Clusterin Expression in Colorectal Carcinomas
Abstract
:1. Introduction
Clusterin and Its Implication in Cancer
2. Objectives and Methods
3. Colorectal Cancer
- Surgery
- Radiotherapy
- Chemotherapy
- Targeted treatments
3.1. Monoclonal Antibodies
3.2. Immune Checkpoint Inhibitors
4. Clusterin as a Therapeutic Target in Colorectal Cancer
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hogg, S.D.; Embery, G. The isolation and partial characterization of a sulphated glycoprotein from human whole saliva which aggregates strains of Streptococcus sanguis but not Streptococcus mutans. Arch. Oral Biol. 1979, 24, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Trougakos, I.P.; Gonos, E.S. Clusterin/apolipoprotein J in human aging and cancer. Int. J. Biochem. Cell Biol. 2002, 34, 1430–1448. [Google Scholar] [CrossRef] [PubMed]
- Pucci, S.; Bettuzzi, S. Chapter 3: The shifting balance between CLU forms during tumor progression. Adv. Cancer Res. 2009, 104, 25–32. [Google Scholar] [PubMed]
- Yu, J.-T.; Tan, L. The role of clusterin in Alzheimer’s disease: Pathways, pathogenesis, and therapy. Mol. Neurobiol. 2012, 45, 314–326. [Google Scholar] [CrossRef] [PubMed]
- Poon, S.; Easterbrook-Smith, S.B.; Rybchyn, M.S.; Carver, J.A.; Wilson, M.R. Clusterin is an ATP-independent chaperone with very broad substrate specificity that stabilizes stressed proteins in a folding-competent state. Biochemistry 2000, 39, 15953–15960. [Google Scholar] [CrossRef]
- Zoubeidi, A.; Martin, G. Small heat shock proteins in cancer therapy and prognosis. Int. J. Biochem. Cell Biol. 2012, 44, 1646–1656. [Google Scholar] [CrossRef]
- Herring, S.K.; Moon, H.-L.; Rawal, P.; Chhibber, A.; Zhao, L. Brain clusterin protein isoforms and mitochondrial localization. eLife 2019, 8, e48255. [Google Scholar] [CrossRef]
- Foster, E.M.; Dangla-Valls, A.; Lovestone, S.; Ribe, E.M.; Buckley, N.J. Clusterin in Alzheimer’s Disease: Mechanisms, Genetics, and Lessons from Other Pathologies. Front. Neurosci. 2019, 13, 164. [Google Scholar] [CrossRef]
- Trougakos, I.P.; Gonos, E.S. Regulation of clusterin/apolipoprotein J, a functional homologue to the small heat shock proteins, by oxidative stress in aging and age-related diseases. Free. Radic. Res. 2006, 40, 1324–1334. [Google Scholar] [CrossRef]
- Bettuzzi, S.; Scorcioni, F.; Astancolle, S.; Davalli, P.; Scaltriti, M.; Corti, A. Clusterin (SGP-2) transient overexpression decreases proliferation rate of SV40-immortalized human prostate epithelial cells by slowing down cell cycle progression. Oncogene 2002, 21, 4328–4334. [Google Scholar] [CrossRef]
- Pucci, S.; Bonanno, E.; Pichiorri, F.; Angeloni, C.; Spagnoli, L.G. Modulation of different clusterin isoforms in human colon tumorigenesis. Oncogene 2004, 23, 2298–2304. [Google Scholar] [CrossRef] [PubMed]
- Caccamo, A.E.; Scaltriti, M.; Caporali, A.; D’Arca, D.; Corti, A.; Corvetta, D.; Sala, A.; Bettuzzi, S. Ca2+ depletion induces nuclear clusterin, a novel effector of apoptosis in immortalized human prostate cells. Cell Death Differ. 2005, 12, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Caccamo, A.E.; Scaltriti, M.; Caporali, A.; D’Arca, D.; Scorcioni, F.; Astancolle, S.; Mangiola, M.; Bettuzzi, S. Cell detachment and apoptosis induction of immortalized human prostate epithelial cells are associated with early accumulation of a 45 kDa nuclear isoform of clusterin. Biochem. J. 2004, 382, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Leskov, K.S.; Klokov, D.Y.; Li, J.; Kinsella, T.J.; Boothman, D.A. Synthesis and functional analyses of nuclear clusterin, a cell death protein. J. Biol. Chem. 2003, 278, 11590–11600. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zoubeidi, A.; Beraldi, E.; Gleave, M.E. GRP78 regulates clusterin stability, retrotranslocation and mitochondrial localization under ER stress in prostate cancer. Oncogene 2013, 32, 1933–1942. [Google Scholar] [CrossRef]
