Probiotics Modulate Host Immune Response and Interact with the Gut Microbiota: Shaping Their Composition and Mediating Antibiotic Resistance
Abstract
:1. Introduction
2. Results
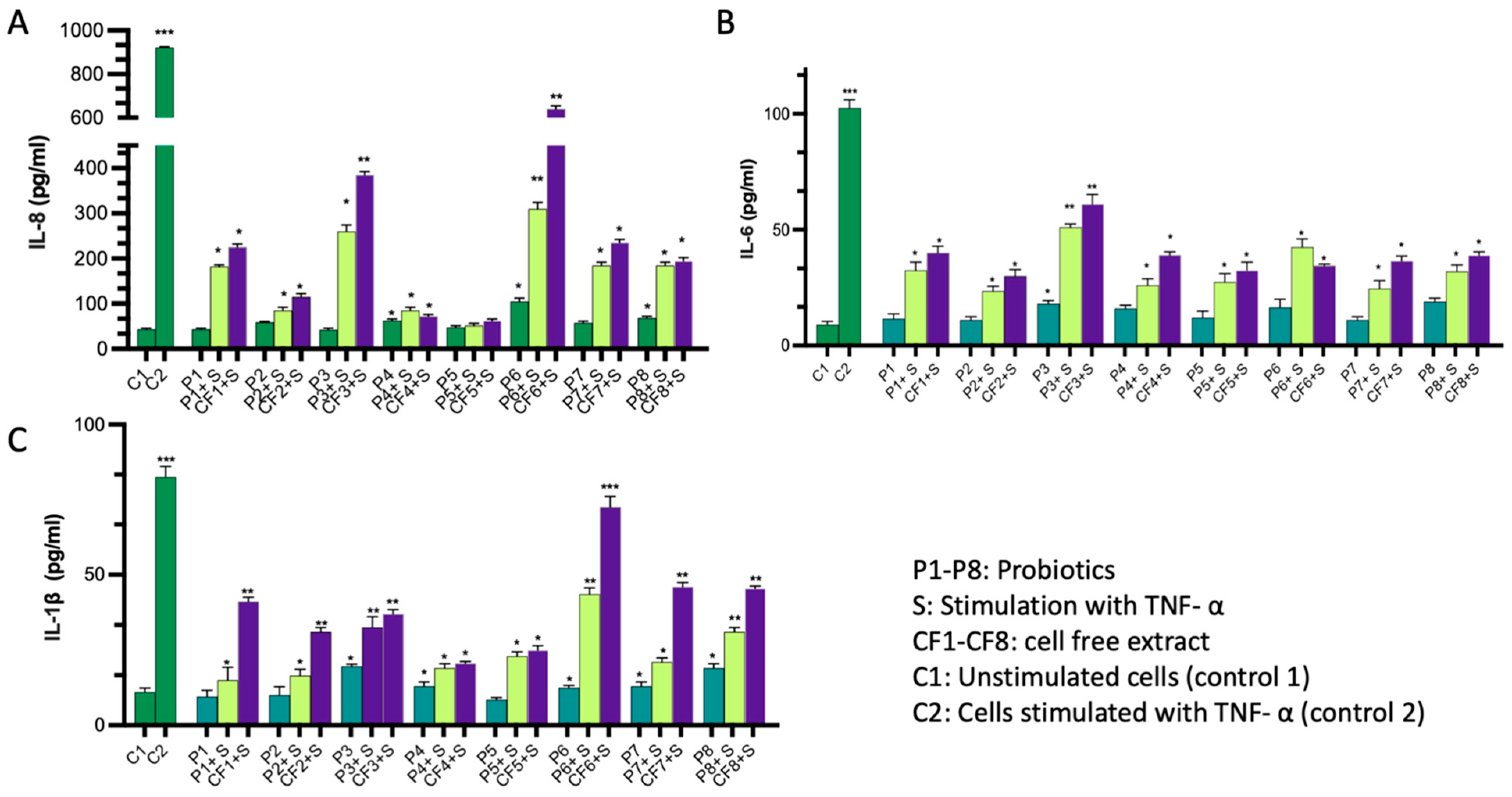
2.1. Immunomodulatory Activity of the Probiotics
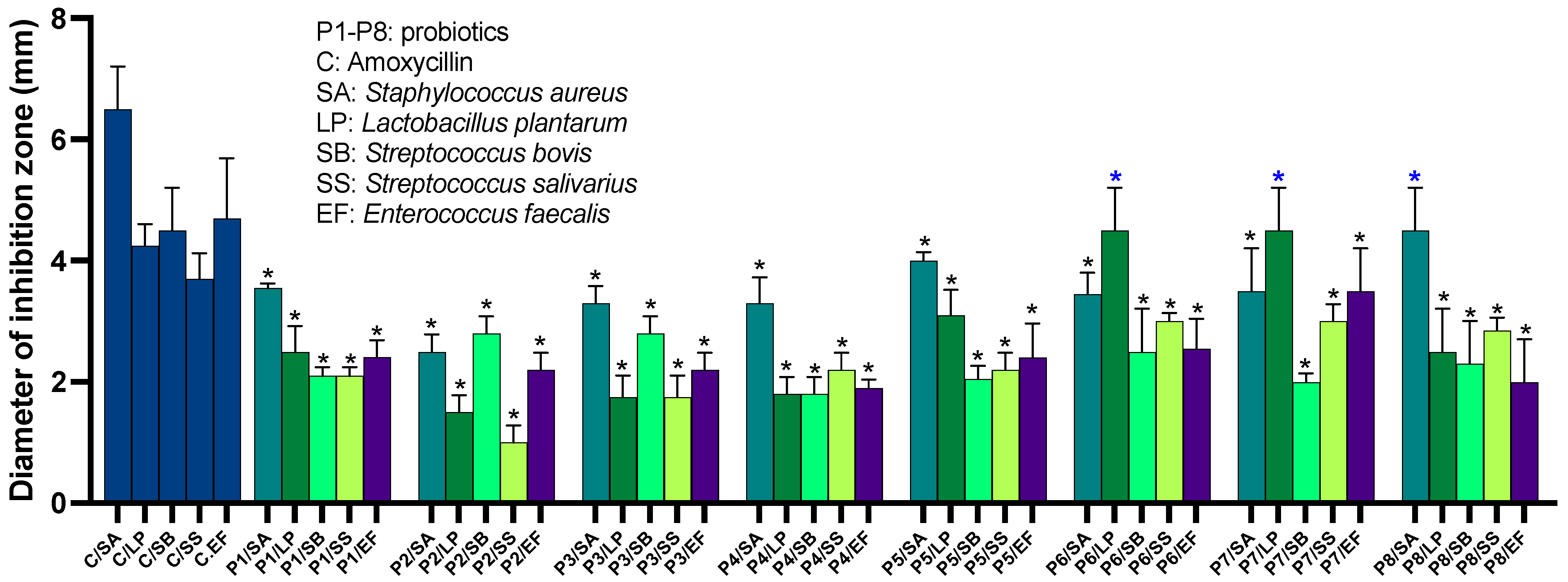
2.2. Probiotics Exhibit Antimicrobial Activity against a Broad Range of Human-Associated Microbes
The Inhibitory Activity of P4 against Gram-Negative Pathogens Is Traceable to a Single Strain from the Probiotic Mixture
2.3. Probiotics Possess Antibiotic Resistance Genes
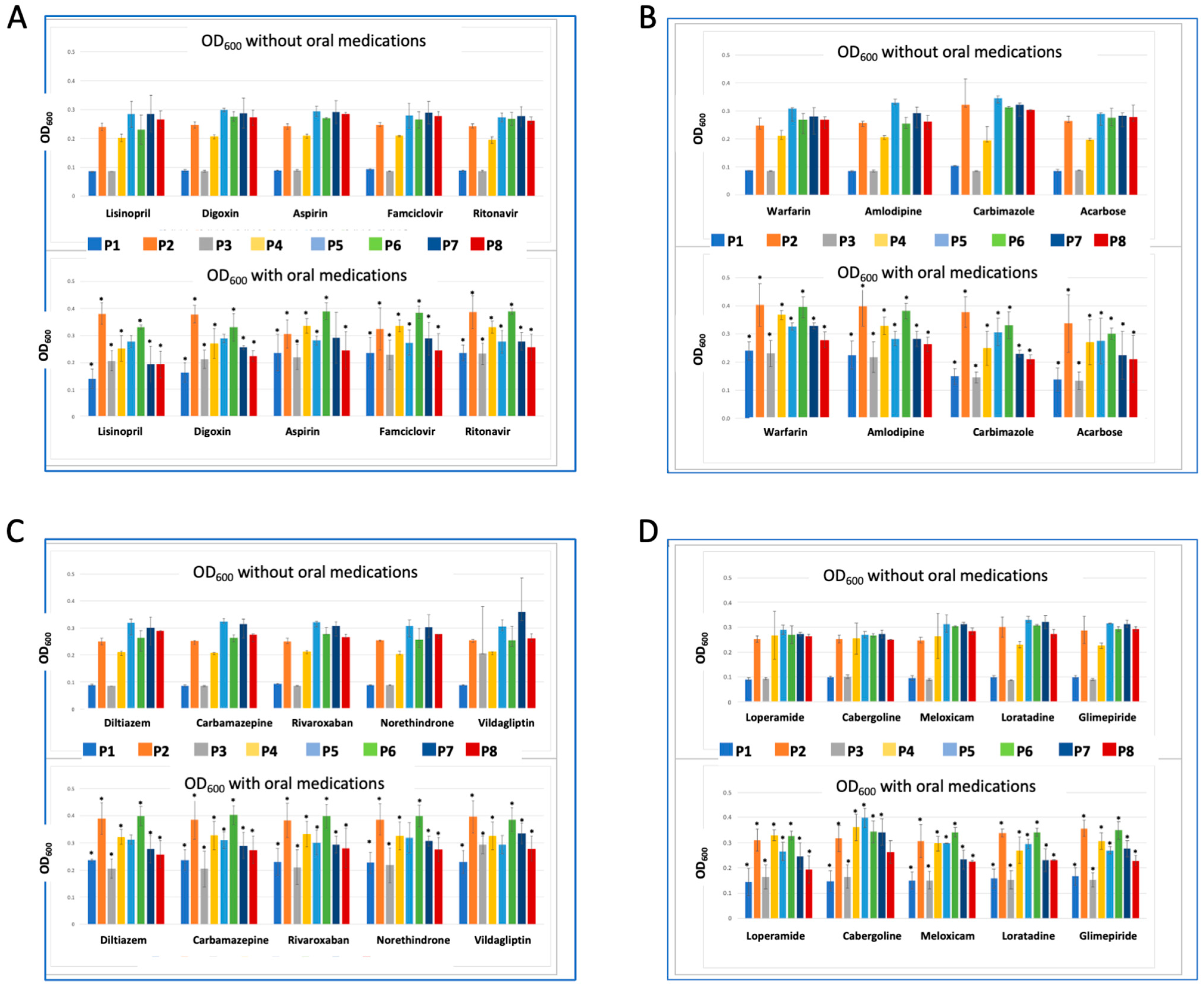
2.4. Synergistic and Antagonistic Effect of Co-Administered Drugs on Probiotics
3. Discussion
3.1. The Interactions between Probiotics, Host, and Microbes Are Evident
3.2. Improved Regulations for the Use of Probiotics
4. Materials and Methods
4.1. Microorganisms and Culture Conditions
4.2. Assessment of Pro/Anti-Inflammatory Activity of Probiotics
4.3. Determination of Antimicrobial Activity of the Probiotics
4.4. DNA Extraction, and Polymerase Chain Reaction Amplification
4.5. Assessing Antibiotic Resistance in Probiotics
4.6. Probiotics/Drugs Co-Incubation Assay
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gilbert, J.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Gacesa, R.; Kurilshikov, A.; Vich Vila, A.; Sinha, T.; Klaassen, M.A.Y.; Bolte, L.A.; Andreu-Sánchez, S.; Chen, L.; Collij, V.; Hu, S.; et al. Environmental factors shaping the gut microbiome in a Dutch population. Nature 2022, 604, 732–739. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Stavropoulou, E.; Bezirtzoglou, E. Probiotics in Medicine: A Long Debate. Front. Immunol. 2020, 11, 2192. [Google Scholar] [CrossRef]
- Sehrawat, N.; Yadav, M.; Singh, M.; Kumar, V.; Sharma, V.R.; Sharma, A.K. Probiotics in microbiome ecological balance providing a therapeutic window against cancer. Semin. Cancer Biol. 2021, 70, 24–36. [Google Scholar] [CrossRef]
- Liao, W.; Chen, C.; Wen, T.; Zhao, Q. Probiotics for the Prevention of Antibiotic-associated Diarrhea in Adults: A Meta-Analysis of Randomized Placebo-Controlled Trials. J. Clin. Gastroenterol. 2021, 55, 469–480. [Google Scholar] [CrossRef]
- Abdi, M.; Lohrasbi, V.; Asadi, A.; Esghaei, M.; Jazi, F.M.; Rohani, M.; Talebi, M. Interesting probiotic traits of mother’s milk Lactobacillus isolates; from bacteriocin to inflammatory bowel disease improvement. Microb. Pathog. 2021, 158, 104998. [Google Scholar] [CrossRef]
- Guglielmetti, S.; Mora, D.; Gschwender, M.; Popp, K. Randomised clinical trial: Bifidobacterium bifidum MIMBb75 significantly alleviates irritable bowel syndrome and improves quality of life—A double-blind, placebo-controlled study. Aliment. Pharmacol. Ther. 2011, 33, 1123–1132. [Google Scholar] [CrossRef]
