Sub-Cellular Dynamic Analysis of BGC823 Cells after Treatment with the Multi-Component Drug CKI Using Raman Spectroscopy
Abstract
:1. Introduction
2. Results and Discussions
2.1. Inhibition of Cellular Proliferation and Viability
2.2. The Nucleic Acid Decrease in the Cell Nucleus
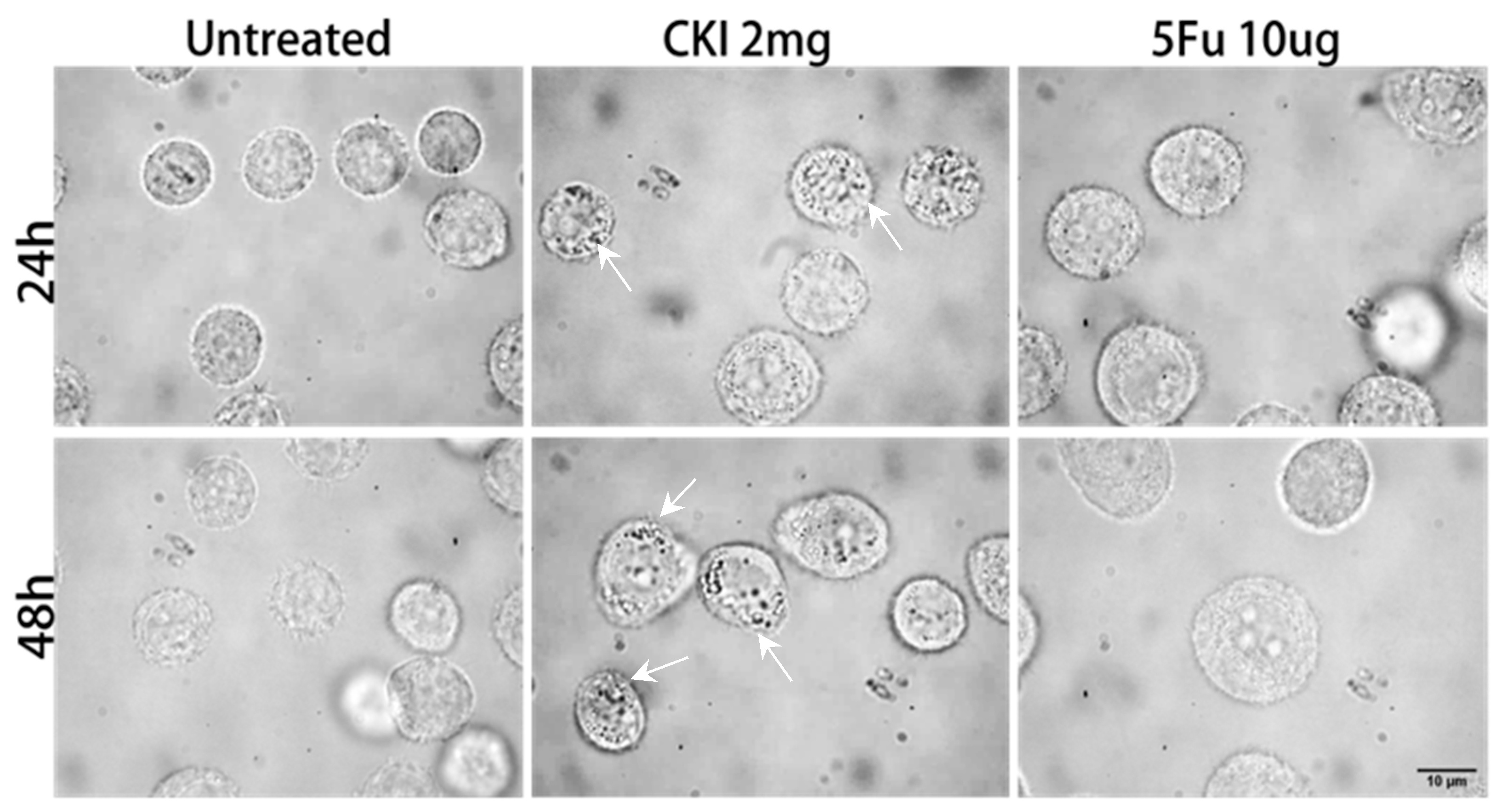
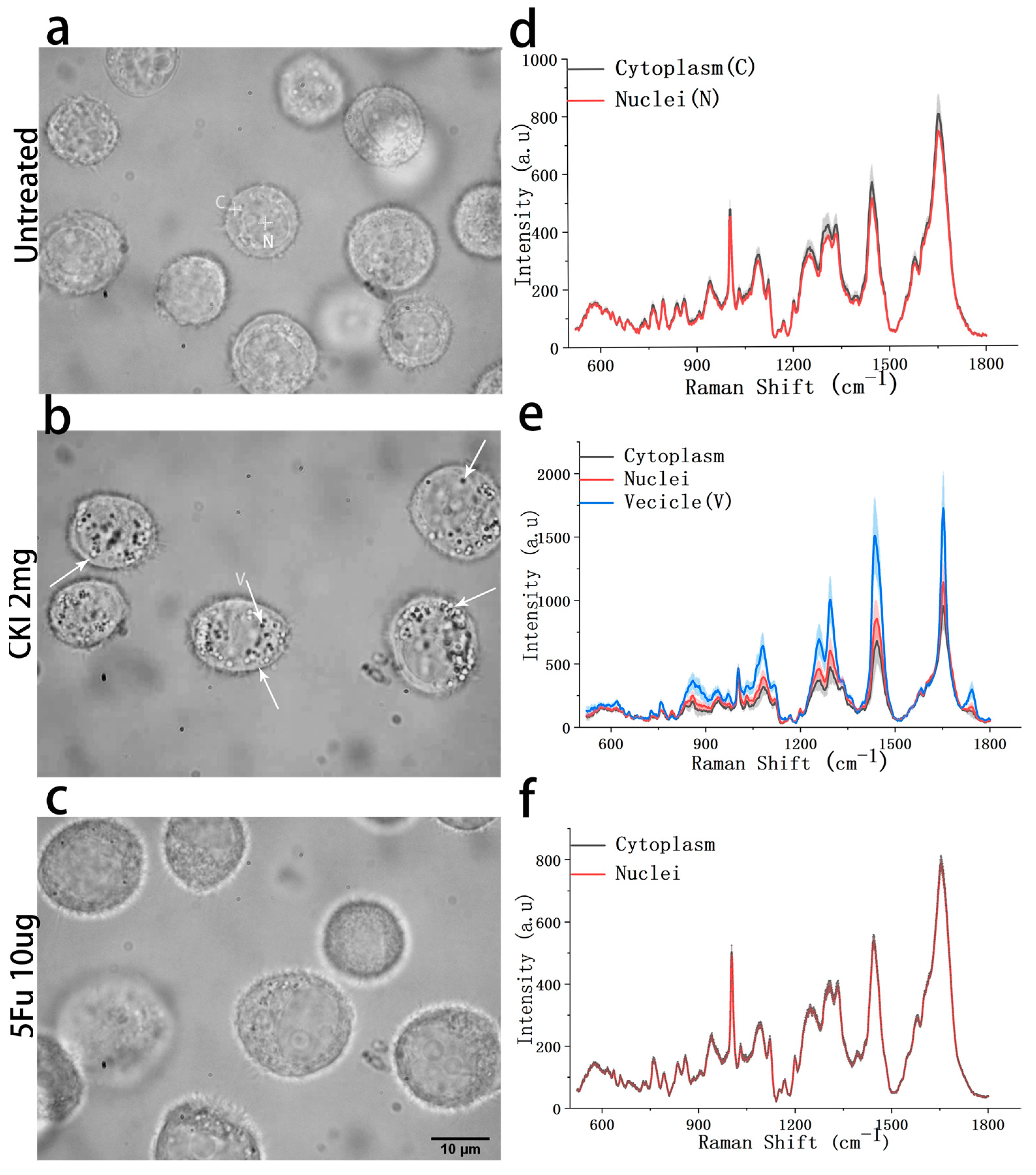
2.3. Intracellular Vesicle Accumulation
2.4. RS of Intracellular Vesicles and Cytoplasm
3. Materials and Methods
3.1. Cell Culture and Drugs
3.2. Cell Viability and Live Cell Counting
3.3. Sub-Cellular Raman Spectroscopy & Cell Bright Field Imaging
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, R.; Zhu, X.; Bai, H.; Ning, K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front. Pharmacol. 2019, 10, 123. [Google Scholar] [CrossRef] [Green Version]
- Bai, M.S.; Wu, Z.P.; Wang, X.C. Advances in research on adjuvant effects applying Traditional Chinese Medicine in cancer chemotherapy. J. Mod. Oncol. 2010, 18, 597–601. [Google Scholar]
- Zhao, Z.; Fan, H.; Higgins, T.; Qi, J.; Haines, D.; Trivett, A.; Oppenheim, J.J.; Wei, H.; Li, J.; Lin, H.; et al. Fufang Kushen injection inhibits sarcoma growth and tumor-induced hyperalgesia via TRPV1 signaling pathways. Cancer Lett. 2014, 355, 232–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Lin, H.S.; Hou, W.; Hua, B.J. Idea and Strategy of Traditional Chinese Medicine Treatment for Cancer. China Cancer 2010, 19, 735–738. [Google Scholar]
- Sun, M.; Cao, H.; Sun, L.; Dong, S.; Bian, Y.; Han, J.; Zhang, L.; Ren, S.; Hu, Y.; Liu, C.; et al. Antitumor activities of kushen: Literature review. Evid.-Based Complement. Altern. Med. 2012, 2012, 373219. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Gao, H.; Liu, J.; Chen, L.; Zhang, Q.; Wang, Z. Identification and Determination of the Chemical Constituents in a Herbal Preparation, Compound Kushen Injection, by Hplc and Lc-Dad-Ms/Ms. J. Liq. Chromatogr. Relat. Technol. 2013, 37, 207–220. [Google Scholar] [CrossRef]
