional antagonism between the subfamily of NGFI-B/Nur77 orphan nuclear receptors and glucocorticoid receptor. Mol. Endocrinol. 2005, 19, 885–897. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, N.; Guo, Y.; Li, J.; Yin, Y.; Dong, Y.; Zhu, J.; Peng, C.; Xu, T.; Liu, J. Integrative analysis reveals structural basis for transcription activation of Nurr1 and Nurr1-RXRalpha heterodimer. Proc. Natl. Acad. Sci. USA 2022, 119, e2206737119. [Google Scholar] [CrossRef]
- Kovalovsky, D.; Refojo, D.; Liberman, A.C.; Hochbaum, D.; Pereda, M.P.; Coso, O.A.; Stalla, G.K.; Holsboer, F.; Arzt, E. Activation and induction of NUR77/NURR1 in corticotrophs by CRH/cAMP: Involvement of calcium, protein kinase A, and MAPK pathways. Mol. Endocrinol. 2002, 16, 1638–1651. [Google Scholar] [CrossRef]
- Davis, I.J.; Lau, L.F. Endocrine and neurogenic regulation of the orphan nuclear receptors Nur77 and Nurr-1 in the adrenal glands. Mol. Cell Biol. 1994, 14, 3469–3483. [Google Scholar] [CrossRef]
- Volakakis, N.; Tiklova, K.; Decressac, M.; Papathanou, M.; Mattsson, B.; Gillberg, L.; Nobre, A.; Bjorklund, A.; Perlmann, T. Nurr1 and Retinoid X Receptor Ligands Stimulate Ret Signaling in Dopamine Neurons and Can Alleviate alpha-Synuclein Disrupted Gene Expression. J. Neurosci. 2015, 35, 14370–14385. [Google Scholar] [CrossRef] [Green Version]
- Loppi, S.; Kolosowska, N.; Karkkainen, O.; Korhonen, P.; Huuskonen, M.; Grubman, A.; Dhungana, H.; Wojciechowski, S.; Pomeshchik, Y.; Giordano, M.; et al. HX600, a synthetic agonist for RXR-Nurr1 heterodimer complex, prevents ischemia-induced neuronal damage. Brain Behav. Immun. 2018, 73, 670–681. [Google Scholar] [CrossRef]
- Wang, J.; Bi, W.; Zhao, W.; Varghese, M.; Koch, R.J.; Walker, R.H.; Chandraratna, R.A.; Sanders, M.E.; Janesick, A.; Blumberg, B.; et al. Selective brain penetrable Nurr1 transactivator for treating Parkinson’s disease. Oncotarget 2016, 7, 7469–7479. [Google Scholar] [CrossRef] [Green Version]
- Spathis, A.D.; Asvos, X.; Ziavra, D.; Karampelas, T.; Topouzis, S.; Cournia, Z.; Qing, X.; Alexakos, P.; Smits, L.M.; Dalla, C.; et al. Nurr1:RXRalpha heterodimer activation as monotherapy for Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2017, 114, 3999–4004. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, F.M.; van Erp, S.; van der Linden, A.J.; von Oerthel, L.; Burbach, J.P.; Smidt, M.P. Pitx3 potentiates Nurr1 in dopamine neuron terminal differentiation through release of SMRT-mediated repression. Development 2009, 136, 531–540. [Google Scholar] [CrossRef] [Green Version]