- Nizard, P.; Tetley, S.; Le Dréan, Y.; Watrin, T.; Le Goff, P.; Wilson, M.R.; Michel, D. Stress-Induced Retrotranslocation of Clusterin/ApoJ into the Cytosol. Traffic 2007, 8, 554–565. [Google Scholar] [CrossRef]
- Trougakos, I.P. The Molecular Chaperone Apolipoprotein J/Clusterin as a Sensor of Oxidative Stress: Implications in Therapeutic Approaches—A Mini-Review. Gerontology 2013, 59, 514–523. [Google Scholar] [CrossRef]
- July, L.V.; Akbari, M.; Zellweger, T.; Jones, E.C.; Goldenberg, S.L.; Gleave, M.E. Clusterin expression is significantly enhanced in prostate cancer cells following androgen withdrawal therapy. Prostate 2002, 50, 179–188. [Google Scholar] [CrossRef]
- Scaltriti, M.; Bettuzzi, S.; Sharrard, R.M.; Caporali, A.; Caccamo, A.E.; Maitland, N.J. Clusterin overexpression in both malignant and nonmalignant prostate epithelial cells induces cell cycle arrest and apoptosis. Br. J. Cancer 2004, 91, 1842–1850. [Google Scholar] [CrossRef]
- Scaltriti, M.; Santamaria, A.; Paciucci, R.; Bettuzzi, S. Intracellular clusterin induces G2-M phase arrest and cell death in PC-3 prostate cancer cells1. Cancer Res. 2004, 64, 6174–6182. [Google Scholar] [CrossRef]
- Collard, M.W.; Griswold, M.D. Biosynthesis, and molecular cloning of sulfated glycoprotein 2 secreted by rat Sertoli cells. Biochemistry 1987, 26, 3297–3303. [Google Scholar] [CrossRef] [PubMed]
- Danik, M.; Chabot, J.G.; Hassan-Gonzalez, D.; Suh, M.; Quirion, R. Localization of sulfated glycoprotein-2/clusterin mRNA in the rat brain by in situ hybridization. J. Comp. Neurol. 1993, 334, 209–227. [Google Scholar] [CrossRef] [PubMed]
- Scaltriti, M.; Brausi, M.; Amorosi, A.; Caporali, A.; D’Arca, D.; Astancolle, S.; Corti, A.; Bettuzzi, S. Clusterin (SGP-2, ApoJ) expression is downregulated in low- and high-grade human prostate cancer. Int. J. Cancer 2004, 108, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Aronow, B.J.; Lund, S.D.; Brown, T.L.; Harmony, J.A.; Witte, D.P. Apolipoprotein J expression at fluid-tissue interfaces: Potential role in barrier cytoprotection. Proc. Natl. Acad. Sci. USA 1993, 90, 725–729. [Google Scholar] [CrossRef]
- Rizzi, F.; Coletta, M.; Bettuzzi, S. Clusterin (CLU): From one gene and two transcripts to many proteins. Adv. Cancer Res. 2009, 104, 9–23. [Google Scholar]
- Park, S.; Mathis, K.W.; Lee, I.K. The physiological roles of apolipoprotein J/clusterin in metabolic and cardiovascular diseases. Rev. Endocr. Metab. Disord. 2014, 15, 45–53. [Google Scholar] [CrossRef]
- Zoubeidi, A.; Chi, K.; Gleave, M. Targeting the cytoprotective chaperone, clusterin, for treatment of advanced cancer. Clin. Cancer Res. 2010, 16, 1088–1093. [Google Scholar] [CrossRef]
- Goetz, E.M.; Shankar, B.; Zou, Y.; Morales, J.C.; Luo, X.; Araki, S.; Bachoo, R.; Mayo, L.D.; Boothman, D.A. ATM dependent IGF-1 induction regulates secretory clusterin expression after DNA damage and in genetic instability. Oncogene. Nat. Publ. Group 2011, 30, 3745–3754. [Google Scholar]
- Prochnow, H.; Gollan, R.; Rohne, P.; Hassemer, M.; Koch-Brandt, C.; Baiersdörfer, M. Non-Secreted Clusterin Isoforms Are Translated in Rare Amounts from Distinct Human mRNA Variants and Do Not Affect BaxMediated Apoptosis or the NF-κB Signaling Pathway. PLoS ONE 2013, 8, e75303. [Google Scholar] [CrossRef]
- Michel, D.; Chatelain, G.; North, S.; Brun, G. Stress-induced transcription of the clusterin/apoJ gene. Biochem. J. 1997, 50, 45–50. [Google Scholar] [CrossRef]
