Long Noncoding RNA ZBED5-AS1 Facilitates Tumor Progression and Metastasis in Lung Adenocarcinoma via ZNF146/ATR/Chk1 Axis
Abstract
:1. Introduction
2. Results
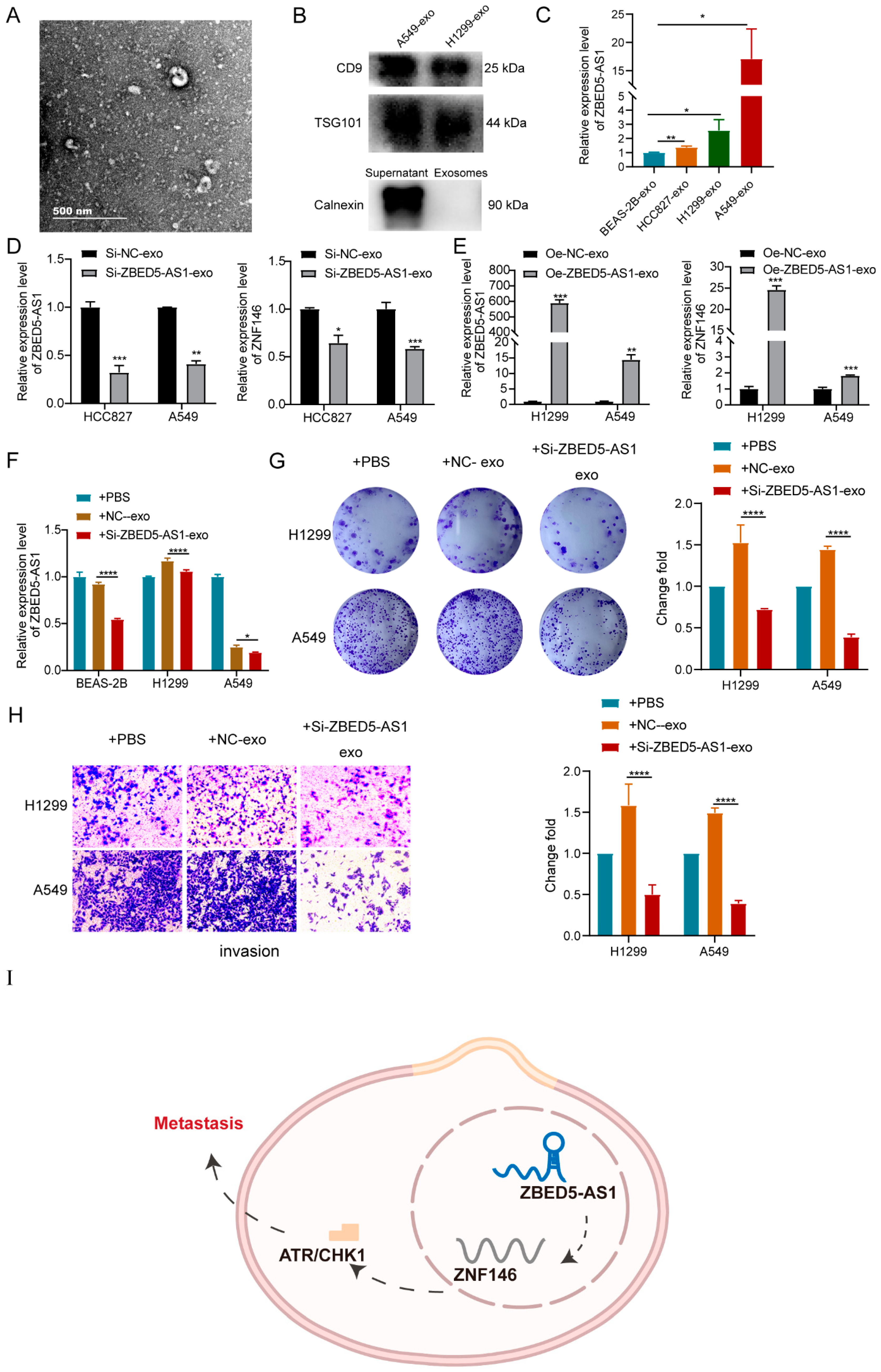
2.1. ZBED5-AS1 Is Highly Expressed in LUAD
2.2. ZBED5-AS1 Promotes Malignant Phenotypes of LUAD Cells
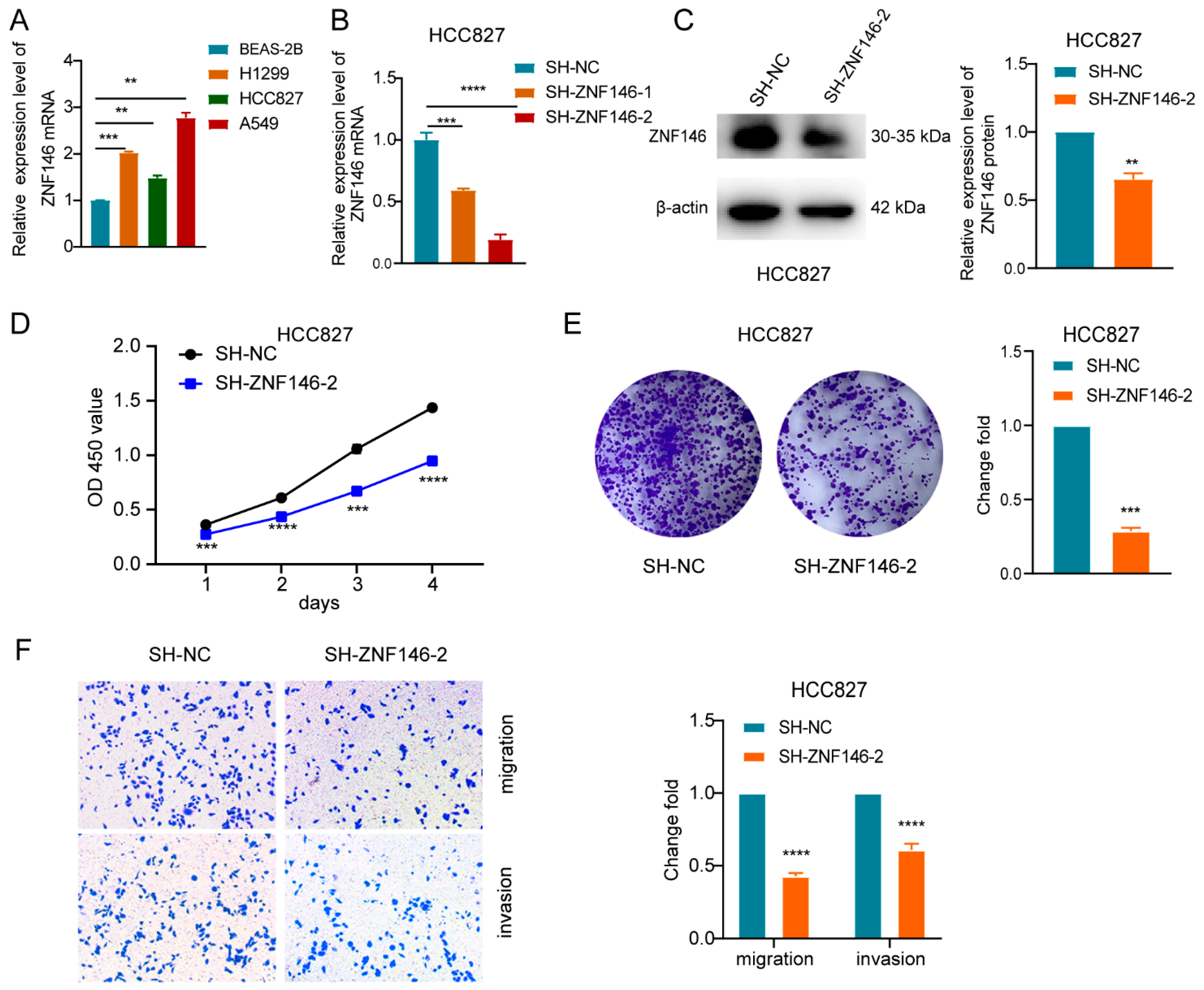
2.3. ZNF146 Enhances the Malignant Phenotypes of LUAD Cells
2.4. ZNF146 Knockdown Inhibits LUAD Cell Proliferation and Invasion Mediated by ZBED5-AS1
2.5. ZBED5-AS1 Silencing Suppresses the Activation of ATR/Chk1 Signaling Pathway
2.6. Si-ZBED5-AS1 Exosomes Inhibite LUAD Cells Proliferation and Metastasis
3. Discussion
4. Materials and Methods
4.1. Patient Samples
4.2. Animal Experiments
4.3. Cell Culture
4.4. Cell Transfection
4.5. Subcellular Fractionation
4.6. Exosome Isolation and Identification
4.7. RNA Extraction and RT-qPCR
4.8. Exosome Treatment
4.9. Cell Proliferation Assay
4.10. Transwell Assay
4.11. Flow Cytometry
4.12. Western Blot
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Denisenko, T.V.; Budkevich, I.N.; Zhivotovsky, B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018, 9, 117. [Google Scholar] [CrossRef]
- Tian, H.; Lian, R.; Li, Y.; Liu, C.; Liang, S.; Li, W.; Tao, T.; Wu, X.; Ye, Y.; Yang, X.; et al. AKT-induced lncRNA VAL promotes EMT-independent metastasis through diminishing Trim16-dependent Vimentin degradation. Nat. Commun. 2020, 11, 5127. [Google Scholar] [CrossRef]
- Shi, Y.; Xu, Y.; Yao, J.; Yan, C.; Su, H.; Zhang, X.; Chen, E.; Ying, K. MTHFD2 promotes tumorigenesis and metastasis in lung adenocarcinoma by regulating AKT/GSK-3β/β-catenin signalling. J. Cell. Mol. Med. 2021, 25, 7013–7027. [Google Scholar] [CrossRef] [PubMed]
- Jathar, S.; Kumar, V.; Srivastava, J.; Tripathi, V. Technological Developments in lncRNA Biology. Adv. Exp. Med. Biol. 2017, 1008, 283–323. [Google Scholar]