- Aung, T.N.; Nourmohammadi, S.; Qu, Z.; Harata-Lee, Y.; Cui, J.; Shen, H.Y.; Yool, A.J.; Pukala, T.; Du, H.; Kortschak, R.D.; et al. Fractional Deletion of Compound Kushen Injection Indicates Cytokine Signaling Pathways are Critical for its Perturbation of the Cell Cycle. Sci. Rep. 2019, 9, 14200. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Qu, Z.; Harata-Lee, Y.; Nwe Aung, T.; Shen, H.; Wang, W.; Adelson, D.L. Cell cycle, energy metabolism and DNA repair pathways in cancer cells are suppressed by Compound Kushen Injection. BMC Cancer 2019, 19, 103. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; You, R.L.; Qin, W.J.; Hai, L.N.; Fang, M.J.; Huang, G.H.; Kang, R.X.; Li, M.H.; Qiao, Y.F.; Li, J.W.; et al. Anti-tumor activities of active ingredients in Compound Kushen Injection. Acta Pharmacol. Sin. 2015, 36, 676–679. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.Q.; Li, Y.M.; Liu, T.; He, W.T.; Chen, Y.T.; Chen, X.H.; Li, X.; Zhou, W.C.; Yi, J.F.; Ren, Z.J. Antitumor effect of matrine in human hepatoma G2 cells by inducing apoptosis and autophagy. World J. Gastroenterol. 2010, 16, 4281–4290. [Google Scholar] [CrossRef]
- Shi, X.J.; Zhu, D.D.; Li, L.; Wang, Z.; Yuan, G.; Yuan, D.G.; Zhu, B. Evaluation of Compound Kushen Injection on the Five CYP Isozymes in Rats using a Cocktail Method. Lat. Am. J. Pharm. 2019, 38, 924–930. [Google Scholar]
- Zhang, D.; Wu, J.; Wang, K.; Duan, X.; Liu, S.; Zhang, B. Which are the best Chinese herbal injections combined with XELOX regimen for gastric cancer?: A PRISMA-compliant network meta-analysis. Medicine 2018, 97, e0127. [Google Scholar] [CrossRef]
- Fang, T.; Shang, W.; Liu, C.; Liu, Y.; Ye, A. Single-Cell Multimodal Analytical Approach by Integrating Raman Optical Tweezers and RNA Sequencing. Anal. Chem. 2020, 92, 10433–10441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, J.; Yu, Y.; Shang, W.; Ye, A. Anti-Cancer Drug Sensitivity Assay with Quantitative Heterogeneity Testing Using Single-Cell Raman Spectroscopy. Molecules 2018, 23, 2903. [Google Scholar] [CrossRef] [Green Version]
- Farhane, Z.; Bonnier, F.; Casey, A.; Byrne, H.J. Raman micro spectroscopy for in vitro drug screening: Subcellular localisation and interactions of doxorubicin. Analyst 2015, 140, 4212–4223. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Jin, L.; Xu, J.; Yu, Y.; Shen, L.; Gao, J.; Ye, A. Dynamic characterization of drug resistance and heterogeneity of the gastric cancer cell BGC823 using single-cell Raman spectroscopy. Analyst 2017, 143, 164–174. [Google Scholar] [CrossRef]