- Serrano, A.; Redondo, M.; Tellez, T.; Castro-Vega, I.; Roldan, M.J.; Mendez, R.; Rueda, A.; Jiménez, E. Regulation of clusterin expression in human cancer via DNA methylation. Tumour Biol. 2009, 30, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Deb, M.; Sengupta, D.; Rath, S.K.; Kar, S.; Parbin, S.; Shilpi, A.; Pradhan, N.; Bhutia, S.K.; Roy, S.; Patra, S.K. Clusterin gene is predominantly regulated by histone modifications in human colon cancer and ectopic expression of the nuclear isoform induces cell death. Biochim. Biophys. Acta 2015, 1852, 1630–1645. [Google Scholar] [CrossRef] [PubMed]
- Rosemblit, N.; Chen, C.L. Regulators for the rat clusterin gene: DNA methylation and cis-acting regulatory elements. J. Mol. Endocrinol. 1994, 13, 69–76. [Google Scholar] [CrossRef] [PubMed]
- July, L.V.; Beraldi, E.; So, A.; Fazli, L.; Evans, K.; English, J.C.; Gleave, M.E. Nucleotide-based therapies targeting clusterin chemosensitize human lung adenocarcinoma cells both in vitro and in vivo. Mol. Cancer Ther. 2004, 3, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.; Guo, A.L.; Lai, Y.R.; Li, B.; Zhong, J.M.; Wu, H.Q.; Xie, Z.; He, Y.L.; Lv, Z.L.; Lau, S.H.; et al. Overexpression of clusterin correlates with tumor progression, metastasis in gastric cancer: A study on tissue microarrays. Neoplasm 2010, 57, 191. [Google Scholar] [CrossRef]
- Xie, D.; Lau, S.H.; Sham, J.S.; Wu, Q.L.; Fang, Y.; Liang, L.Z.; Che, L.H.; Zeng, Y.X.; Guan, X.Y. Up-regulated expression of cytoplasmic clusterin in human ovarian carcinoma. Cancer 2005, 103, 277–283. [Google Scholar] [CrossRef]
- Redondo, M.; Villar, E.; Torres-Muñoz, J.; Tellez, T.; Morell, M.; Petito, C.K. Overexpression of clusterin in human breast carcinoma. Am. J. Pathol. 2000, 157, 393–439. [Google Scholar] [CrossRef]
- Redondo, M.; Tellez, T.; Roldan, M.J. The role of clusterin (CLU) in malignant transformation and drug resistance in breast carcinomas. Adv. Cancer Res. 2009, 105, 21–43. [Google Scholar]
- Mazzarelli, P.; Pucci, S.; Spagnoli, L.G. CLU and colon cancer. The dual face of CLU: From normal to malignant phenotype. Adv. Cancer Res. 2009, 105, 45–61. [Google Scholar]
- Redondo, M.; Rodrigo, I.; Alcaide, J.; Tellez, T.; Roldan, M.J.; Funez, R.; Diaz-Martin, A.; Rueda, A.; Jiménez, E. Clusterin expression is associated with decreased disease-free survival of patients with colorectal carcinomas. Histopathology 2010, 56, 932–936. [Google Scholar] [CrossRef]
- Miyake, H.; Gleave, M.; Kamidono, S.; Hara, I. Overexpression of clusterin in transitional cell carcinoma of the bladder is related to disease progression and recurrence. Urology 2002, 59, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Patarat, R.; Riku, S.; Kunadirek, P.; Chuaypen, N.; Tangkijvanich, P.; Mutirangura, A.; Puttipanyalears, C. The expression of FLNA and CLU in PBMCs as a novel screening marker for hepatocellular carcinoma. Sci. Rep. 2021, 11, 14838. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.J.; Motoo, Y.; Su, S.B.; Mouri, H.; Ohtsubo, K.; Matsubara, F.; Sawabu, N. Expression of clusterin in human pancreatic cancer. Pancreas 2002, 25, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Hoeller, C.; Pratscher, B.; Thallinger, C.; Winter, D.; Fink, D.; Kovacic, B.; Sexl, V.; Wacheck, V.; Gleave, M.E.; Pehamberger, H.; et al. Clusterin regulates drug-resistance in melanoma cells. J. Investig. Dermatol. 2005, 124, 1300–1307. [Google Scholar] [CrossRef] [PubMed]
- He, L.R.; Liu, M.Z.; Li, B.K.; Rao, H.L.; Liao, Y.J.; Zhang, L.J.; Guan, X.Y.; Zeng, Y.X.; Xie, D. Clusterin as a predictor for chemoradiotherapy sensitivity and patient survival in esophageal squamous cell carcinoma. Cancer Sci. 2009, 100, 2354–2360. [Google Scholar] [CrossRef]
- Wellmann, A.; Thieblemont, C.; Pittaluga, S.; Sakai, A.; Jaffe, E.S.; Siebert, P.; Raffeld, M. Detection of differentially expressed genes in lymphomas using cDNA arrays: Identification of clusterin as a new diagnostic marker for anaplastic large-cell lymphomas. Blood 2000, 96, 398–404. [Google Scholar] [CrossRef]