- Didari, T.; Mozaffari, S.; Nikfar, S.; Abdollahi, M. Effectiveness of probiotics in irritable bowel syndrome: Updated systematic review with meta-analysis. World J. Gastroenterol. 2015, 21, 3072–3084. [Google Scholar] [CrossRef]
- Sun, J.R.; Kong, C.F.; Qu, X.K.; Deng, C.; Lou, Y.N.; Jia, L.Q. Efficacy and safety of probiotics in irritable bowel syndrome: A systematic review and meta-analysis. Saudi J. Gastroenterol. 2020, 26, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Vallianou, N.; Stratigou, T.; Christodoulatos, G.S.; Tsigalou, C.; Dalamaga, M. Probiotics, Prebiotics, Synbiotics, Postbiotics, and Obesity: Current Evidence, Controversies, and Perspectives. Curr. Obes. Rep. 2020, 9, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Lohrasbi, V.; Abdi, M.; Asadi, A.; Rohani, M.; Esghaei, M.; Talebi, M.; Amirmozafari, N. The effect of improved formulation of chitosan-alginate microcapsules of Bifidobacteria on serum lipid profiles in mice. Microb. Pathog. 2020, 149, 104585. [Google Scholar] [CrossRef]
- Asadi, A.; Lohrasbi, V.; Abdi, M.; Mirkalantari, S.; Esghaei, M.; Kashanian, M.; Oshaghi, M.; Talebi, M. The probiotic properties and potential of vaginal Lactobacillus spp. isolated from healthy women against some vaginal pathogens. Lett. Appl. Microbiol. 2022, 74, 752–764. [Google Scholar] [CrossRef]
- Yue, Y.; Ye, K.; Lu, J.; Wang, X.; Zhang, S.; Liu, L.; Yang, B.; Nassar, K.; Xu, X.; Pang, X.; et al. Probiotic strain Lactobacillus plantarum YYC-3 prevents colon cancer in mice by regulating the tumour microenvironment. Biomed. Pharmacother. 2020, 127, 110159. [Google Scholar] [CrossRef] [PubMed]
- Learn-Han, L.; Vengadesh, L.; Jodi Woan-Fei, L.; Yatinesh, K.; Sivakumar, T.; Loh Teng-Hern, T. IDDF2022-ABS-0241 Exploring the potential role of probiotics in alleviating insomnia. Gut 2022, 71, A65. [Google Scholar] [CrossRef]
- Kong, G.Y.-E.; Letchumanan, V.; Tan, L.T.-H.; Law, J.W.-F. Gut Microbiome in Obsessive Compulsive Disorder: Potential of Probiotics as an Adjuvant Therapy. Prog. Microbes Mol. Biol. 2022, 5. [Google Scholar] [CrossRef]
- Binda, S.; Hill, C.; Johansen, E.; Obis, D.; Pot, B.; Sanders, M.E.; Tremblay, A.; Ouwehand, A.C. Criteria to Qualify Microorganisms as “Probiotic” in Foods and Dietary Supplements. Front. Microbiol. 2020, 11, 1662. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Pang, B.; Li, N.; Jin, H.; Li, J.; Wu, W.; Ai, C.; Jiang, C.; Shi, J. Therapeutic effect of Lactobacillus rhamnosus SHA113 on intestinal infection by multi-drug-resistant Staphylococcus aureus and its underlying mechanisms. Food Funct. 2020, 11, 6226–6239. [Google Scholar] [CrossRef]
- Wei, Y.; Yang, F.; Wu, Q.; Gao, J.; Liu, W.; Liu, C.; Guo, X.; Suwal, S.; Kou, Y.; Zhang, B.; et al. Protective Effects of Bifidobacterial Strains Against Toxigenic Clostridium difficile. Front. Microbiol. 2018, 9, 888. [Google Scholar] [CrossRef]