- Nawaz, H.; Bonnier, F.; Meade, A.D.; Lyng, F.M.; Byrne, H.J. Comparison of subcellular responses for the evaluation and prediction of the chemotherapeutic response to cisplatin in lung adenocarcinoma using Raman spectroscopy. Analyst 2011, 136, 2450–2463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawaz, H.; Garcia, A.; Meade, A.D.; Lyng, F.M.; Byrne, H.J. Raman micro spectroscopy study of the interaction of vincristine with A549 cells supported by expression analysis of bcl-2 protein. Analyst 2013, 138, 6177–6184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farhane, Z.; Bonnier, F.; Howe, O.; Casey, A.; Byrne, H.J. Doxorubicin kinetics and effects on lung cancer cell lines using in vitro Raman micro-spectroscopy: Binding signatures, drug resistance and DNA repair. J. Biophotonics 2018, 11, e201700060. [Google Scholar] [CrossRef] [Green Version]
- El-Mashtoly, S.F.; Yosef, H.K.; Petersen, D.; Mavarani, L.; Maghnouj, A.; Hahn, S.A.; Kötting, C.; Gerwert, K. Label-Free Raman Spectroscopic Imaging Monitors the Integral Physiologically Relevant Drug Responses in Cancer Cells. Anal. Chem. 2015, 87, 7297–7304. [Google Scholar]
- Yosef, H.K.; Mavarani, L.; Maghnouj, A.; Hahn, S.A.; El-Mashtoly, S.F.; Gerwert, K. In vitro prediction of the efficacy of molecularly targeted cancer therapy by Raman spectral imaging. Anal. Bioanal. Chem. 2015, 407, 8321–8331. [Google Scholar] [PubMed]
- Qu, Z.; Cui, J.; Harata-Lee, Y.; Aung, T.N.; Feng, Q.; Raison, J.M.; Kortschak, R.D.; Adelson, D.L. Identification of candidate anti-cancer molecular mechanisms of Compound Kushen Injection using functional genomics. Oncotarget 2016, 7, 66003–66019. [Google Scholar] [CrossRef] [Green Version]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Qu, Z.; Harata-Lee, Y.; Aung, T.N.; Cui, J.; Wang, W.; Kortschak, R.D.; Adelson, D.L. Understanding the Mechanistic Contribution of Herbal Extracts in Compound Kushen Injection With Transcriptome Analysis. Front. Oncol. 2019, 9, 632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, J.; Qu, Z.; Harata-Lee, Y.; Shen, H.; Aung, T.N.; Wang, W.; Kortschak, R.D.; Adelson, D.L. The effect of compound kushen injection on cancer cells: Integrated identification of candidate molecular mechanisms. PLoS ONE 2020, 15, e0236395. [Google Scholar] [CrossRef]