- Ma, J.; Gao, W.; Gao, J. sCLU as prognostic biomarker and therapeutic target in osteosarcoma. Bioengineered 2019, 10, 229–239. [Google Scholar] [CrossRef]
- Rizzi, F.; Bettuzzi, S. Clusterin (CLU) and prostate cancer. Adv. Cancer Res. 2009, 105, 1–19. [Google Scholar]
- Praharaj, P.P.; Patra, S.; Panigrahi, D.P.; Patra, S.K.; Bhutia, S.K. Clusterin as modulator of carcinogenesis: A potential avenue for targeted cancer therapy. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188500. [Google Scholar] [CrossRef]
- Liu, X.; Su, K.; Sun, X.; Jiang, Y.; Wang, L.; Hu, C.; Zhang, C.; Lu, M.; Du, X.; Xing, B. Sec62 promotes stemness and chemoresistance of human colorectal cancer through activating Wnt/β-catenin pathway. J. Exp. Clin. Cancer Res. CR 2021, 40, 132. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Nogueira, L.; Devasia, T.; Mariotto, A.B.; Yabroff, K.R.; Jemal, A.; Kramer, J.; Siegel, R.L. Cancer treatment and survivorship statistics, 2022. CA Cancer J. Clin. 2022, 72, 409–436. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Zarcos-Pedrinaci, I.; Fernández-López, A.; Téllez, T.; Rivas-Ruiz, F.; Rueda, A.; Morales Suarez-Varela, M.M.; Briones, E.; Baré, M.; Escobar, A.; Sarasqueta, C.; et al. CARESS-CCR Study Group. Factors that influence treatment delay in patients with colorectal cancer. Oncotarget 2017, 8, 36728–36742. [Google Scholar] [CrossRef] [PubMed]
- Padilla-Ruiz, M.; Morales-Suárez-Varela, M.; Rivas-Ruiz, F.; Alcaide, J.; Varela-Moreno, E.; Zarcos-Pedrinaci, I.; Téllez, T.; Fernández-de Larrea-Baz, N.; Baré, M.; Bilbao, A.; et al. On Behalf of Caress–Ccr Study Group. Influence of Diagnostic Delay on Survival Rates for Patients with Colorectal Cancer. Int. J. Environ. Res. Public Health 2022, 19, 3626. [Google Scholar] [CrossRef]
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef]
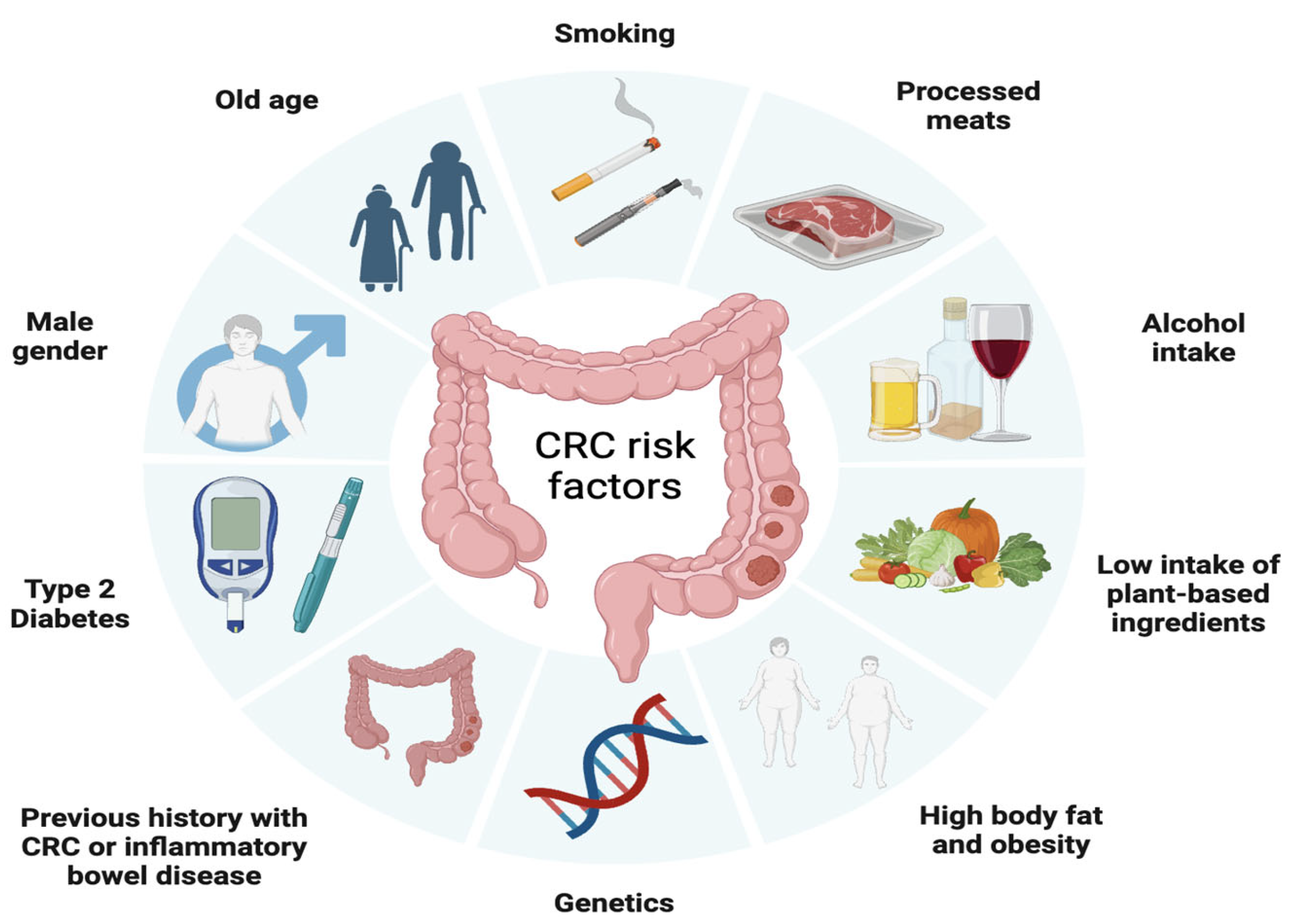
- Sninsky, J.A.; Shore, B.M.; Lupu, G.V.; Crockett, S.D. Risk Factors for Colorectal Polyps and Cancer. Gastrointest. Endosc. Clin. N. Am. 2022, 32, 195–213. [Google Scholar] [CrossRef]
- Li, J.; Ma, X.; Chakravarti, D.; Shalapour, S.; DePinho, R.A. Genetic and biological hallmarks of colorectal cancer. Genes Dev. 2021, 35, 787–820. [Google Scholar] [CrossRef]
- Shah, S.C.; Itzkowitz, S.H. Colorectal Cancer in Inflammatory Bowel Disease: Mechanisms and Management. Gastroenterology 2022, 162, 715–730.e3. [Google Scholar] [CrossRef]
- Ciardiello, F.; Ciardiello, D.; Martini, G.; Napolitano, S.; Tabernero, J.; Cervantes, A. Clinical management of metastatic colorectal cancer in the era of precision medicine. CA Cancer J. Clin. 2022, 72, 372–401. [Google Scholar] [CrossRef]
- Sullivan, B.A.; Noujaim, M.; Roper, J. Cause, Epidemiology, and Histology of Polyps and Pathways to Colorectal Cancer. Gastrointest. Endosc. Clin. N. Am. 2022, 32, 177–194. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.L.; Dawson, S.N.; Arends, M.J.; Guttula, K.; Hall, N.; Cameron, E.A.; Huang, T.H.; Brenton, J.D.; Tavaré, S.; Bienz, M.; et al. Boosting Wnt activity during colorectal cancer progression through selective hypermethylation of Wnt signaling antagonists. BMC Cancer 2014, 14, 891. [Google Scholar] [CrossRef] [PubMed]
- Jass, J.R. Classification of colorectal cancer based on correlation of clinical, morphological, and molecular features. Histopathology 2007, 50, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef] [PubMed]
- Bettington, M.; Walker, N.; Clouston, A.; Brown, I.; Leggett, B.; Whitehall, V. The serrated pathway to colorectal carcinoma: Current concepts and challenges. Histopathology 2013, 62, 367–386. [Google Scholar] [CrossRef]