- Charles Richard, J.L.; Eichhorn, P.J.A. Platforms for Investigating LncRNA Functions. SLAS Technol. 2018, 23, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhou, F.; Ren, W.; Guo, J.; Huang, X.; Pu, J.; Niu, X.; Jiang, X. LncRNA-AC02278.4 Is a Novel Prognostic Biomarker That Promotes Tumor Growth and Metastasis in Lung Adenocarcinoma. Front. Oncol. 2022, 12, 860961. [Google Scholar] [CrossRef]
- Li, N.; Zhan, X. Identification of clinical trait-related lncRNA and mRNA biomarkers with weighted gene co-expression network analysis as useful tool for personalized medicine in ovarian cancer. EPMA J. 2019, 10, 273–290. [Google Scholar] [CrossRef]
- Huang, X.; Zhou, H.; Yang, X.; Shi, W.; Hu, L.; Wang, J.; Zhang, F.; Shao, F.; Zhang, M.; Jiang, F.; et al. Construction and analysis of expression profile of exosomal lncRNAs in pleural effusion in lung adenocarcinoma. J. Clin. Lab. Anal. 2022, 36, e24777. [Google Scholar] [CrossRef]
- Ma, Y.; Cong, X.; Zhang, Y.; Yin, X.; Zhu, Z.; Xue, Y. CircPIP5K1A facilitates gastric cancer progression via miR-376c-3p/ZNF146 axis. Cancer Cell Int. 2020, 20, 81. [Google Scholar] [CrossRef]
- Ferbus, D.; Bovin, C.; Validire, P.; Goubin, G. The zinc finger protein OZF (ZNF146) is overexpressed in colorectal cancer. J. Pathol. 2003, 200, 177–182. [Google Scholar] [CrossRef]
- Fernandez, P.C.; Frank, S.R.; Wang, L.; Schroeder, M.; Liu, S.; Greene, J.; Cocito, A.; Amati, B. Genomic targets of the human c-Myc protein. Genes Dev. 2003, 17, 1115–1129. [Google Scholar] [CrossRef]
- Byers, L.A.; Wang, J.; Nilsson, M.B.; Fujimoto, J.; Saintigny, P.; Yordy, J.; Giri, U.; Peyton, M.; Fan, Y.H.; Diao, L.; et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2012, 2, 798–811. [Google Scholar] [CrossRef]
- Manic, G.; Obrist, F.; Sistigu, A.; Vitale, I. Trial Watch: Targeting ATM-CHK2 and ATR-CHK1 pathways for anticancer therapy. Mol. Cell. Oncol. 2015, 2, e1012976. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Tho, L.M.; Xu, N.; Gillespie, D.A. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 2010, 108, 73–112. [Google Scholar]
- Wang, W.Y.; Cao, Y.X.; Zhou, X.; Wei, B.; Zhan, L.; Fu, L.T. HMGA2 gene silencing reduces epithelial-mesenchymal transition and lymph node metastasis in cervical cancer through inhibiting the ATR/Chk1 signaling pathway. Am. J. Transl. Res. 2018, 10, 3036–3052. [Google Scholar] [PubMed]