- Niu, H.-L.; Xiao, J.-Y. The efficacy and safety of probiotics in patients with irritable bowel syndrome: Evidence based on 35 randomized controlled trials. Int. J. Surg. 2020, 75, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Ng, Q.X.; Peters, C.; Ho, C.Y.X.; Lim, D.Y.; Yeo, W.S. A meta-analysis of the use of probiotics to alleviate depressive symptoms. J. Affect. Disord. 2018, 228, 13–19. [Google Scholar] [CrossRef]
- Un-Nisa, A.; Khan, A.; Zakria, M.; Siraj, S.; Ullah, S.; Tipu, M.K.; Ikram, M.; Kim, M.O. Updates on the Role of Probiotics against Different Health Issues: Focus on Lactobacillus. Int. J. Mol. Sci. 2023, 24, 142. [Google Scholar] [CrossRef] [PubMed]
- Tette, F.M.; Kwofie, S.K.; Wilson, M.D. Therapeutic Anti-Depressant Potential of Microbial GABA Produced by Lactobacillus rhamnosus Strains for GABAergic Signaling Restoration and Inhibition of Addiction-Induced HPA Axis Hyperactivity. Curr. Issues Mol. Biol. 2022, 44, 1434–1451. [Google Scholar] [CrossRef]
- Tan, A.H.; Hor, J.W.; Chong, C.W.; Lim, S.Y. Probiotics for Parkinson’s disease: Current evidence and future directions. JGH Open 2021, 5, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sun, Z.; Jiang, S.; Bai, X.; Ma, C.; Peng, Q.; Chen, K.; Chang, H.; Fang, T.; Zhang, H. Probiotic Bifidobacterium lactis V9 Regulates the Secretion of Sex Hormones in Polycystic Ovary Syndrome Patients through the Gut-Brain Axis. mSystems 2019, 4, e00017-19. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liang, Q.; Lu, B.; Shen, H.; Liu, S.; Shi, Y.; Leptihn, S.; Li, H.; Wei, J.; Liu, C.; et al. Whole-genome analysis of probiotic product isolates reveals the presence of genes related to antimicrobial resistance, virulence factors, and toxic metabolites, posing potential health risks. BMC Genom. 2021, 22, 210. [Google Scholar] [CrossRef]
- Zhao, Y.; Zeng, Y.; Zeng, D.; Wang, H.; Zhou, M.; Sun, N.; Xin, J.; Khalique, A.; Rajput, D.S.; Pan, K.; et al. Probiotics and MicroRNA: Their Roles in the Host-Microbe Interactions. Front. Microbiol. 2020, 11, 604462. [Google Scholar] [CrossRef]
- Oh, J.K.; Vasquez, R.; Kim, S.H.; Hwang, I.C.; Song, J.H.; Park, J.H.; Kim, I.H.; Kang, D.K. Multispecies probiotics alter fecal short-chain fatty acids and lactate levels in weaned pigs by modulating gut microbiota. J. Anim. Sci. Technol. 2021, 63, 1142–1158. [Google Scholar] [CrossRef]
- Ruíz-Ramírez, Y.; Guadarrama-Mendoza, P.C.; Escalante, A.; Giles-Gómez, M.; Valadez-Blanco, R. Probiotic activity traits in vitro and production of antimicrobial peptides by Lactobacillaceae isolates from pulque using Lactobacillus acidophilus NCFM as control. Braz. J. Microbiol. 2022, 53, 921–933. [Google Scholar] [CrossRef]
- Tóth, A.G.; Csabai, I.; Judge, M.F.; Maróti, G.; Becsei, Á.; Spisák, S.; Solymosi, N. Mobile Antimicrobial Resistance Genes in Probiotics. Antibiotics 2021, 10, 1287. [Google Scholar] [CrossRef]