- Joseph, B.; Wallen-Mackenzie, A.; Benoit, G.; Murata, T.; Joodmardi, E.; Okret, S.; Perlmann, T. p57(Kip2) cooperates with Nurr1 in developing dopamine cells. Proc. Natl. Acad. Sci. USA 2003, 100, 15619–15624. [Google Scholar] [CrossRef]
- Daffern, N.; Radhakrishnan, I. A Novel Mechanism of Coactivator Recruitment by the Nurr1 Nuclear Receptor. J. Mol. Biol. 2022, 434, 167718. [Google Scholar] [CrossRef]
- van Tiel, C.M.; Kurakula, K.; Koenis, D.S.; van der Wal, E.; de Vries, C.J. Dual function of Pin1 in NR4A nuclear receptor activation: Enhanced activity of NR4As and increased Nur77 protein stability. Biochim. Biophys. Acta 2012, 1823, 1894–1904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurakula, K.; van der Wal, E.; Geerts, D.; van Tiel, C.M.; de Vries, C.J. FHL2 protein is a novel co-repressor of nuclear receptor Nur77. J. Biol. Chem. 2011, 286, 44336–44343. [Google Scholar] [CrossRef] [Green Version]
- Sekiya, T.; Kashiwagi, I.; Inoue, N.; Morita, R.; Hori, S.; Waldmann, H.; Rudensky, A.Y.; Ichinose, H.; Metzger, D.; Chambon, P.; et al. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat. Commun. 2011, 2, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saijo, K.; Winner, B.; Carson, C.T.; Collier, J.G.; Boyer, L.; Rosenfeld, M.G.; Gage, F.H.; Glass, C.K. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell 2009, 137, 47–59. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Wang, P.; Ren, H.; Fan, J.; Wang, G. NGFI-B nuclear orphan receptor Nurr1 interacts with p53 and suppresses its transcriptional activity. Mol. Cancer Res. 2009, 7, 1408–1415. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Sanchez, E.; Lopez-Ramirez, A.M.; Ruiz-Chow, A.; Calvillo, M.; Resendiz-Albor, A.A.; Anguiano, B.; Rojas, P. Variability in Behavioral Phenotypes after Forced Swimming-Induced Stress in Rats Is Associated with Expression of the Glucocorticoid Receptor, Nurr1, and IL-1beta in the Hippocampus. Int. J. Mol. Sci. 2021, 22, 12700. [Google Scholar] [CrossRef] [PubMed]
- Eskandarian Boroujeni, M.; Aliaghaei, A.; Maghsoudi, N.; Gardaneh, M. Complementation of dopaminergic signaling by Pitx3-GDNF synergy induces dopamine secretion by multipotent Ntera2 cells. J. Cell. Biochem. 2020, 121, 200–212. [Google Scholar] [CrossRef]
