The Transcription Factor CsgD Contributes to Engineered Escherichia coli Resistance by Regulating Biofilm Formation and Stress Responses
Abstract
:1. Introduction
2. Results and Discussion
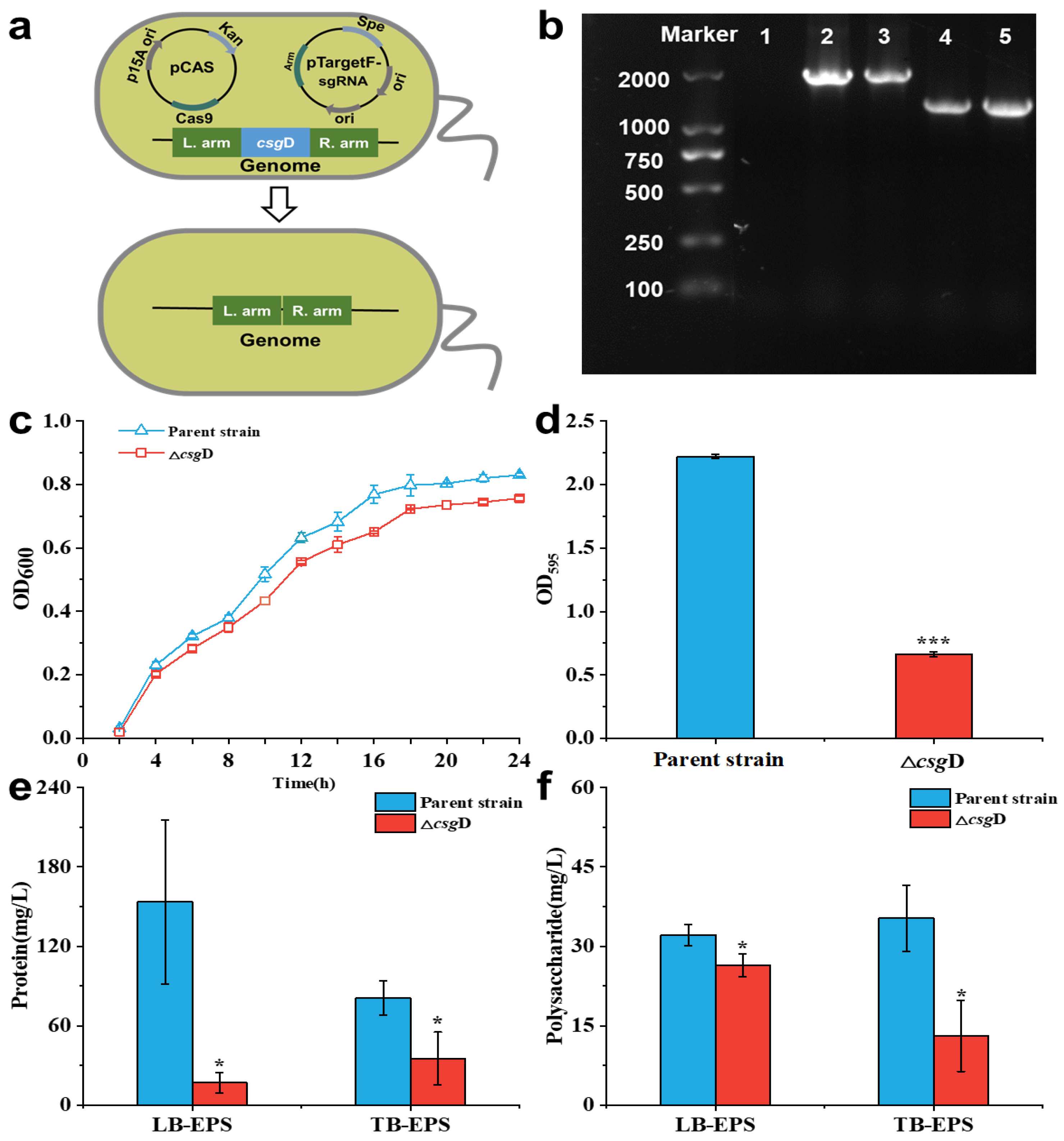
2.1. Deletion of the csgD Gene Downregulated E. coli Biofilm Formation
2.2. Deletion of the csgD Gene Influences the E. coli Morphotype
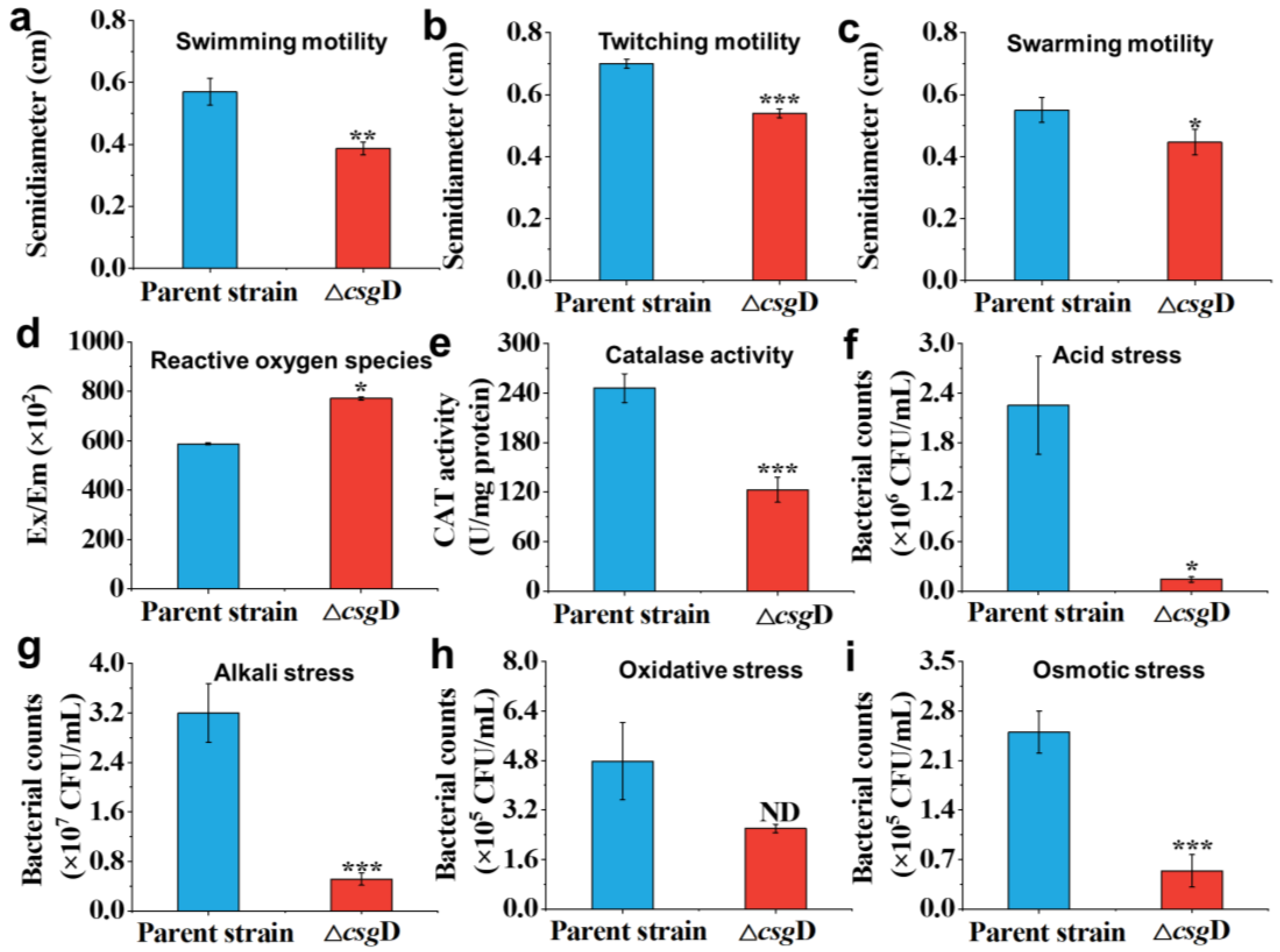
2.3. Motility and Environmental Stress Resistance of the csgD Gene Deletion Strain
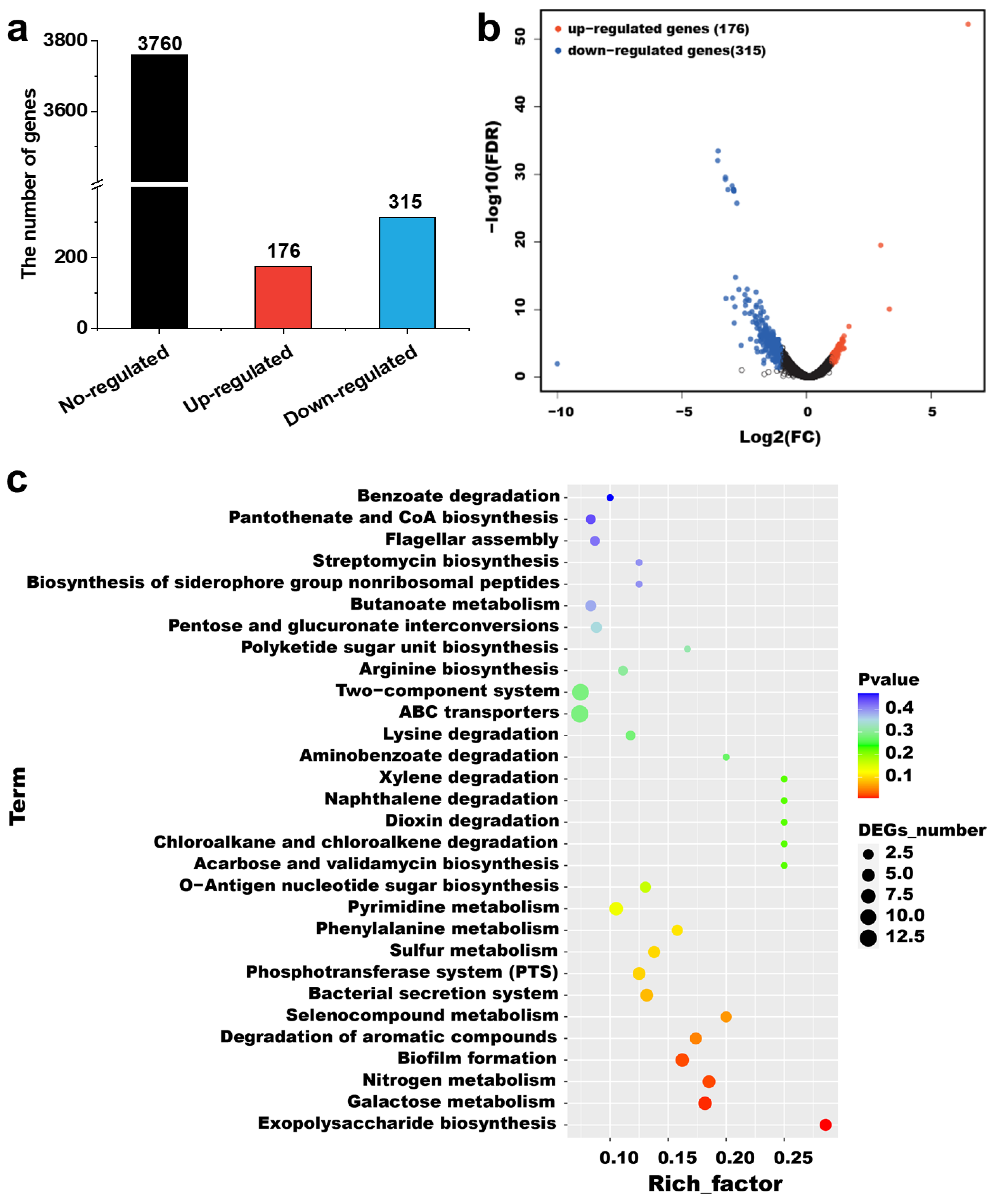
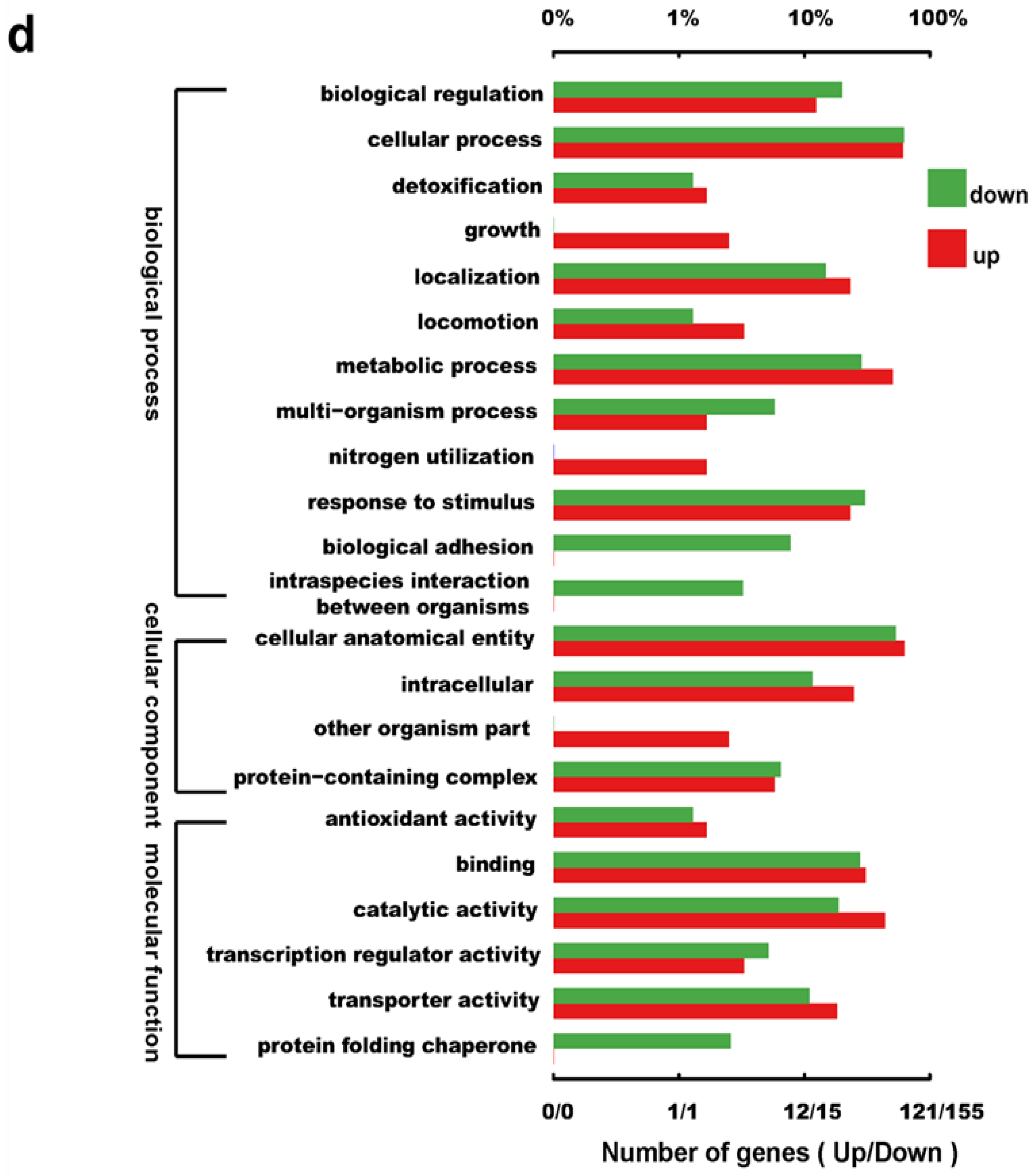
2.4. Transcriptome Analysis of the csgD-Deficient Mutant Strain
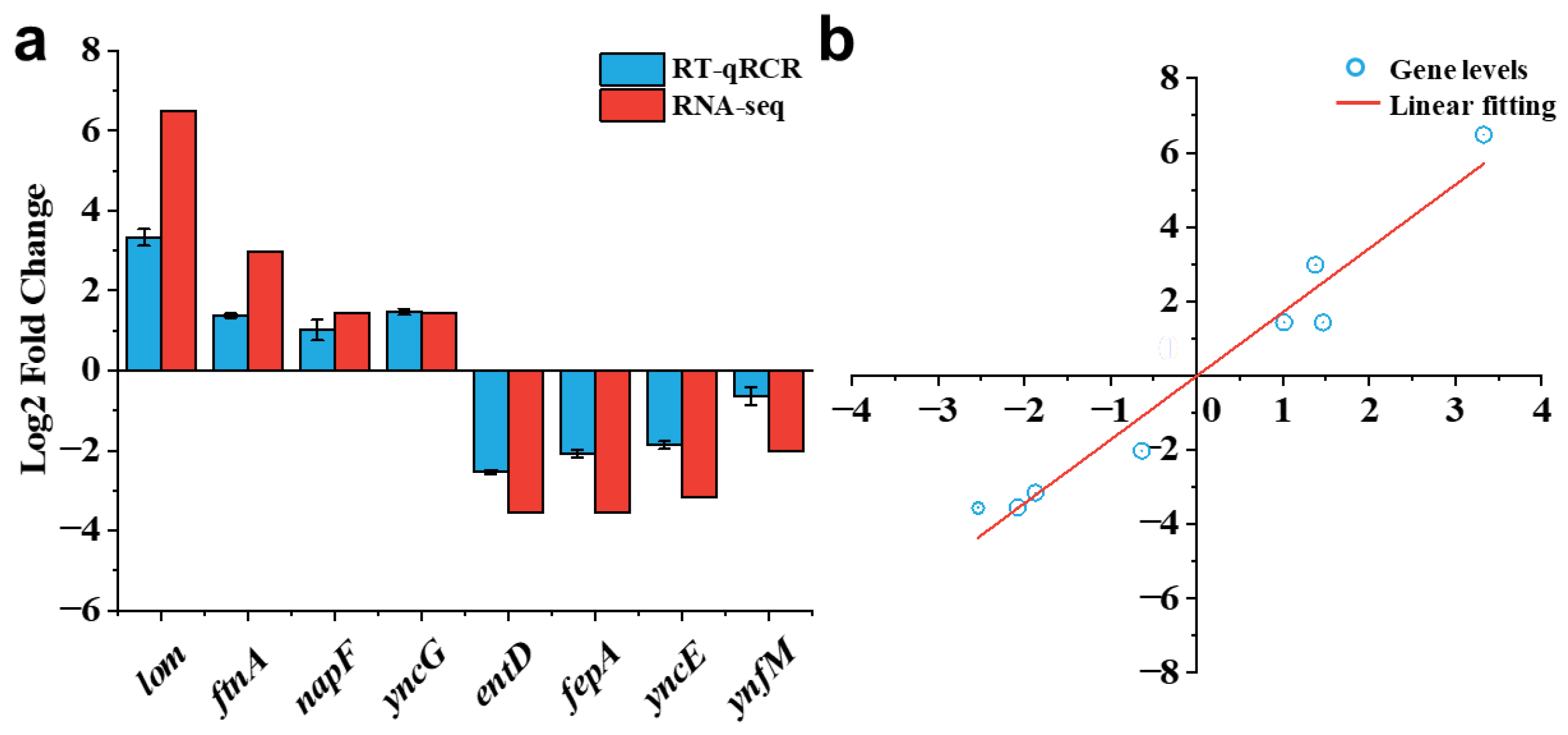
2.5. Validation of Transcriptome Analysis
2.6. Deletion of the csgD Gene Downregulated the Expression of Biofilms and Stress Response Genes
3. Materials and Methods
3.1. Bacterial Strains, Plasmids and Growth Conditions
3.2. Construction of the csgD Deletion Strain
3.3. Growth Curves and Biofilm Formation Assay
3.4. Extracellular Polymeric Substance Analysis
3.5. Motility Analysis of Mutant Strain
3.6. Environmental Tolerance Analysis
3.7. Colony Morphology and Biofilm Characterization
3.8. RNA-Sequencing Analysis
3.9. RNA Isolation and RT-qPCR
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Del Pozo, J.L. Biofilm-related disease. Expert Rev. Anti-Infect. Ther. 2018, 16, 51–65. [Google Scholar] [CrossRef]
- Mohsin, M.Z.; Omer, R.; Huang, J.F.; Mohsin, A.; Guo, M.J.; Qian, J.C.; Zhuang, Y.O. Advances in engineered Bacillus subtilis biofilms and spores, and their applications in bioremediation, biocatalysis, and biomaterials. Synth. Syst. Biotechnol. 2021, 6, 180–191. [Google Scholar] [CrossRef] [PubMed]
- Gunes, B. A critical review on biofilm-based reactor systems for enhanced syngas fermentation processes. Renew. Sustain. Energy Rev. 2021, 143, 110950. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yan, C.H.; Zhan, Y.F.; Geng, L.T.; Zhu, L.L.; Gong, L.C.; Wang, J. Boron derivatives accelerate biofilm formation of recombinant Escherichia coli via increasing quorum sensing system autoinducer-2 activity. Int. J. Mol. Sci. 2022, 23, 8059. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Nag, M.; Lahiri, D.; Sarkar, T.; Pati, S.; Kari, Z.A.; Nirmal, N.P.; Edinur, H.A.; Ray, R.R. Engineered biofilm: Innovative nextgen strategy for quality enhancement of fermented foods. Front. Nutr. 2022, 9, 808630. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, H.R.; Hong, J.; Malroy, J.; Zhu, X.J. An E. coli-based biosynthetic platform expands the structural diversity of natural benzoxazoles. ACS Synth. Biol. 2021, 10, 2151–2158. [Google Scholar] [CrossRef]
- Krol, J.E.; Hall, D.C.; Balashov, S.; Pastor, S.; Sibert, J.; McCaffrey, J.; Lang, S.; Ehrlich, R.L.; Earl, J.; Mell, J.C.; et al. Genome rearrangements induce biofilm formation in Escherichia coli C—An old model organism with a new application in biofilm research. BMC Genom. 2019, 20, 767. [Google Scholar] [CrossRef]
- Liu, L.N.; Ma, X.L.; Bilal, M.; Wei, L.L.; Tang, S.J.; Luo, H.Z.; Zhao, Y.P.; Duan, X.G. Mechanistic insight into phenolic compounds toxicity and state-of-the-art strategies for enhancing the tolerance of Escherichia coli to phenolic compounds. Biotechnol. Bioprocess Eng. 2022, 27, 533–542. [Google Scholar] [CrossRef]
- Ahan, R.E.; Saltepe, B.; Apaydin, O.; Seker, U.O.S. Cellular biocatalysts using synthetic genetic circuits for prolonged and durable enzymatic activity. ChemBioChem 2019, 20, 1799–1809. [Google Scholar] [CrossRef]
- Tong, X.; Barberi, T.T.; Botting, C.H.; Sharma, S.V.; Simmons, M.J.H.; Overton, T.W.; Goss, R.J.M. Rapid enzyme regeneration results in the striking catalytic longevity of an engineered, single species, biocatalytic biofilm. Microb. Cell Factories 2016, 15, 180. [Google Scholar] [CrossRef] [PubMed]
- Brombacher, E.; Baratto, A.; Dorel, C.; Landini, P. Gene expression regulation by the curli activator CsgD protein: Modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J. Bacteriol. 2006, 188, 2027–2037. [Google Scholar] [CrossRef] [PubMed]