- Imperial, I.C.V.J.; Ibana, J.A. Addressing the Antibiotic Resistance Problem with Probiotics: Reducing the Risk of Its Double-Edged Sword Effect. Front. Microbiol. 2016, 7, 1983. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Yang, X.; Qin, J.; Lu, N.; Cheng, G.; Wu, N.; Pan, Y.; Li, J.; Zhu, L.; Wang, X.; et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 2013, 4, 2151. [Google Scholar] [CrossRef] [PubMed]
- Rokon-Uz-Zaman, M.; Bushra, A.; Pospo, T.A.; Runa, M.A.; Tasnuva, S.; Parvin, M.S.; Islam, M.T. Detection of antimicrobial resistance genes in Lactobacillus spp. from poultry probiotic products and their horizontal transfer among Escherichia coli. Vet. Anim. Sci. 2023, 20, 100292. [Google Scholar] [CrossRef] [PubMed]
- Montassier, E.; Valdés-Mas, R.; Batard, E.; Zmora, N.; Dori-Bachash, M.; Suez, J.; Elinav, E. Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner. Nat. Microbiol. 2021, 6, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Massip, C.; Branchu, P.; Bossuet-Greif, N.; Chagneau, C.V.; Gaillard, D.; Martin, P.; Boury, M.; Sécher, T.; Dubois, D.; Nougayrède, J.P.; et al. Deciphering the interplay between the genotoxic and probiotic activities of Escherichia coli Nissle 1917. PLoS Pathog. 2019, 15, e1008029. [Google Scholar] [CrossRef]
- Dore, M.P.; Bibbò, S.; Fresi, G.; Bassotti, G.; Pes, G.M. Side Effects Associated with Probiotic Use in Adult Patients with Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Nutrients 2019, 11, 2913. [Google Scholar] [CrossRef]
- Derwa, Y.; Gracie, D.J.; Hamlin, P.J.; Ford, A.C. Systematic review with meta-analysis: The efficacy of probiotics in inflammatory bowel disease. Aliment. Pharmacol. Ther. 2017, 46, 389–400. [Google Scholar] [CrossRef]
- Land, M.H.; Rouster-Stevens, K.; Woods, C.R.; Cannon, M.L.; Cnota, J.; Shetty, A.K. Lactobacillus sepsis associated with probiotic therapy. Pediatrics 2005, 115, 178–181. [Google Scholar] [CrossRef]
- Kulkarni, H.S.; Khoury, C.C. Sepsis associated with Lactobacillus bacteremia in a patient with ischemic colitis. Indian J. Crit. Care Med. 2014, 18, 606–608. [Google Scholar] [CrossRef]
- Guenther, K.; Straube, E.; Pfister, W.; Guenther, A.; Huebler, A. Sever sepsis after probiotic treatment with Escherichia coli NISSLE 1917. Pediatr. Infect. Dis. J. 2010, 29, 188–189. [Google Scholar] [CrossRef] [PubMed]
- Kunz, A.N.; Noel, J.M.; Fairchok, M.P. Two Cases of Lactobacillus Bacteremia During Probiotic Treatment of Short Gut Syndrome. J. Pediatr. Gastroenterol. Nutr. 2004, 38, 457–458. [Google Scholar] [CrossRef] [PubMed]
- Franko, B.; Vaillant, M.; Recule, C.; Vautrin, E.; Brion, J.P.; Pavese, P. Lactobacillus paracasei endocarditis in a consumer of probiotics. Med. Mal. Infect. 2013, 43, 171–173. [Google Scholar] [CrossRef] [PubMed]
- Boumis, E.; Capone, A.; Galati, V.; Venditti, C.; Petrosillo, N. Probiotics and infective endocarditis in patients with hereditary hemorrhagic telangiectasia: A clinical case and a review of the literature. BMC Infect. Dis. 2018, 18, 65. [Google Scholar] [CrossRef] [PubMed]