- Chang, K.; Willis, J.A.; Reumers, J.; Taggart, M.W.; San Lucas, F.A.; Thirumurthi, S.; Kanth, P.; Delker, D.A.; Hagedorn, C.H.; Lynch, P.M.; et al. Colorectal premalignancy is associated with consensus molecular subtypes 1 and 2. Ann. Oncol. 2018, 29, 2061–2067. [Google Scholar] [CrossRef]
- Toyota, M.; Ahuja, N.; Ohe-Toyota, M.; Herman, J.G.; Baylin, S.B.; Issa, J.P.J. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. USA 1999, 96, 8681–8686. [Google Scholar] [CrossRef]
- Bird, A.P. CpG-rich islands and the function of DNA methylation. Nature 1986, 321, 209–213. [Google Scholar] [CrossRef]
- Delattre, J.F.; Selcen Oguz Erdogan, A.; Cohen, R.; Shi, Q.; Emile, J.F.; Taieb, J.; Tabernero, J.; André, T.; Meyerhardt, J.A.; Nagtegaal, I.D.; et al. A comprehensive overview of tumor deposits in colorectal cancer: Towards a next TNM classification. Cancer Treat. Rev. 2022, 103, 102325. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aranda Aguilar, E.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef]
- Kormi, S.M.A.; Ardehkhani, S.; Kerachian, M.A. New insights into colorectal cancer screening and early detection tests. Color. Cancer 2017, 6, 63–68. [Google Scholar] [CrossRef]
- Hwang, T.J.; Lehmann, L.S.; Kesselheim, A.S. Precision medicine and the FDA’s draft guidance on laboratory-developed tests. Nat. Biotechnol. 2015, 33, 449–451. [Google Scholar] [CrossRef]
- Johdi, N.A.; Sukor, N.F. Colorectal Cancer Immunotherapy: Options and Strategies. Front. Immunol. 2020, 11, 1624. [Google Scholar] [CrossRef]
- Park, S.C.; Sohn, D.K.; Kim, M.J.; Chang, H.J.; Chang, H.J.; Han, K.S.; Hyun, J.H.; Joo, J.; Oh, J.H. Phase II Clinical Trial to Evaluate the Efficacy of Transanal Endoscopic Total Mesorectal Excision for Rectal Cancer. Dis. Colon Rectum 2018, 61, 554–560. [Google Scholar] [CrossRef]
- Ogura, A.; Konishi, T.; Cunningham, C.; Garcia-Aguilar, J.; Iversen, H.; Toda, S.; Lee, I.K.; Lee, H.X.; Uehara, K.; Lee, P.; et al. Neoadjuvant (Chemo)radiotherapy With Total Mesorectal Excision Only Is Not Sufficient to Prevent Lateral Local Recurrence in Enlarged Nodes: Results of the Multicenter Lateral Node Study of Patients with Low cT3/4 Rectal Cancer. J. Clin. Oncol. 2019, 37, 33–43. [Google Scholar] [CrossRef]
- Kulka, U.; Schaffer, M.; Siefert, A.; Schaffer, P.M.; Olsner, A.; Kasseb, K.; Hofstetter, A.; Duhmke, E.; Jori, G. Photofrin as a radiosensitizer in an in vitro cell survival assay. Biochem. Biography. Jt. Res. 2003, 311, 98–103. [Google Scholar] [CrossRef]
- Kuipers, E.J.; Grady, W.M.; Lieberman, D.; Seufferlein, T.; Sung, J.J.; Boelens, P.G.; van de Velde, C.J.; Watanabe, T. Colorectal cancer. Nat. Rev. Dis. Primers 2015, 1, 15065. [Google Scholar] [CrossRef]
- Duschinsky, R.; Pleven, E.; Heidelberger, C. The Synthesis of 5-Fluoropyrimidines. J. Am. Chem. Soc. 1957, 79, 4559–4560. [Google Scholar] [CrossRef]
- Kroemer, G.; Galluzzi, L.; Kepp, O.; Zitvogel, L. Immunogenic Cell Death in Cancer Therapy. Annu. Rev. Immunol. 2013, 31, 51–72. [Google Scholar] [CrossRef]
- Grothey, A.; Sargent, D.; Goldberg, R.M.; Schmoll, H.J. Survival of Patients with Advanced Colorectal Cancer Improves with the Availability of Fluorouracil-Leucovorin, Irinotecan, and Oxaliplatin in the Course of Treatment. J. Clin. Oncol. 2004, 22, 1209–1214. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Jiang, T.; Xiao, Y.; Wang, Q.; Zeng, Z.; Cai, P.; Zhao, Y.; Zhao, Z.; Wu, D.; Lin, H.; et al. Good Tumor Response to Chemoradioimmunotherapy in dMMR/MSI-H Advanced Colorectal Cancer: A Case Series. Front. Immunol. 2021, 12, 784336. [Google Scholar] [CrossRef] [PubMed]
- Brenner, H.; Kloor, M.; Pox, C.P. Colorectal cancer. Lancet 2014, 383, 1490–1502. [Google Scholar] [CrossRef] [PubMed]