- Zhou, W.; Wu, J.; Zhu, Y.; Meng, Z.; Liu, X.; Liu, S.; Ni, M.; Jia, S.; Zhang, J.; Guo, S. Study on the mechanisms of compound Kushen injection for the treatment of gastric cancer based on network pharmacology. BMC Complement. Med. Ther. 2020, 20, 6. [Google Scholar] [CrossRef] [Green Version]
- Batista de Carvalho, A.L.; Pilling, M.; Gardner, P.; Doherty, J.; Cinque, G.; Wehbe, K.; Kelley, C.; Batista de Carvalho, L.A.; Marques, M.P. Chemotherapeutic response to cisplatin-like drugs in human breast cancer cells probed by vibrational microspectroscopy. Faraday Discuss. 2016, 187, 273–298. [Google Scholar] [CrossRef] [Green Version]
- Notingher, I.; Hench, L.L. Raman microspectroscopy: A noninvasive tool for studies of individual living cells in vitro. Expert Rev. Med. Devices 2006, 3, 215–234. [Google Scholar] [CrossRef]
- Notingher, I. Raman Spectroscopy cell-based Biosensors. Sensors 2007, 7, 1343–1358. [Google Scholar] [CrossRef] [Green Version]
- Kruglik, S.G.; Royo, F.; Guigner, J.M.; Palomo, L.; Seksek, O.; Turpin, P.Y.; Tatischeff, I.; Falcon-Perez, J.M. Raman tweezers microspectroscopy of circa 100 nm extracellular vesicles. Nanoscale 2019, 11, 1661–1679. [Google Scholar] [CrossRef] [Green Version]
- De Gelder, J.; De Gussem, K.; Vandenabeele, P.; Moens, L. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 2007, 38, 1133–1147. [Google Scholar] [CrossRef]
- Verrier, S.; Zoladek, A.; Notingher, I. Raman micro-spectroscopy as a non-invasive cell viability test. Methods Mol. Biol. 2011, 740, 179–189. [Google Scholar] [PubMed]
- Henry, C.M.; Hollville, E.; Martin, S.J. Measuring apoptosis by microscopy and flow cytometry. Methods 2013, 61, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Rangan, S.; Kamal, S.; Konorov, S.O.; Schulze, H.G.; Blades, M.W.; Turner, R.F.B.; Piret, J.M. Types of cell death and apoptotic stages in Chinese Hamster Ovary cells distinguished by Raman spectroscopy. Biotechnol. Bioeng. 2018, 115, 401–412. [Google Scholar] [CrossRef]