- Kalliomäki, M.; Salminen, S.; Arvilommi, H.; Kero, P.; Koskinen, P.; Isolauri, E. Probiotics in primary prevention of atopic disease: A randomised placebo-controlled trial. Lancet 2001, 357, 1076–1079. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.L.; Dunstan, J.A.; Prescott, S.L. Probiotic supplementation for the first 6 months of life fails to reduce the risk of atopic dermatitis and increases the risk of allergen sensitization in high-risk children: A randomized controlled trial. J. Allergy Clin. Immunol. 2007, 119, 184–191. [Google Scholar] [CrossRef]
- Borase, H.; Dwivedi, M.K.; Krishnamurthy, R.; Patil, S. Chapter 30—Probiotics: Health safety considerations. In Probiotics in the Prevention and Management of Human Diseases; Dwivedi, M.K., Amaresan, N., Sankaranarayanan, A., Kemp, E.H., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 449–463. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Q. Relationship between tetracycline antibiotic susceptibility and genotype in oral cavity Lactobacilli clinical isolates. Antimicrob. Resist. Infect. Control 2019, 8, 27. [Google Scholar] [CrossRef]
- Piwowar, H.; Priem, J.; Larivière, V.; Alperin, J.P.; Matthias, L.; Norlander, B.; Farley, A.; West, J.; Haustein, S. The state of OA: A large-scale analysis of the prevalence and impact of Open Access articles. PeerJ 2018, 6, e4375. [Google Scholar] [CrossRef]
- Maier, L.; Pruteanu, M.; Kuhn, M.; Zeller, G.; Telzerow, A.; Anderson, E.E.; Brochado, A.R.; Fernandez, K.C.; Dose, H.; Mori, H.; et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018, 555, 623–628. [Google Scholar] [CrossRef]
- Tang, T.; Wang, J.; Jiang, Y.; Zhu, X.; Zhang, Z.; Wang, Y.; Shu, X.; Deng, Y.; Zhang, F. Bifidobacterium lactis TY-S01 Prevents Loperamide-Induced Constipation by Modulating Gut Microbiota and Its Metabolites in Mice. Front. Nutr. 2022, 9, 890314. [Google Scholar] [CrossRef]
- Aljeldah, M.M. Antimicrobial Resistance and Its Spread Is a Global Threat. Antibiotics 2022, 11, 1082. [Google Scholar] [CrossRef]
- Vernier, C.L.; Chin, I.M.; Adu-Oppong, B.; Krupp, J.J.; Levine, J.; Dantas, G.; Ben-Shahar, Y. The gut microbiome defines social group membership in honey bee colonies. Sci. Adv. 2020, 6, eabd3431. [Google Scholar] [CrossRef] [PubMed]
- Vijay, A.; Valdes, A.M. Role of the gut microbiome in chronic diseases: A narrative review. Eur. J. Clin. Nutr. 2022, 76, 489–501. [Google Scholar] [CrossRef]
- Corthésy, B.; Gaskins, H.R.; Mercenier, A. Cross-Talk between Probiotic Bacteria and the Host Immune System1,2. J. Nutr. 2007, 137, 781S–790S. [Google Scholar] [CrossRef] [PubMed]
- Stadlbauer, V. Immunosuppression and probiotics: Are they effective and safe? Benef. Microbes 2015, 6, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Ramírez, L.M.; Pérez-Solano, R.A.; Castañón-Alonso, S.L.; Moreno Guerrero, S.S.; Ramírez Pacheco, A.; García Garibay, M.; Eslava, C. Probiotic Lactobacillus Strains Stimulate the Inflammatory Response and Activate Human Macrophages. J. Immunol. Res. 2017, 2017, 4607491. [Google Scholar] [CrossRef]