- Uhlich, G.A.; Chen, C.; Cottrell, B.J.; Andreozzi, E.; Irwin, P.L.; Ly-Huong, N. Genome amplification and promoter mutation expand the range of csgD-dependent biofilm responses in an STEC population. Microbiology 2017, 163, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Azam, M.W.W.; Khan, A.U.U. CRISPRi-mediated suppression of E. coli Nissle 1917 virulence factors: A strategy for creating an engineered probiotic using csgD gene suppression. Front. Nutr. 2022, 9, 938989. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.K.N.; Minh-Duy, P.; Peters, K.M.; Lo, A.W.; Forde, B.M.; Chong, T.M.; Yin, W.; Chan, K.; Chromek, M.; Brauner, A.; et al. Discovery of new genes involved in curli production by a uropathogenic Escherichia coli strain from the highly virulent O45:K1:H7 lineage. mBio 2018, 9, e01462-18. [Google Scholar] [CrossRef]
- Uhlich, G.A.; Chen, C.; Cottrell, B.J.; Irwin, P.L.; Phillips, J.G. Peroxide resistance in Escherichia coli serotype O157:H7 biofilms is regulated by both RpoS-dependent and -independent mechanisms. Microbiology 2012, 158, 2225–2234. [Google Scholar] [CrossRef]
- Gualdi, L.; Tagliabue, L.; Landini, P. Biofilm formation-gene expression relay system in Escherichia coli: Modulation of sigma(s)-dependent gene expression by the CsgD regulatory protein via sigma(s) protein stabilization. J. Bacteriol. 2007, 189, 8034–8043. [Google Scholar] [CrossRef]
- Gualdi, L.; Tagliabue, L.; Bertagnoli, S.; Ierano, T.; De Castro, C.; Landini, P. Cellulose modulates biofilm formation by counteracting curil-mediated colonization of solid surfaces in Escherichia coli. Microbiology 2008, 154, 2017–2024. [Google Scholar] [CrossRef]
- Simoes, L.C.; Simoes, M. Contribution to understanding the mechanisms involved in biofilm formation, tolerance and control. Int. J. Mol. Sci. 2023, 24, 9475. [Google Scholar] [CrossRef]
- Romling, U.; Galperin, M.Y. Bacterial cellulose biosynthesis: Diversity of operons, subunits, products, and functions. Trends Microbiol. 2015, 23, 545–557. [Google Scholar] [CrossRef]