- Sacchetti, P.; Carpentier, R.; Segard, P.; Olive-Cren, C.; Lefebvre, P. Multiple signaling pathways regulate the transcriptional activity of the orphan nuclear receptor NURR1. Nucleic Acids Res. 2006, 34, 5515–5527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajan, S.; Toh, H.T.; Ye, H.; Wang, Z.; Basil, A.H.; Parnaik, T.; Yoo, J.Y.; Lim, K.L.; Yoon, H.S. Prostaglandin A2 Interacts with Nurr1 and Ameliorates Behavioral Deficits in Parkinson’s Disease Fly Model. NeuroMol. Med. 2022, 24, 469–478. [Google Scholar] [CrossRef]
- Chen, H.Z.; Li, L.; Wang, W.J.; Du, X.D.; Wen, Q.; He, J.P.; Zhao, B.X.; Li, G.D.; Zhou, W.; Xia, Y.; et al. Prolyl isomerase Pin1 stabilizes and activates orphan nuclear receptor TR3 to promote mitogenesis. Oncogene 2012, 31, 2876–2887. [Google Scholar] [CrossRef]
- Darragh, J.; Soloaga, A.; Beardmore, V.A.; Wingate, A.D.; Wiggin, G.R.; Peggie, M.; Arthur, J.S. MSKs are required for the transcription of the nuclear orphan receptors Nur77, Nurr1 and Nor1 downstream of MAPK signalling. Biochem. J. 2005, 390, 749–759. [Google Scholar] [CrossRef] [Green Version]
- Ye, F.; Alvarez-Carbonell, D.; Nguyen, K.; Leskov, K.; Garcia-Mesa, Y.; Sreeram, S.; Valadkhan, S.; Karn, J. Recruitment of the CoREST transcription repressor complexes by Nerve Growth factor IB-like receptor (Nurr1/NR4A2) mediates silencing of HIV in microglial cells. PLoS Pathog. 2022, 18, e1010110. [Google Scholar] [CrossRef]
- Liu, D.; Jia, H.; Holmes, D.I.; Stannard, A.; Zachary, I. Vascular endothelial growth factor-regulated gene expression in endothelial cells: KDR-mediated induction of Egr3 and the related nuclear receptors Nur77, Nurr1, and Nor1. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 2002–2007. [Google Scholar] [CrossRef] [Green Version]
- Nordzell, M.; Aarnisalo, P.; Benoit, G.; Castro, D.S.; Perlmann, T. Defining an N-terminal activation domain of the orphan nuclear receptor Nurr1. Biochem. Biophys. Res. Commun. 2004, 313, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Houslay, M.D.; Kolch, W. Cell-type specific integration of cross-talk between extracellular signal-regulated kinase and cAMP signaling. Mol. Pharmacol. 2000, 58, 659–668. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Jia, N.; Fei, E.; Wang, P.; Liao, Z.; Ding, L.; Yan, M.; Nukina, N.; Zhou, J.; Wang, G. Nurr1 is phosphorylated by ERK2 in vitro and its phosphorylation upregulates tyrosine hydroxylase expression in SH-SY5Y cells. Neurosci. Lett. 2007, 423, 118–122. [Google Scholar] [CrossRef]