- Jiayu, H.; Hanke, Z.; Ying, G. The Role of Exosomes in Diseases Related to Infertility. Curr. Stem Cell Res. Ther. 2019, 14, 437–441. [Google Scholar] [CrossRef]
- Qiu, J.J.; Lin, X.J.; Tang, X.Y.; Zheng, T.T.; Lin, Y.Y.; Hua, K.Q. Exosomal Metastasis-Associated Lung Adenocarcinoma Transcript 1 Promotes Angiogenesis and Predicts Poor Prognosis in Epithelial Ovarian Cancer. Int. J. Biol. Sci. 2018, 14, 1960–1973. [Google Scholar] [CrossRef]
- Chen, J.; Cheuk, I.W.; Siu, M.T.; Yang, W.; Cheng, A.S.; Shin, V.Y.; Kwong, A. Human haptoglobin contributes to breast cancer oncogenesis through glycolytic activity modulation. Am. J. Cancer Res. 2020, 10, 2865–2877. [Google Scholar]
- Tan, Z.; Zhang, M.; Han, Q.; Wen, J.; Luo, K.; Lin, P.; Zhang, L.; Yang, H.; Fu, J. A novel blood tool of cancer prognosis in esophageal squamous cell carcinoma: The Fibrinogen/Albumin Ratio. J. Cancer 2017, 8, 1025–1029. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Hu, W.; Xue, X.; Fu, W.; Dai, L.; Jiang, Z.; Zhong, S.; Deng, B.; Zhao, J. Epigenetic activation of hepatocyte growth factor is associated with epithelial-mesenchymal transition and clinical outcome in non-small cell lung cancer. J. Cancer 2019, 10, 5070–5081. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhou, D.; Chen, F.; Yang, Z.; Gu, W.; Zhang, K. SIX5-activated LINC01468 promotes lung adenocarcinoma progression by recruiting SERBP1 to regulate SERPINE1 mRNA stability and recruiting USP5 to facilitate PAI1 protein deubiquitylation. Cell Death Dis. 2022, 13, 312. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Han, C.; Liu, T.; Ma, Z.; Qiu, M.; Wang, J.; You, Q.; Zheng, X.; Xu, W.; Xia, W.; et al. FAM83H-AS1 is a noncoding oncogenic driver and therapeutic target of lung adenocarcinoma. Clin. Transl. Med. 2021, 11, e316. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, S.Q.; Wang, L.; Lin, H.; Zhu, J.B.; Chen, R.; Li, L.F.; Cheng, Y.D.; Duan, C.J.; Zhang, C.F. m6A methyltransferase METTL3-induced lncRNA SNHG17 promotes lung adenocarcinoma gefitinib resistance by epigenetically repressing LATS2 expression. Cell Death Dis. 2022, 13, 657. [Google Scholar] [CrossRef]