- Harimawan, A.; Ting, Y.P. Investigation of extracellular polymeric substances (EPS) properties of P. aeruginosa and B. subtillis and their role in bacterial adhesion. Colloids Surf. B Biointerfaces 2016, 146, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gu, Y.; Zheng, Y.X.; Wang, Y.; Nie, L.L.; Qiao, R.F.; He, Y.F. Deletion of luxS gene mediated by λ-Red gene recombination technology reduces biofilm formation and stress resistance of Lactobacillus fermentum. Food Biosci. 2022, 49, 101892. [Google Scholar] [CrossRef]
- Gu, L.H.; Chen, Q.; Guo, A.L.; Liu, W.K.; Ruan, Y.; Zhang, X.S.; Nou, X.W. Differential effects of growth medium salinity on biofilm formation of two Salmonella enterica strains. J. Food Prot. 2020, 83, 196–203. [Google Scholar] [CrossRef]
- Simm, R.; Ahmad, I.; Rhen, M.; Le Guyon, S.; Romling, U. Regulation of biofilm formation in Salmonella enterica serovar Typhimurium. Future Microbiol. 2014, 9, 1261–1282. [Google Scholar] [CrossRef]
- Sharma, V.K.; Bearson, B.L. Hha controls Escherichia coli O157:H7 biofilm formation by differential regulation of global transcriptional regulators FlhDC and CsgD. Appl. Environ. Microbiol. 2013, 79, 2384–2396. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, H.; Yamamoto, K.; Ishihama, A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J. Bacteriol. 2011, 193, 2587–2597. [Google Scholar] [CrossRef]
- White, A.P.; Weljie, A.M.; Apel, D.; Zhang, P.; Shaykhutdinov, R.; Vogel, H.J.; Surette, M.G. A global metabolic shift is linked to salmonella multicellular development. PLoS ONE 2010, 5, e11814. [Google Scholar] [CrossRef]
- Varghese, A.; Ray, S.; Verma, T.; Nandi, D. Multicellular string-like structure formation by Salmonella typhimurium depends on cellulose production: Roles of diguanylate cyclases, YedQ and YfiN. Front. Microbiol. 2020, 11, 613704. [Google Scholar] [CrossRef]
- Middlemiss, A.D.; Haycocks, J.R.J.; Stringer, A.M.; Piddock, L.J.V.; Wade, J.T.; Grainger, D.C. Mapping direct and indirect MarA/SoxS/Rob/RamA regulons in Salmonella Typhimurium reveals repression of csgD and biofilm formation. Microbiology 2023, 169, 001330. [Google Scholar] [CrossRef]