- Ciardiello, F.; Tortora, G. EGFR Antagonists in Cancer Treatment. N. Engl. J. Med. 2008, 358, 1160–1174. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Pei, L.; Xia, H.; Tang, Q.; Bi, F. Role of oncogenic KRAS in the prognosis, diagnosis and treatment of colorectal cancer. Mol. Cancer 2021, 20, 143. [Google Scholar] [CrossRef]
- Chan, D.L.H.; Segelov, E.; Wong, R.S.; Smith, A.; Herbertson, R.A.; Li, B.T.; Tebbutt, N.; Price, T.; Pavlakis, N. Epidermal growth factor receptor (EGFR) inhibitors for metastatic colorectal cancer. Cochrane Database Syst. Rev. 2017, 6, CD007047. [Google Scholar] [CrossRef]
- Halder, S.; Basu, S.; Lall, S.P.; Ganti, A.K.; Batra, S.K.; Seshacharyulu, P. Targeting the EGFR signaling pathway in cancer therapy: What’s new in 2023? Expert Opin. Ther. Targets 2023, 27, 305–324. [Google Scholar] [CrossRef]
- Dias Carvalho, P.; Guimarães, C.F.; Cardoso, A.P.; Mendonça, S.; Costa, Â.M.; Oliveira, M.J.; Velho, S. KRAS Oncogenic Signaling Extends beyond Cancer Cells to Orchestrate the Microenvironment. Cancer Res. 2018, 78, 7–14. [Google Scholar] [CrossRef]
- Nassar, A.H.; Adib, E.; Kwiatkowski, D.J. Distribution of KRAS G12C Somatic Mutations across Race, Sex, and Cancer Type. N. Engl. J. Med. 2021, 384, 185–187. [Google Scholar] [CrossRef]
- Ciombor, K.K.; Strickler, J.H.; Bekaii-Saab, T.S.; Yaeger, R. BRAF-Mutated Advanced Colorectal Cancer: A Rapidly Changing Therapeutic Landscape. J. Clin. Oncol. 2022, 40, 2706–2715. [Google Scholar] [CrossRef]
- Cohen, R.; Liu, H.; Fiskum, J.; Adams, R.; Chibaudel, B.; Maughan, T.S.; Van Cutsem, E.; Venook, A.; Douillard, J.Y.; Heinemann, V.; et al. BRAF V600E Mutation in First-Line Metastatic Colorectal Cancer: An Analysis of Individual Patient Data from the ARCAD Database. JNCI J. Natl. Cancer Inst. 2021, 113, 1386–1395. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Niida, A.; Uchi, R.; Hirata, H.; Komatsu, H.; Sakimura, S.; Hayashi, S.; Nambara, S.; Kuroda, Y.; Ito, S.; et al. A temporal shift of the evolutionary principle shaping intratumor heterogeneity in colorectal cancer. Nat. Commun. 2018, 9, 2884. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, H.; Fehrenbacher, L.; Novotny, W.; Cartwright, T.; Hainsworth, J.; Heim, W.; Berlin, J.; Baron, A.; Griffing, S.; Holmgren, E.; et al. Bevacizumab plus Irinotecan, Fluorouracil, and Leucovorin for Metastatic Colorectal Cancer. N. Engl. J. Med. 2004, 350, 2335–2342. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, J.; Clarke, S.; Díaz-Rubio, E.; Scheithauer, W.; Figer, A.; Wong, R.; Koski, S.; Rittweger, K.; Gilberg, F.; Saltz, L. XELOX vs. FOLFOX-4 as first-line therapy for metastatic colorectal cancer: NO16966 updated results. Br. J. Cancer 2011, 105, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Kemeny, N.E.; Jarnagin, W.R.; Capanu, M.; Fong, Y.; Gewirtz, A.N.; DeMatteo, R.P.; D’Angelica, M.I. Randomized Phase II Trial of Adjuvant Hepatic Arterial Arterial Infusion and Systemic Chemotherapy with or without Bevacizumab in Patients with Resected Hepatic Metastases from Colorectal Cancer. J. Clin. Oncol. 2011, 29, 884–889. [Google Scholar] [CrossRef]
- Giantonio, B.J.; Catalano, P.J.; Meropol, N.J.; O’Dwyer, P.J.; Mitchell, E.P.; Alberts, S.R.; Schwartz, M.A.; Benson, A.B., 3rd. Eastern Cooperative Oncology Group Study E3200. Bevacizumab in Combination with Oxaliplatin, Fluorouracil, and Leucovorin (FOLFOX4) for Previously Treated Metastatic Colorectal Cancer: Results from the Eastern Cooperative Oncology Group Study E3200. J. Clin. Oncol. 2007, 25, 1539–1544. [Google Scholar] [CrossRef]
- Tang, P.A.; Cohen, S.J.; Kollmannsberger, C.; Bjarnason, G.; Virik, K.; MacKenzie, M.J.; Lourenco, L.; Wang, L.; Chen, A.; Moore, M.J. Phase II Clinical and Pharmacokinetic Study of Aflibercept in Patients with Previously Treated Metastatic Colorectal Cancer. Clin. Cancer Res. 2012, 18, 6023–6031. [Google Scholar] [CrossRef]