- Onisiforou, A.; Spyrou, G.M. Immunomodulatory effects of microbiota-derived metabolites at the crossroad of neurodegenerative diseases and viral infection: Network-based bioinformatics insights. Front. Immunol. 2022, 13, 843128. [Google Scholar] [CrossRef]
- Cristofori, F.; Dargenio, V.N.; Dargenio, C.; Miniello, V.L.; Barone, M.; Francavilla, R. Anti-Inflammatory and Immunomodulatory Effects of Probiotics in Gut Inflammation: A Door to the Body. Front. Immunol. 2021, 12, 578386. [Google Scholar] [CrossRef]
- Ruiz, L.; Flórez, A.B.; Sánchez, B.; Moreno-Muñoz, J.A.; Rodriguez-Palmero, M.; Jiménez, J.; Gavilán, C.; Gueimonde, M.; Ruas-Madiedo, P.; Margolles, A. Bifidobacteriumlongum subsp. infantis CECT7210 (B. infantis IM-1(®)) Displays In Vitro Activity against Some Intestinal Pathogens. Nutrients 2020, 12, 3259. [Google Scholar] [CrossRef]
- Chichlowski, M.; De Lartigue, G.; German, J.B.; Raybould, H.E.; Mills, D.A. Bifidobacteria isolated from infants and cultured on human milk oligosaccharides affect intestinal epithelial function. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 321–327. [Google Scholar] [CrossRef]
- Chen, J.; Chen, X.; Ho, C.L. Recent Development of Probiotic Bifidobacteria for Treating Human Diseases. Front. Bioeng. Biotechnol. 2021, 9, 770248. [Google Scholar] [CrossRef]
- Enan, G.; Abdel-Shafi, S.; Ouda, S.; Negm, S. Novel antibacterial activity of lactococcus lactis subspecies lactis z11 isolated from zabady. Int. J. Biomed. Sci. 2013, 9, 174–180. [Google Scholar]
- van Zyl, W.F.; Deane, S.M.; Dicks, L.M.T. Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria. Gut Microbes 2020, 12, 1831339. [Google Scholar] [CrossRef]
- Amiri, S.; Rezaei Mokarram, R.; Sowti Khiabani, M.; Rezazadeh Bari, M.; Alizadeh Khaledabad, M. Characterization of antimicrobial peptides produced by Lactobacillus acidophilus LA-5 and Bifidobacterium lactis BB-12 and their inhibitory effect against foodborne pathogens. LWT 2022, 153, 112449. [Google Scholar] [CrossRef]
- Silva, D.R.; Sardi, J.d.C.O.; Pitangui, N.d.S.; Roque, S.M.; Silva, A.C.B.d.; Rosalen, P.L. Probiotics as an alternative antimicrobial therapy: Current reality and future directions. J. Funct. Foods 2020, 73, 104080. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, P.; Zhang, X. Probiotics Regulate Gut Microbiota: An Effective Method to Improve Immunity. Molecules 2021, 26, 6076. [Google Scholar] [CrossRef] [PubMed]
- Hemarajata, P.; Versalovic, J. Effects of probiotics on gut microbiota: Mechanisms of intestinal immunomodulation and neuromodulation. Ther. Adv. Gastroenterol. 2013, 6, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Duche, R.; Singh, A.; Wandhare, A.G.; Sangwan, V.; Sihag, M.K.; Nwagu, T.; Panwar, H.; Ezeogu, L. Antibiotic Resistance in Potential Probiotic Lactobacillary Strains of Fermented Foods and Human Origin From Nigeria. Res. Sq. 2022, 23, 142. [Google Scholar] [CrossRef]