- Fu, D.D.; Wu, J.M.; Wu, X.Y.; Shao, Y.; Song, X.J.; Tu, J.; Qi, K.Z. The two-component system histidine kinase EnvZ contributes to Avian pathogenic Escherichia coli pathogenicity by regulating biofilm formation and stress responses. Poult. Sci. 2023, 102, 102388. [Google Scholar] [CrossRef]
- Alvarez-Ordonez, A.; Alvseike, O.; Omer, M.K.; Heir, E.; Axelsson, L.; Holck, A.; Prieto, M. Heterogeneity in resistance to food-related stresses and biofilm formation ability among verocytotoxigenic Escherichia coli strains. Int. J. Food Microbiol. 2013, 161, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Serra, D.O.; Mika, F.; Richter, A.M.; Hengge, R. The green tea polyphenol EGCG inhibits E. coli biofilm formation by impairing amyloid curli fibre assembly and downregulating the biofilm regulator CsgD via the sigma(E)-dependent sRNA RybB. Mol. Microbiol. 2016, 101, 136–151. [Google Scholar] [CrossRef]
- Ding, X.Y.; Wang, X.Y.; Kong, Y.H.; Zhao, C.X.; Qin, S.; Sun, X.; Li, M.W. Comparative transcriptome analysis of Bombyx mori (Lepidoptera) larval hemolymph in response to Autographa californica nucleopolyhedrovirus in differentially resistant strains. Processes 2021, 9, 1401. [Google Scholar] [CrossRef]
- Wang, X.; Dubey, A.K.; Suzuki, K.; Baker, C.S.; Babitzke, P.; Romeo, T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 2005, 56, 1648–1663. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Rice, J.D.; Goller, C.; Pannuri, A.; Taylor, J.; Meisner, J.; Beveridge, T.J.; Preston, J.F.I.; Romeo, T. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J. Bacteriol. 2008, 190, 3670–3680. [Google Scholar] [CrossRef]
- Yang, Q.N.; Wang, L.F.; Gao, J.X.; Liu, X.B.; Feng, Y.Q.; Wu, Q.; Baloch, A.B.; Cui, L.; Xia, X.D. Tannin-rich fraction from pomegranate rind inhibits quorum sensing in chromobacterium violaceum and biofilm formation in Escherichia coli. Foodborne Pathog. Dis. 2016, 13, 28–35. [Google Scholar] [CrossRef]