- Zhuang, D.; Liang, L.; Zhang, H.; Feng, X. miR-202 Suppresses Hepatocellular Carcinoma Progression via Downregulating BCL2 Expression. Oncol. Res. 2020, 28, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Xiong, Y.; Qiao, T.; Li, X.; Jia, L.; Han, Y. Lactate dehydrogenase A: A key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 2018, 7, 6124–6136. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhu, Y.; Liu, L. Elevated pretreatment plasma D-dimer levels and platelet counts predict poor prognosis in pancreatic adenocarcinoma. OncoTargets Ther. 2015, 8, 1335–1340. [Google Scholar] [CrossRef]
- Bao, L.; Wang, M.; Fan, Q. Hsa_circ_NOTCH3 regulates ZNF146 through sponge adsorption of miR-875-5p to promote tumorigenesis of hepatocellular carcinoma. J. Gastrointest. Oncol. 2021, 12, 2388–2402. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Chen, C.Y.; Hao, Y. LncRNA KCNQ1OT1 acts as miR-216b-5p sponge to promote colorectal cancer progression via up-regulating ZNF146. J. Mol. Histol. 2021, 52, 479–490. [Google Scholar] [CrossRef]
- Block, W.D.; Yu, Y.; Lees-Miller, S.P. Phosphatidyl inositol 3-kinase-like serine/threonine protein kinases (PIKKs) are required for DNA damage-induced phosphorylation of the 32 kDa subunit of replication protein A at threonine 21. Nucleic Acids Res. 2004, 32, 997–1005. [Google Scholar] [CrossRef]
- Zou, L. Single- and double-stranded DNA: Building a trigger of ATR-mediated DNA damage response. Genes Dev. 2007, 21, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.R.; Yang, Z.Z.; Wang, S.J.; Zhang, L.; Luo, J.R.; Feng, Y.; Yu, X.L.; Chen, X.X.; Guo, X.M. The Chk1 inhibitor MK-8776 increases the radiosensitivity of human triple-negative breast cancer by inhibiting autophagy. Acta Pharmacol. Sin. 2017, 38, 513–523. [Google Scholar] [CrossRef]