- Piawah, S.; Venook, A.P. Targeted therapy for colorectal cancer metastases: A review of current methods of molecularly targeted therapy and the use of tumor biomarkers in the treatment of metastatic colorectal cancer. Cancer 2019, 125, 4139–4147. [Google Scholar] [CrossRef]
- Yarom, N.; Jonker, D.J. The role of the epidermal growth factor receptor in the mechanism and treatment of colorectal cancer. Discov. Med. 2011, 11, 95–105. [Google Scholar]
- Sharma, P.; Allison, J.P. Immune Checkpoint Targeting in Cancer Therapy: Toward Combination Strategies with Curative Potential. Cell 2015, 161, 205–214. [Google Scholar] [CrossRef]
- Parseghian, C.M.; Napolitano, S.; Loree, J.M.; Kopetz, S. Mechanisms of Innate and Acquired Resistance to Anti-EGFR Therapy: A Review of Current Knowledge with a Focus on Rechallenge Therapies. Clin. Cancer Res. 2019, 25, 6899–6908. [Google Scholar] [CrossRef] [PubMed]
- Biller, L.H.; Schrag, D. Diagnosis and Treatment of Metastatic Colorectal Cancer. JAMA 2021, 325, 669. [Google Scholar] [CrossRef] [PubMed]
- Delord, J.P.; Robert, C.; Nyakas, M.; McArthur, G.A.; Kudchakar, R.; Mahipal, A.; Yamada, Y.; Sullivan, R.; Arance, A.; Kefford, R.F.; et al. Phase I Dose—Escalation and Expansion Study of the BRAF Inhibitor Encorafenib (LGX818) in Metastatic BRAF—Mutant Melanoma. Clin. Cancer Res. 2017, 23, 5339–5348. [Google Scholar] [CrossRef]
- Franke, A.J.; Skelton, W.P.; Starr, J.S.; Parekh, H.; Lee, J.J.; Overman, M.J.; Allegra, C.; George, T.J. Immunotherapy for Colorectal Cancer: A Review of Current and Novel Therapeutic Approaches. JNCI J. Natl. Cancer Inst. 2019, 111, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Pardollm, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Saunders, P.A.; Hendrycks, V.R.; Lidinsky, W.A.; Woods, M.L. PD-L2:PD-1 involvement in T cell proliferation, cytokine production, and integrin-mediated adhesion. Eur. J. Immunol. 2005, 35, 3561–3569. [Google Scholar] [CrossRef]
- Price, T.J.; Thavaneswaran, S.; Burge, M.; Segelov, E.; Haller, D.G.; Punt, C.J.; Arnold, D.; Karapetis, C.S.; Tebbutt, N.C.; Pavlakis, N.; et al. Update on optimal treatment for metastatic colorectal cancer from the ACTG/AGITG expert meeting: ECCO 2015. Expert Rev. Anticancer Ther. 2016, 16, 557–571. [Google Scholar] [CrossRef]
- Duan, F.; Duitama, J.; Al Seesi, S.; Ayres, C.M.; Corcelli, S.A.; Pawashe, A.P.; Blanchard, T.; McMahon, D.; Sidney, J.; Sette, A.; et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J. Exp. Med. 2014, 211, 2231–2248. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef]
- Overman, M.J.; Lonardi, S.; Wong, K.Y.M.; Lenz, H.J.; Gelsomino, F.; Aglietta, M.; Morse, M.A.; Van Cutsem, E.; McDermott, R.; Hill, A.; et al. Durable Clinical Benefit with Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. J. Clin. Oncol. 2018, 36, 773–779. [Google Scholar] [CrossRef]
- Segal, N.H.; Saltz, L.B. Translational Considerations on the Outlook of Immunotherapy for Colorectal Cancer. Curr. Colorectal Cancer Rep. 2015, 11, 92–97. [Google Scholar] [CrossRef]
- Bertuzzi, M.; Marelli, C.; Bagnati, R.; Colombi, A.; Fanelli, R.; Saieva, C.; Ceroti, M.; Bendinelli, B.; Caini, S.; Airoldi, L.; et al. Plasma clusterin as a candidate prediagnostic marker of colorectal cancer risk in the Florence cohort of the European Prospective Investigation into Cancer and Nutrition: A pilot study. BMC Cancer 2015, 15, 56. [Google Scholar] [CrossRef]
- Artemaki, P.I.; Sklirou, A.D.; Kontos, C.K.; Liosi, A.A.; Gianniou, D.D.; Papadopoulos, I.N.; Trougakos, I.P.; Scorilas, A. High levels of clusterin mRNA (CLU) expression in tumors of colorectal cancer patients predict poor prognostic outcome. Clin. Biochem. 2020, 75, 62–69. [Google Scholar] [CrossRef]