- Hu, J.G.; Gu, Y.; Lu, H.Q.; Raheem, M.A.; Yu, F.H.; Niu, X.P.; Zuo, J.K.; Yin, H.F.; Huang, C.Q.; Song, X.J. Identification of novel biofilm genes in avian pathogenic Escherichia coli by Tn5 transposon mutant library. World J. Microbiol. Biotechnol. 2022, 38, 130. [Google Scholar] [CrossRef]
- Battesti, A.; Hoskins, J.R.; Tong, S.; Milanesio, P.; Mann, J.M.; Kravats, A.; Tsegaye, Y.M.; Bougdour, A.; Wickner, S.; Gottesman, S. Anti-adaptors provide multiple modes for regulation of the RssB adaptor protein. Genes Dev. 2013, 27, 2722–2735. [Google Scholar] [CrossRef]
- Ma, Q.; Wood, T.K. OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ. Microbiol. 2009, 11, 2735–2746. [Google Scholar] [CrossRef]
- Zhou, G.; Wang, Y.S.; Peng, H.; Li, S.J.; Sun, T.L.; Shen, P.F.; Xie, X.B.; Shi, Q.S. Roles of ompA of Citrobacter werkmanii in bacterial growth, biocide resistance, biofilm formation and swimming motility. Appl. Microbiol. Biotechnol. 2021, 105, 2841–2854. [Google Scholar] [CrossRef]
- Si, M.R.; Su, T.; Chen, C.; Wei, Z.F.; Gong, Z.J.; Li, G.Z. OsmC in Corynebacterium glutamicum was a thiol-dependent organic hydroperoxide reductase. Int. J. Biol. Macromol. 2019, 136, 642–652. [Google Scholar] [CrossRef]
- Bolstad, H.M.; Wood, M.J. An in vivo method for characterization of protein interactions within sulfur trafficking systems of E. coli. J. Proteome Res. 2010, 9, 6740–6751. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.X.; Li, Y.M.; Zhan, W.E.; Wood, T.K.; Wang, X.X. Resistance to oxidative stress by inner membrane protein ElaB is regulated by OxyR and RpoS. Microb. Biotechnol. 2019, 12, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Alfaro, T.; Elmore, J.R.; Stromberg, Z.R.; Hutchison, J.R.; Hess, B.M. Engineering Citrobacter freundii using CRISPR/Cas9 system. J. Microbiol. Methods 2022, 200, 106533. [Google Scholar] [CrossRef] [PubMed]
- Shamim, S.; Rehman, A.; Qazi, M.H. Swimming, swarming, twitching, and chemotactic responses of Cupriavidus metallidurans CH34 and Pseudomonas putida mt2 in the presence of cadmium. Arch. Environ. Contam. Toxicol. 2014, 66, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, L.; Rodriguez, M.; Prats, D. Effect of different extraction methods on bound EPS from MBR sludges. Part I: Influence of extraction methods over three-dimensional EEM fluorescence spectroscopy fingerprint. Desalination 2010, 261, 19–26. [Google Scholar] [CrossRef]