- Malewicz, M.; Kadkhodaei, B.; Kee, N.; Volakakis, N.; Hellman, U.; Viktorsson, K.; Leung, C.Y.; Chen, B.; Lewensohn, R.; van Gent, D.C.; et al. Essential role for DNA-PK-mediated phosphorylation of NR4A nuclear orphan receptors in DNA double-strand break repair. Genes Dev. 2011, 25, 2031–2040. [Google Scholar] [CrossRef] [Green Version]
- Wingate, A.D.; Campbell, D.G.; Peggie, M.; Arthur, J.S. Nur77 is phosphorylated in cells by RSK in response to mitogenic stimulation. Biochem. J. 2006, 393, 715–724. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Zhao, S.; Gao, G.; Sun, X.; Zhao, H.; Yang, H. DJ-1/PARK7, But Not Its L166P Mutant Linked to Autosomal Recessive Parkinsonism, Modulates the Transcriptional Activity of the Orphan Nuclear Receptor Nurr1 In Vitro and In Vivo. Mol. Neurobiol. 2016, 53, 7363–7374. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Sun, X.; Liu, Y.; Zhao, H.; Zhao, S.; Yang, H. DJ-1 upregulates tyrosine hydroxylase gene expression by activating its transcriptional factor Nurr1 via the ERK1/2 pathway. Int. J. Biochem. Cell Biol. 2012, 44, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.K.; Nikodem, V.M. Differential role of ERK in cAMP-induced Nurr1 expression in N2A and C6 cells. Neuroreport 2004, 15, 99–102. [Google Scholar] [CrossRef]
- Wang, A.; Rud, J.; Olson, C.M., Jr.; Anguita, J.; Osborne, B.A. Phosphorylation of Nur77 by the MEK-ERK-RSK cascade induces mitochondrial translocation and apoptosis in T cells. J. Immunol. 2009, 183, 3268–3277. [Google Scholar] [CrossRef] [Green Version]
- Cavanaugh, J.E.; Jaumotte, J.D.; Lakoski, J.M.; Zigmond, M.J. Neuroprotective role of ERK1/2 and ERK5 in a dopaminergic cell line under basal conditions and in response to oxidative stress. J. Neurosci. Res. 2006, 84, 1367–1375. [Google Scholar] [CrossRef]
- Grassi, D.; Diaz-Perez, N.; Volpicelli-Daley, L.A.; Lasmezas, C.I. Palpha-syn* mitotoxicity is linked to MAPK activation and involves tau phosphorylation and aggregation at the mitochondria. Neurobiol. Dis. 2019, 124, 248–262. [Google Scholar] [CrossRef]
- Argyrofthalmidou, M.; Spathis, A.D.; Maniati, M.; Poula, A.; Katsianou, M.A.; Sotiriou, E.; Manousaki, M.; Perier, C.; Papapanagiotou, I.; Papadopoulou-Daifoti, Z.; et al. Nurr1 repression mediates cardinal features of Parkinson’s disease in alpha-synuclein transgenic mice. Hum. Mol. Genet. 2021, 30, 1469–1483. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Jin, J.; Yang, B.; Zhang, W.; Hu, J.; Zhang, Y.; Chen, N.H. Overexpressed alpha-synuclein regulated the nuclear factor-kappaB signal pathway. Cell Mol. Neurobiol. 2008, 28, 21–33. [Google Scholar] [CrossRef]