- Suzuki, M.; Yamamori, T.; Bo, T.; Sakai, Y.; Inanami, O. MK-8776, a novel Chk1 inhibitor, exhibits an improved radiosensitizing effect compared to UCN-01 by exacerbating radiation-induced aberrant mitosis. Transl. Oncol. 2017, 10, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Doerr, F.; George, J.; Schmitt, A.; Beleggia, F.; Rehkämper, T.; Hermann, S.; Walter, V.; Weber, J.P.; Thomas, R.K.; Wittersheim, M.; et al. Targeting a non-oncogene addiction to the ATR/CHK1 axis for the treatment of small cell lung cancer. Sci. Rep. 2017, 7, 15511. [Google Scholar] [CrossRef] [PubMed]
- Vanni, I.; Alama, A.; Grossi, F.; Dal Bello, M.G.; Coco, S. Exosomes: A new horizon in lung cancer. Drug Discov. Today 2017, 22, 927–936. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Pathania, A.S.; Challagundla, K.B. Exosomal Long Non-coding RNAs: Emerging Players in the Tumor Microenvironment. Mol. Ther. Nucleic Acids 2021, 23, 1371–1383. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhao, H.; Chen, Y.; Li, K.; Li, T.; Chen, J.; Zhang, B.; Guo, C.; Qing, L.; Shen, J.; et al. Exosomal long noncoding RNA HOXD-AS1 promotes prostate cancer metastasis via miR-361-5p/FOXM1 axis. Cell Death Dis. 2021, 12, 1129. [Google Scholar] [CrossRef]
- Li, Y.; Yin, Z.; Fan, J.; Zhang, S.; Yang, W. The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal Transduct. Target. Ther. 2019, 4, 47. [Google Scholar] [CrossRef]
- Lee, S.H.; Oh, H.J.; Kim, M.J.; Lee, B.C. Exosomes derived from oviduct cells mediate the EGFR/MAPK signaling pathway in cumulus cells. J. Cell. Physiol. 2020, 235, 1386–1404. [Google Scholar] [CrossRef]
- Xu, H.; Ling, M.; Xue, J.; Dai, X.; Sun, Q.; Chen, C.; Liu, Y.; Zhou, L.; Liu, J.; Luo, F.; et al. Exosomal microRNA-21 derived from bronchial epithelial cells is involved in aberrant epithelium-fibroblast cross-talk in COPD induced by cigarette smoking. Theranostics 2018, 8, 5419–5433. [Google Scholar] [CrossRef] [PubMed]




| Characteristics | Low Expression | High Expression | p Value |
|---|---|---|---|
| n | 28 | 28 | |
| gender, n (%) | 1.0000 | ||
| female | 15 (26.8%) | 15 (26.8%) | |