- Cui, H.Y.; Zhang, C.H.; Li, C.Z.; Lin, L. Inhibition of Escherichia coli O157:H7 biofilm on vegetable surface by solid liposomes of clove oil. LWT-Food Sci. Technol. 2020, 117, 108656. [Google Scholar] [CrossRef]
- Saeki, E.K.; Yamada, A.Y.; de Araujo, L.A.; Anversa, L.; Garcia, D.D.; de Souza, R.L.B.; Martins, H.M.; Kobayashi, R.K.T.; Nakazato, G. Subinhibitory concentrations of biogenic silver nanoparticles affect motility and biofilm formation in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2021, 11, 656984. [Google Scholar] [CrossRef]
- Misba, L.; Zaidi, S.; Khan, A.U. A comparison of antibacterial and antibiofilm efficacy of phenothiazinium dyes between Gram positive and Gram negative bacterial biofilm. Photodiagnosis Photodyn. Ther. 2017, 18, 24–33. [Google Scholar] [CrossRef]
- Zhu, C.T.; Mei, Y.Y.; Zhu, L.L.; Xu, Y.; Sheng, S.; Wang, J. Recombinant Escherichia coli BL21-pET28a-egfp cultivated with nanomaterials in a modified microchannel for biofilm formation. Int. J. Mol. Sci. 2018, 19, 2590. [Google Scholar] [CrossRef]
- Zhang, F.; Zhu, C.T.; Peng, Q.M.; Wang, F.Q.; Sheng, S.; Wu, Q.Y.; Wang, J. Enhanced permeability of recombinant E. coli cells with deep eutectic solvent for transformation of rutin. J. Chem. Technol. Biotechnol. 2020, 95, 384–393. [Google Scholar] [CrossRef]
- Zhi, F.J.; Zhou, D.; Chen, J.L.; Fang, J.Y.; Zheng, W.F.; Li, J.M.; Hao, M.Y.; Shi, Y.; Jin, Y.P.; Wang, A.H. An ArsR transcriptional regulator facilitates Brucella sp. survival via regulating self and outer membrane protein. Int. J. Mol. Sci. 2021, 11, 10860. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 45. [Google Scholar] [CrossRef] [PubMed]


| Sample | Total Reads | GC Content (%) | Q30 (%) | Mapped Reads | Mapped Ratio (%) |
|---|---|---|---|---|---|
| Parent strain-1 | 33,823,474 | 52.74 | 96.97 | 31,023,820 | 91.72 |