- Pattingre, S.; Tassa, A.; Qu, X.; Garuti, R.; Liang, X.H.; Mizushima, N.; Packer, M.; Schneider, M.D.; Levine, B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005, 122, 927–939. [Google Scholar] [CrossRef] [Green Version]
- Krampe, B.; Al-Rubeai, M. Cell death in mammalian cell culture: Molecular mechanisms and cell line engineering strategies. Cytotechnology 2010, 62, 175–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, T.; Shang, W.; Liu, C.; Xu, J.; Zhao, D.; Liu, Y.; Ye, A. Nondestructive Identification and Accurate Isolation of Single Cells through a Chip with Raman Optical Tweezers. Anal. Chem. 2019, 91, 9932–9939. [Google Scholar] [CrossRef]
- Jaishy, B.; Abel, E.D. Lipids, lysosomes, and autophagy. J. Lipid Res. 2016, 57, 1619–1635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dall’Armi, C.; Devereaux, K.A.; Di Paolo, G. The role of lipids in the control of autophagy. Curr. Biol. 2013, 23, R33–R45. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Navarro, J.A.; Cuervo, A.M. Autophagy and lipids: Tightening the knot. Semin. Immunopathol. 2010, 32, 343–353. [Google Scholar] [CrossRef]
- Christian, P.; Sacco, J.; Adeli, K. Autophagy: Emerging roles in lipid homeostasis and metabolic control. Biochim. Biophys. Acta 2013, 1831, 819–824. [Google Scholar] [CrossRef]
- Beloribi-Djefaflia, S.; Vasseur, S.; Guillaumond, F. Lipid metabolic reprogramming in cancer cells. Oncogenesis 2016, 5, e189. [Google Scholar] [CrossRef] [Green Version]
- Strober, W. Trypan Blue Exclusion Test of Cell Viability. Curr. Protoc. Immunol. 2015, 111, A3.B.1–A3.B.3. [Google Scholar] [CrossRef]
- Hongfei, M.A.; Zhang, Y.A. Single-cell discrimination based on optical tweezers Raman spectroscopy. Chin. Sci. Bull. 2013, 58, 2594–2600. [Google Scholar]
- Yong, Z.; Anpei, Y.; Cheng, W. A Micro-Raman Spectroscopy Combing with Optical Tweezers. Acta Opt. Sin. 2010, 30, 491–497. [Google Scholar] [CrossRef]
- Austin, L.A.; Osseiran, S.; Evans, C.L. Raman technologies in cancer diagnostics. Analyst 2016, 141, 476–503. [Google Scholar] [CrossRef]
- Konorov, S.; Jardon, M.; Piret, M.; Blades, M.W.; Turner, R.F.B. Raman microspectroscopy of live cells under autophagy-inducing conditions. Analyst 2012, 137, 4662–4668. [Google Scholar] [CrossRef]