- Devirgiliis, C.; Zinno, P.; Perozzi, G. Update on antibiotic resistance in foodborne Lactobacillus and Lactococcus species. Front. Microbiol. 2013, 4, 301. [Google Scholar] [CrossRef]
- Duche, R.T.; Singh, A.; Wandhare, A.G.; Sangwan, V.; Sihag, M.K.; Nwagu, T.N.T.; Panwar, H.; Ezeogu, L.I. Antibiotic resistance in potential probiotic lactic acid bacteria of fermented foods and human origin from Nigeria. BMC Microbiol. 2023, 23, 142. [Google Scholar] [CrossRef]
- Li, B.; Chen, D.; Lin, F.; Wu, C.; Cao, L.; Chen, H.; Hu, Y.; Yin, Y. Genomic Island-Mediated Horizontal Transfer of the Erythromycin Resistance Gene erm(X) among Bifidobacteria. Appl. Environ. Microbiol. 2022, 88, e0041022. [Google Scholar] [CrossRef]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Toole, P.W. Genus-Wide Assessment of Antibiotic Resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2019, 85, e01738-18. [Google Scholar] [CrossRef]
- Sotoudegan, F.; Daniali, M.; Hassani, S.; Nikfar, S.; Abdollahi, M. Reappraisal of probiotics’ safety in human. Food Chem. Toxicol. 2019, 129, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Ng, Q.X.; Lim, Y.L.; Yaow, C.Y.L.; Ng, W.K.; Thumboo, J.; Liew, T.M. Effect of Probiotic Supplementation on Gut Microbiota in Patients with Major Depressive Disorders: A Systematic Review. Nutrients 2023, 15, 1351. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, C.; Wang, Q.; Zhang, Z.; Nie, W.; Shang, L. Armored probiotics for oral delivery. Smart Med. 2023, e20230019. [Google Scholar] [CrossRef]
- Han, S.; Lu, Y.; Xie, J.; Fei, Y.; Zheng, G.; Wang, Z.; Liu, J.; Lv, L.; Ling, Z.; Berglund, B.; et al. Probiotic Gastrointestinal Transit and Colonization After Oral Administration: A Long Journey. Front. Cell Infect. Microbiol. 2021, 11, 609722. [Google Scholar] [CrossRef]
- Kothari, D.; Patel, S.; Kim, S.-K. Probiotic supplements might not be universally-effective and safe: A review. Biomed. Pharmacother. 2019, 111, 537–547. [Google Scholar] [CrossRef]
- Sanders, M.E.; Akkermans, L.M.; Haller, D.; Hammerman, C.; Heimbach, J.; Hörmannsperger, G.; Huys, G.; Levy, D.D.; Lutgendorff, F.; Mack, D.; et al. Safety assessment of probiotics for human use. Gut Microbes 2010, 1, 164–185. [Google Scholar] [CrossRef]
- Hradicka, P.; Adamkova, P.; Lenhardt, L.; Gancarcikova, S.; Iannaccone, S.F.; Demeckova, V. Addressing safety concerns of long-term probiotic use: In vivo evidence from a rat model. J. Funct. Foods 2023, 104, 105521. [Google Scholar] [CrossRef]