| Parent strain-2 | 28,198,342 | 52.81 | 97.27 | 23,954,412 | 84.95 |
| Parent strain-3 | 17,748,764 | 51.49 | 96.98 | 15,749,686 | 88.74 |
| ΔcsgD-1 | 25,372,190 | 53.08 | 93.61 | 23,002,214 | 90.66 |
| ΔcsgD-2 | 25,189,478 | 52.91 | 97.44 | 23,089,232 | 91.66 |
| ΔcsgD-3 | 28,254,794 | 52.84 | 96.83 | 26,430,290 | 93.54 |
| Classification | Gene ID | Gene | Description |
|---|---|---|---|
| Biological adhesion | HO396_05355 | pgaA | partially deacetylated poly-β-1,6-N-acetyl-D-glucosamine export outer membrane porin PgaA |
| HO396_09555 | motB | flagellar motor protein MotB | |
| HO396_02470 | fimA | type 1 fimbrial major subunit FimA | |
| HO396_02475 | fimC | type 1 fimbriae periplasmic chaperone FimC | |
| Response to stimulus | HO396_01705 | iraP | anti-adapter protein IraP |
| HO396_05005 | ompA | outer membrane protein OmpA | |
| HO396_07480 | osmC | peroxiredoxin OsmC | |
| HO396_08475 | sufE | cysteine desulfuration protein SufE | |
| HO396_11145 | elaB | stress response protein ElaB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, C.-H.; Chen, F.-H.; Yang, Y.-L.; Zhan, Y.-F.; Herman, R.A.; Gong, L.-C.; Sheng, S.; Wang, J. The Transcription Factor CsgD Contributes to Engineered Escherichia coli Resistance by Regulating Biofilm Formation and Stress Responses. Int. J. Mol. Sci. 2023, 24, 13681. https://doi.org/10.3390/ijms241813681
Yan C-H, Chen F-H, Yang Y-L, Zhan Y-F, Herman RA, Gong L-C, Sheng S, Wang J. The Transcription Factor CsgD Contributes to Engineered Escherichia coli Resistance by Regulating Biofilm Formation and Stress Responses. International Journal of Molecular Sciences. 2023; 24(18):13681. https://doi.org/10.3390/ijms241813681
Chicago/Turabian StyleYan, Cheng-Hai, Fang-Hui Chen, Yu-Lu Yang, Yu-Fan Zhan, Richard A. Herman, Lu-Chan Gong, Sheng Sheng, and Jun Wang. 2023. "The Transcription Factor CsgD Contributes to Engineered Escherichia coli Resistance by Regulating Biofilm Formation and Stress Responses" International Journal of Molecular Sciences 24, no. 18: 13681. https://doi.org/10.3390/ijms241813681