- Robertson, H.; Hayes, J.D.; Sutherland, C. A partnership with the proteasome; the destructive nature of GSK3. Biochem. Pharmacol. 2018, 147, 77–92. [Google Scholar] [CrossRef]
- Han, Q.W.; Shao, Q.H.; Wang, X.T.; Ma, K.L.; Chen, N.H.; Yuan, Y.H. CB2 receptor activation inhibits the phagocytic function of microglia through activating ERK/AKT-Nurr1 signal pathways. Acta Pharmacol. Sin. 2022, 43, 2253–2266. [Google Scholar] [CrossRef] [PubMed]
- Dohmen, R.J. SUMO protein modification. Biochim. Biophys. Acta 2004, 1695, 113–131. [Google Scholar] [CrossRef] [Green Version]
- Dodat, F.; Cotnoir-White, D.; Dianati, E.; Vallet, A.; Mader, S.; Levesque, D. Complex regulation of orphan nuclear receptor Nur77 (Nr4a1) transcriptional activity by SUMO2 and PIASgamma. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118908. [Google Scholar] [CrossRef] [PubMed]
- Dodat, F.; Mader, S.; Levesque, D. Minireview: What is Known about SUMOylation Among NR4A Family Members? J. Mol. Biol. 2021, 433, 167212. [Google Scholar] [CrossRef]
- Oh, M.; Kim, S.Y.; Gil, J.E.; Byun, J.S.; Cha, D.W.; Ku, B.; Lee, W.; Kim, W.K.; Oh, K.J.; Lee, E.W.; et al. Nurr1 performs its anti-inflammatory function by regulating RasGRP1 expression in neuro-inflammation. Sci. Rep. 2020, 10, 10755. [Google Scholar] [CrossRef] [PubMed]
- Oka, M.; Yoneda, Y. Importin alpha: Functions as a nuclear transport factor and beyond. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2018, 94, 259–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cautain, B.; Hill, R.; de Pedro, N.; Link, W. Components and regulation of nuclear transport processes. FEBS J. 2015, 282, 445–462. [Google Scholar] [CrossRef]
- Lowe, A.R.; Tang, J.H.; Yassif, J.; Graf, M.; Huang, W.Y.; Groves, J.T.; Weis, K.; Liphardt, J.T. Importin-beta modulates the permeability of the nuclear pore complex in a Ran-dependent manner. Elife 2015, 4. [Google Scholar] [CrossRef]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef] [Green Version]
- Dingwall, C.; Sharnick, S.V.; Laskey, R.A. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell 1982, 30, 449–458. [Google Scholar] [CrossRef]
- Fischer, U.; Huber, J.; Boelens, W.C.; Mattaj, I.W.; Luhrmann, R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 1995, 82, 475–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalderon, D.; Roberts, B.L.; Richardson, W.D.; Smith, A.E. A short amino acid sequence able to specify nuclear location. Cell 1984, 39, 499–509. [Google Scholar] [CrossRef]