| male | 13 (23.2%) | 13 (23.2%) | |
| age, n (%) | 0.5842 | ||
| ≤65 | 18 (32.1%) | 16 (28.6%) | |
| >65 | 10 (17.9%) | 12 (21.4%) | |
| T, n (%) | 0.3881 | ||
| T1 | 22 (39.3%) | 21 (37.5%) | |
| T2 | 4 (7.1%) | 5 (8.9%) | |
| T4 | 2 (3.6%) | 0 (0%) | |
| T3 | 0 (0%) | 1 (1.8%) | |
| X | 0 (0%) | 1 (1.8%) | |
| N.stage, n (%) | 0.0860 | ||
| N0 | 22 (39.3%) | 26 (46.4%) | |
| N1 | 6 (10.7%) | 1 (1.8%) | |
| X | 0 (0%) | 1 (1.8%) | |
| M.stage, n (%) | 0.9999 | ||
| M0 | 27 (48.2%) | 27 (48.2%) | |
| M1 | 1 (1.8%) | 0 (0%) | |
| X | 0 (0%) | 1 (1.8%) | |
| Tumor size, n (%) | 0.1764 | ||
| >2 | 9 (16.1%) | 14 (25%) | |
| ≤2 | 19 (33.9%) | 13 (23.2%) | |
| X | 0 (0%) | 1 (1.8%) | |
| CEA (μg/L), median (IQR) | 2.4 (1.35, 4.75) | 2.45 (1.9, 6.3) | 0.4775 |
| NSE (ng/mL), median (IQR) | 12.6 (9.95, 19.45) | 13.1 (11, 16.2) | 0.6602 |
| ProGRP (ng/L), mean ± sd | 48.708 ± 14.977 | 51.05 ± 23.442 | 0.7021 |
| D-dimer, median (IQR) | 0.345 (0.3, 0.72) | 0.91 (0.37, 1.06) | 0.0245 |
| LDH (U/L), median (IQR) | 178 (162, 189.5) | 187 (178.5, 236) | 0.0480 |
| PLT counts, median (IQR) | 231.5 (185.5, 276.5) | 253 (222.5, 294) | 0.2158 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, F.; Huang, X.; Ling, L.; Tang, S.; Zhou, H.; Cai, X.; Wang, Y. Long Noncoding RNA ZBED5-AS1 Facilitates Tumor Progression and Metastasis in Lung Adenocarcinoma via ZNF146/ATR/Chk1 Axis. Int. J. Mol. Sci. 2023, 24, 13925. https://doi.org/10.3390/ijms241813925
Jiang F, Huang X, Ling L, Tang S, Zhou H, Cai X, Wang Y. Long Noncoding RNA ZBED5-AS1 Facilitates Tumor Progression and Metastasis in Lung Adenocarcinoma via ZNF146/ATR/Chk1 Axis. International Journal of Molecular Sciences. 2023; 24(18):13925. https://doi.org/10.3390/ijms241813925
Chicago/Turabian StyleJiang, Feng, Xiaolu Huang, Liqun Ling, Shiyi Tang, Huixin Zhou, Xueding Cai, and Yumin Wang. 2023. "Long Noncoding RNA ZBED5-AS1 Facilitates Tumor Progression and Metastasis in Lung Adenocarcinoma via ZNF146/ATR/Chk1 Axis" International Journal of Molecular Sciences 24, no. 18: 13925. https://doi.org/10.3390/ijms241813925