- Ho, C.S.; Jean, N.; Hogan, C.A.; Blackmon, L.; Jeffrey, S.S.; Holodniy, M.; Banaei, N.; Saleh, A.A.E.; Ermon, S.; Dionne, J. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 2019, 10, 4927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, L.F.; Bonnier, F.; O’Callaghan, K.; O’Sullivan, J.; Flint, S.; Byrne, H.J.; Lyng, F.M. Raman micro-spectroscopy for rapid screening of oral squamous cell carcinoma. Exp. Mol. Pathol. 2015, 98, 502–509. [Google Scholar] [CrossRef]
- Brauchle, E.; Thude, S.; Brucker, S.Y.; Schenke-Layland, K. Cell death stages in single apoptotic and necrotic cells monitored by Raman microspectroscopy. Sci. Rep. 2014, 4, 4698. [Google Scholar]
- Eberhardt, K.; Beleites, C.; Marthandan, S.; Matthäus, C.; Diekmann, S.; Popp, J. Raman and Infrared Spectroscopy Distinguishing Replicative Senescent from Proliferating Primary Human Fibroblast Cells by Detecting Spectral Differences Mainly Due to Biomolecular Alterations. Anal. Chem. 2017, 89, 2937–2947. [Google Scholar] [CrossRef] [PubMed]
- Liendl, L.; Grillari, J.; Schosserer, M. Raman fingerprints as promising markers of cellular senescence and aging. Geroscience 2019, 42, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; Ilin, Y.; Kraft, M.L.; Harley, B.A.C. Tracing Hematopoietic Progenitor Cell Neutrophilic Differentiation via Raman Spectroscopy. Bioconjugate Chem. 2018, 29, 3121–3128. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, W.; Ye, A.; Tong, Y.-K. Sub-Cellular Dynamic Analysis of BGC823 Cells after Treatment with the Multi-Component Drug CKI Using Raman Spectroscopy. Int. J. Mol. Sci. 2023, 24, 12750. https://doi.org/10.3390/ijms241612750
Shang W, Ye A, Tong Y-K. Sub-Cellular Dynamic Analysis of BGC823 Cells after Treatment with the Multi-Component Drug CKI Using Raman Spectroscopy. International Journal of Molecular Sciences. 2023; 24(16):12750. https://doi.org/10.3390/ijms241612750
Chicago/Turabian StyleShang, Wenhao, Anpei Ye, and Yu-Kai Tong. 2023. "Sub-Cellular Dynamic Analysis of BGC823 Cells after Treatment with the Multi-Component Drug CKI Using Raman Spectroscopy" International Journal of Molecular Sciences 24, no. 16: 12750. https://doi.org/10.3390/ijms241612750