- Venugopalan, V.; Shriner, K.A.; Wong-Beringer, A. Regulatory Oversight and Safety of Probiotic Use. Emerg. Infect. Dis. J. 2010, 16, 1661. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Zhang, R.; Tian, X.; Zhou, X.; Pan, X.; Wong, A. Assessing the Risk of Probiotic Dietary Supplements in the Context of Antibiotic Resistance. Front. Microbiol. 2017, 8, 908. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Ku, S.; Kim, S.Y.; Lee, H.H.; Jin, H.; Kang, S.; Li, R.; Johnston, T.V.; Park, M.S.; Ji, G.E. Safety Evaluations of Bifidobacterium bifidum BGN4 and Bifidobacterium longum BORI. Int. J. Mol. Sci. 2018, 19, 1422. [Google Scholar] [CrossRef] [PubMed]
- Rosander, A.; Connolly, E.; Roos, S. Removal of Antibiotic Resistance Gene-Carrying Plasmids from Lactobacillus reuteri ATCC 55730 and Characterization of the Resulting Daughter Strain, L. reuteri DSM 17938. Appl. Environ. Microbiol. 2008, 74, 6032–6040. [Google Scholar] [CrossRef]
- Sanders, M.E.; Guarner, F.; Guerrant, R.; Holt, P.R.; Quigley, E.M.; Sartor, R.B.; Sherman, P.M.; Mayer, E.A. An update on the use and investigation of probiotics in health and disease. Gut 2013, 62, 787–796. [Google Scholar] [CrossRef]
- Bazireh, H.; Shariati, P.; Azimzadeh Jamalkandi, S.; Ahmadi, A.; Boroumand, M.A. Isolation of Novel Probiotic Lactobacillus and Enterococcus Strains From Human Salivary and Fecal Sources. Front. Microbiol. 2020, 11, 597946. [Google Scholar] [CrossRef]
- Fijan, S.; Kocbek, P.; Steyer, A.; Vodičar, P.M.; Strauss, M. The Antimicrobial Effect of Various Single-Strain and Multi-Strain Probiotics, Dietary Supplements or Other Beneficial Microbes against Common Clinical Wound Pathogens. Microorganisms 2022, 10, 2518. [Google Scholar] [CrossRef]
- Gwee, K.A.; Lee, W.W.; Ling, K.L.; Ooi, C.J.; Quak, S.H.; Dan, Y.Y.; Siah, K.T.; Huang, J.G.; Chua, A.S.B.; Hilmi, I.N.; et al. Consensus and contentious statements on the use of probiotics in clinical practice: A south east Asian gastro-neuro motility association working team report. J. Gastroenterol. Hepatol. 2018, 33, 1707–1716. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mousa, W.K.; Mousa, S.; Ghemrawi, R.; Obaid, D.; Sarfraz, M.; Chehadeh, F.; Husband, S. Probiotics Modulate Host Immune Response and Interact with the Gut Microbiota: Shaping Their Composition and Mediating Antibiotic Resistance. Int. J. Mol. Sci. 2023, 24, 13783. https://doi.org/10.3390/ijms241813783
Mousa WK, Mousa S, Ghemrawi R, Obaid D, Sarfraz M, Chehadeh F, Husband S. Probiotics Modulate Host Immune Response and Interact with the Gut Microbiota: Shaping Their Composition and Mediating Antibiotic Resistance. International Journal of Molecular Sciences. 2023; 24(18):13783. https://doi.org/10.3390/ijms241813783
Chicago/Turabian StyleMousa, Walaa K., Sara Mousa, Rose Ghemrawi, Dana Obaid, Muhammad Sarfraz, Fadia Chehadeh, and Shannon Husband. 2023. "Probiotics Modulate Host Immune Response and Interact with the Gut Microbiota: Shaping Their Composition and Mediating Antibiotic Resistance" International Journal of Molecular Sciences 24, no. 18: 13783. https://doi.org/10.3390/ijms241813783