- Lee, B.J.; Cansizoglu, A.E.; Suel, K.E.; Louis, T.H.; Zhang, Z.; Chook, Y.M. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 2006, 126, 543–558. [Google Scholar] [CrossRef] [Green Version]
- Wen, W.; Meinkoth, J.L.; Tsien, R.Y.; Taylor, S.S. Identification of a signal for rapid export of proteins from the nucleus. Cell 1995, 82, 463–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojeda, V.; Fuentealba, J.A.; Galleguillos, D.; Andres, M.E. Rapid increase of Nurr1 expression in the substantia nigra after 6-hydroxydopamine lesion in the striatum of the rat. J. Neurosci. Res. 2003, 73, 686–697. [Google Scholar] [CrossRef]
- Volakakis, N.; Kadkhodaei, B.; Joodmardi, E.; Wallis, K.; Panman, L.; Silvaggi, J.; Spiegelman, B.M.; Perlmann, T. NR4A orphan nuclear receptors as mediators of CREB-dependent neuroprotection. Proc. Natl. Acad. Sci. USA 2010, 107, 12317–12322. [Google Scholar] [CrossRef]
- Perlmann, T.; Wallen-Mackenzie, A. Nurr1, an orphan nuclear receptor with essential functions in developing dopamine cells. Cell Tissue Res. 2004, 318, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Erdo, F.; Trapp, T.; Mies, G.; Hossmann, K.A. Immunohistochemical analysis of protein expression after middle cerebral artery occlusion in mice. Acta Neuropathol. 2004, 107, 127–136. [Google Scholar] [CrossRef]
- Wei, X.; Gao, H.; Zou, J.; Liu, X.; Chen, D.; Liao, J.; Xu, Y.; Ma, L.; Tang, B.; Zhang, Z.; et al. Contra-directional Coupling of Nur77 and Nurr1 in Neurodegeneration: A Novel Mechanism for Memantine-Induced Anti-inflammation and Anti-mitochondrial Impairment. Mol. Neurobiol. 2016, 53, 5876–5892. [Google Scholar] [CrossRef] [PubMed]
- Boldingh Debernard, K.A.; Mathisen, G.H.; Paulsen, R.E. Differences in NGFI-B, Nurr1, and NOR-1 expression and nucleocytoplasmic translocation in glutamate-treated neurons. Neurochem. Int. 2012, 61, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Shen, T.; Chitranshi, N.; Gupta, V.; Basavarajappa, D.; Sarkar, S.; Mirzaei, M.; You, Y.; Krezel, W.; Graham, S.L.; et al. Retinoid X Receptor: Cellular and Biochemical Roles of Nuclear Receptor with a Focus on Neuropathological Involvement. Mol. Neurobiol. 2022, 59, 2027–2050. [Google Scholar] [CrossRef] [PubMed]



| Partners | Partner-Binding Domain | NURR1-Binding Motif | Effect | Reference |
|---|---|---|---|---|
| RXRα; RXRγ | I-box (LBD) Glu390, Glu394 | I-box (LBD) Lys554-Leu555-Leu556 | Transcription activity | [21,60] |
| GR | Full-length protein (A/B, DBD and LBD) | Transactivation domain A/B (first 58 aas) | Transcription activity | [19] |
| PITX3 | Unknown | Unknown | Transcription activity | [73] |
| p57Kip2 | Unknown | Transactivation domain A/B (Full-length) | Transcription activity | [74] |
| SRCs | PAS-B domain | AF1 domain (Transactivation A/B) | Transcription activity | [75] |
| PIN1 | WW domain | Transactivation domain A/B and DBD | Transcription activity | [76] |
| FHL2 | Full-length protein (LIMs domains) | Transactivation domain A/B and DBD | Transcription activity | [77] |
| RUNX1 | Unknown | Unknown | Differentiation CD4+ T cells | [78] |
| CoREST | Unknown | DBD | Transrepression | [79] |
| p53 | COOH-terminal | DBD | Anti-apoptotic | [80] |
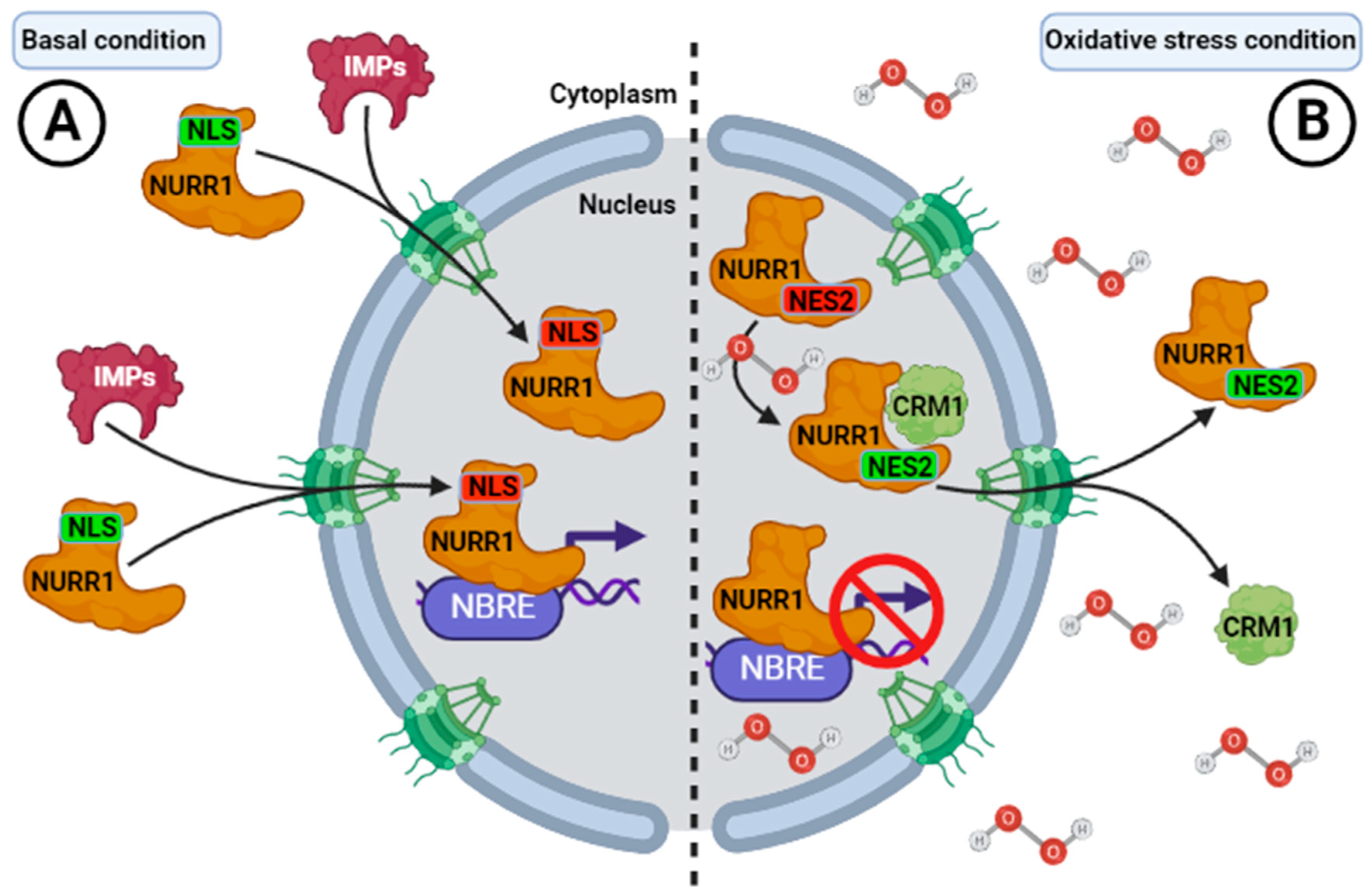
| Importins | Cargo-recognition motif | DBD (NLS) | Import to nucleus | [18] |
| Exportins (CRM1) | Cargo-recognition motif | LBD (NES) | Export to nucleus | [18] |
| Kinase | Phospho-Motifs | Effect | Reference |
| ERK1/2 | 124-PSpS(126)PPTPSpT(132)P-133 | Transcription activity | [91] |
| GSK-3β | 124-PS pS(126)PPpT(129)PSpT(132)P-133 | Degradation | [16] |
| ERK5 | 166-RKpT(168)PVSRLSLFpS(177)FK-179 | Transcription activity | [83] |
| DNA-PK | 335-TDpS(337)LKG-340 | Double-strand break repair | [92] |
| AKT | 344-LPpS(347)KP-349 | Degradation | [15] |
| RSK | 344-LPpS(347)KP-349 | Unknown | [93] |
| SUMO-E3 Ligases | SUMO-Motifs | Effect | Reference |
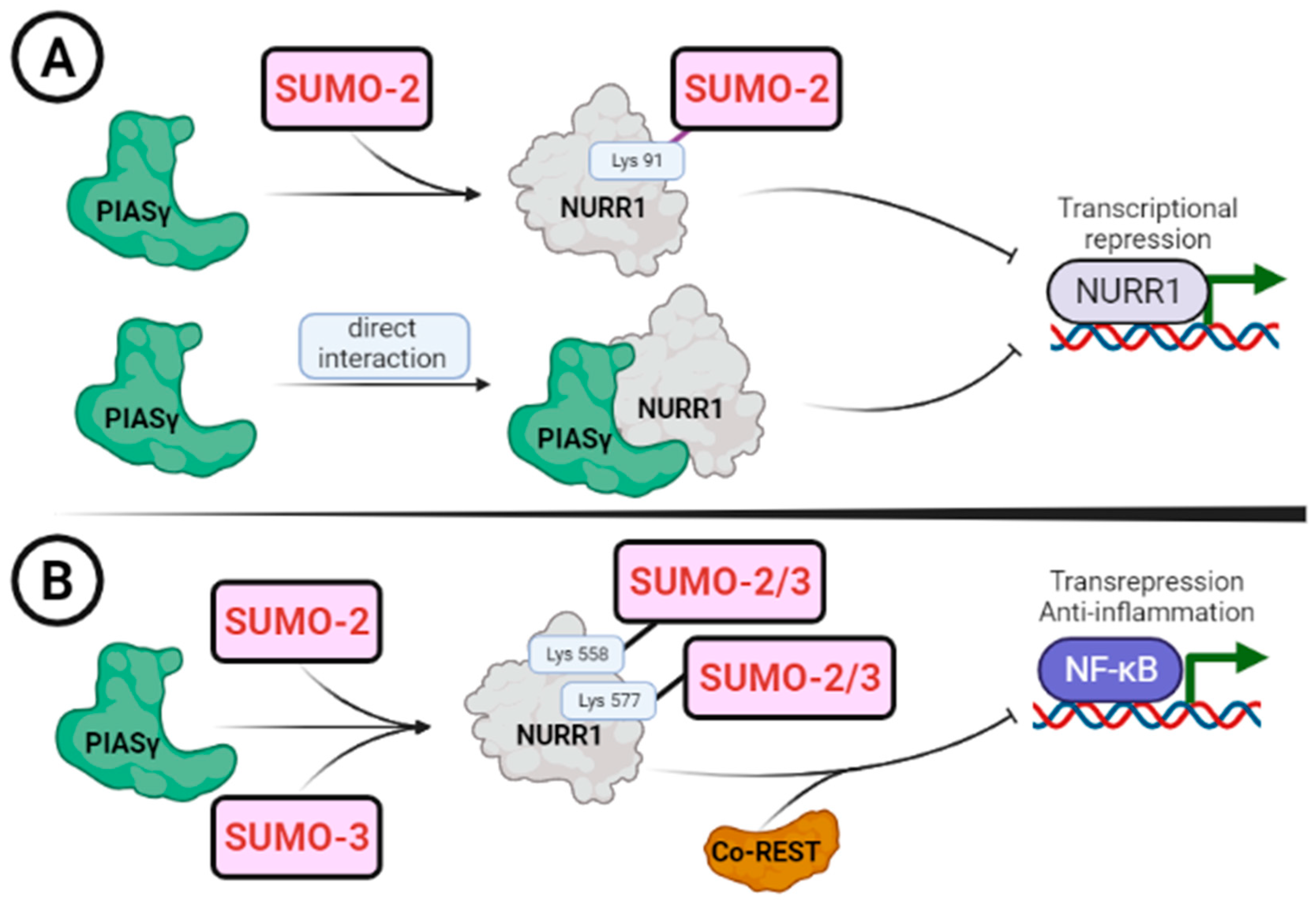
| PIASγ/PIAS4 | 85-GQQSSIKSUMO-2(91)VEDIQMH-98 | Transcription repression | [22,23] |
| PIASγ/PIAS4 | 571-QRIFYLKSUMO-2/3(577)LEDLVPP-584 | Transcription repression | [23] |
| PIASγ/PIAS4 | 552-LSKLLGKSUMO-2/3(558)LPELRTL-565 | Transrepression | [23,79] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Yagüe, Á.J.; Cuadrado, A. Mechanisms of NURR1 Regulation: Consequences for Its Biological Activity and Involvement in Pathology. Int. J. Mol. Sci. 2023, 24, 12280. https://doi.org/10.3390/ijms241512280
García-Yagüe ÁJ, Cuadrado A. Mechanisms of NURR1 Regulation: Consequences for Its Biological Activity and Involvement in Pathology. International Journal of Molecular Sciences. 2023; 24(15):12280. https://doi.org/10.3390/ijms241512280
Chicago/Turabian StyleGarcía-Yagüe, Ángel Juan, and Antonio Cuadrado. 2023. "Mechanisms of NURR1 Regulation: Consequences for Its Biological Activity and Involvement in Pathology" International Journal of Molecular Sciences 24, no. 15: 12280. https://doi.org/10.3390/ijms241512280