- Kopylov, A.T.; Stepanov, A.A.; Malsagova, K.A.; Soni, D.; Kushlinsky, N.E.; Enikeev, D.V.; Potoldykova, N.V.; Lisitsa, A.V.; Kaysheva, A.L. Unveiling proteomic markers for early-stage colorectal cancer. Molecules 2020, 25, 619. [Google Scholar] [CrossRef]
- Pucci, S.; Mazzarelli, P.; Rabitti, C.; Giai, M.; Gallucci, M.; Flammia, G.; Alcini, A.; Altomare, V.; Fazio, V.M. Tumor specific modulation of Ku70/80 DNA binding activity in breast and bladder human tumor biopsies. Oncogene 2001, 20, 739–747. [Google Scholar] [CrossRef]
- Komuro, Y.; Watanabe, T.; Hosoi, Y.; Matsumoto, Y.; Nakagawa, K.; Tsuno, N.; Kazama, S.; Kitayama, J.; Suzuki, N.; Nagawa, H. The expression pattern of Ku correlates with tumor radiosensitivity and disease-free survival in patients with rectal carcinoma. Cancer 2002, 95, 1199–1205. [Google Scholar] [CrossRef]
- Komuro, Y.; Watanabe, T.; Hosoi, Y.; Matsumoto, Y.; Nakagawa, K.; Saito, S.; Ishihara, S.; Kazama, S.; Tsuno, N.; Kitayama, J.; et al. Prediction of tumor radiosensitivity in rectal carcinoma based on p53 and Ku70 expression. J. Exp. Clin. Cancer Res. 2003, 22, 223–228. [Google Scholar]
- Mazzarelli, P.; Parrella, P.; Seripa, D.; Signori, E.; Perrone, G.; Rabitti, C.; Borzomati, D.; Gabbrielli, A.; Matera, M.G.; Gravina, C.; et al. DNA end binding activity and Ku70/80 heterodimer expression in human colorectal tumor. World J. Gastroenterol. 2005, 11, 6694–6700. [Google Scholar] [CrossRef]
- Pucci, S.; Polidoro, C.; Joubert, A.; Mastrangeli, F.; Tolu, B.; Benassi, M.; Fiaschetti, V.; Greco, L.; Miceli, R.; Floris, R.; et al. Ku70, Ku80, and sClusterin: A Cluster of Predicting Factors for Response to Neoadjuvant Chemoradiation Therapy in Patients with Locally Advanced Rectal Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2017, 97, 381–388. [Google Scholar] [CrossRef]
- Kevans, D.; Gorman, S.; Tosetto, M.; Sheahan, K.; O’Donoghue, D.; Mulcahy, H.; O’Sullivan, J. Clusterin and chemotherapy sensitivity under normoxic and graded hypoxic conditions in colorectal cancer. J. Gastrointest. Cancer 2012, 43, 305–313. [Google Scholar] [CrossRef]
- Engel, R.M.; Chan, W.H.; Nickless, D.; Hlavca, S.; Richards, E.; Kerr, G.; Oliva, K.; McMurrick, P.J.; Jardé, T.; Abud, H.E. Patient-Derived Colorectal Cancer Organoids Upregulate Revival Stem Cell Marker Genes following Chemotherapeutic Treatment. J. Clin. Med. 2020, 9, 128. [Google Scholar] [CrossRef]
- Grosgeorges, M.; Picque Lasorsa, L.; Pastor, B.; Prévostel, C.; Crapez, E.; Sanchez, C.; Frayssinoux, F.; Jarlier, M.; Pezzella, V.; Monard, L.; et al. A straightforward method to quantify circulating mRNAs as biomarkers of colorectal cancer. Sci. Rep. 2023, 13, 2739. [Google Scholar] [CrossRef]
- Barbazán, J.; Muinelo-Romay, L.; Vieito, M.; Candamio, S.; Díaz-López, A.; Cano, A.; Gómez-Tato, A.; de los Ángeles Casares de Cal, M.; Abal, M.; López-López, R. A multimarker panel for circulating tumor cells detection predicts patient outcome and therapy response in metastatic colorectal cancer. Int. J. Cancer. 2014, 135, 2633–2643. [Google Scholar] [CrossRef]
- Insua, Y.V.; Cámara, J.; Vázquez, E.B.; Fernández, A.; Rivera, F.V.; Silva, M.J.V.V.; Barbazán, J.; Muinelo-Romay, L.; Folgar, S.C.; Abalo, A.; et al. Predicting Outcome and Therapy Response in mCRC Patients Using an Indirect Method for CTCs Detection by a Multigene Expression Panel: A Multicentric Prospective Validation Study. Int. J. Mol. Sci. 2017, 18, 1265. [Google Scholar] [CrossRef]
- Du, X.; Qi, H.; Ji, W.; Li, P.; Hua, R.; Hu, W.; Qi, F. Construction of a Colorectal Cancer Prognostic Risk Model and Screening of Prognostic Risk Genes Using Machine-Learning Algorithms. Comput. Math Methods Med. 2022, 2022, 9408839. [Google Scholar] [CrossRef]
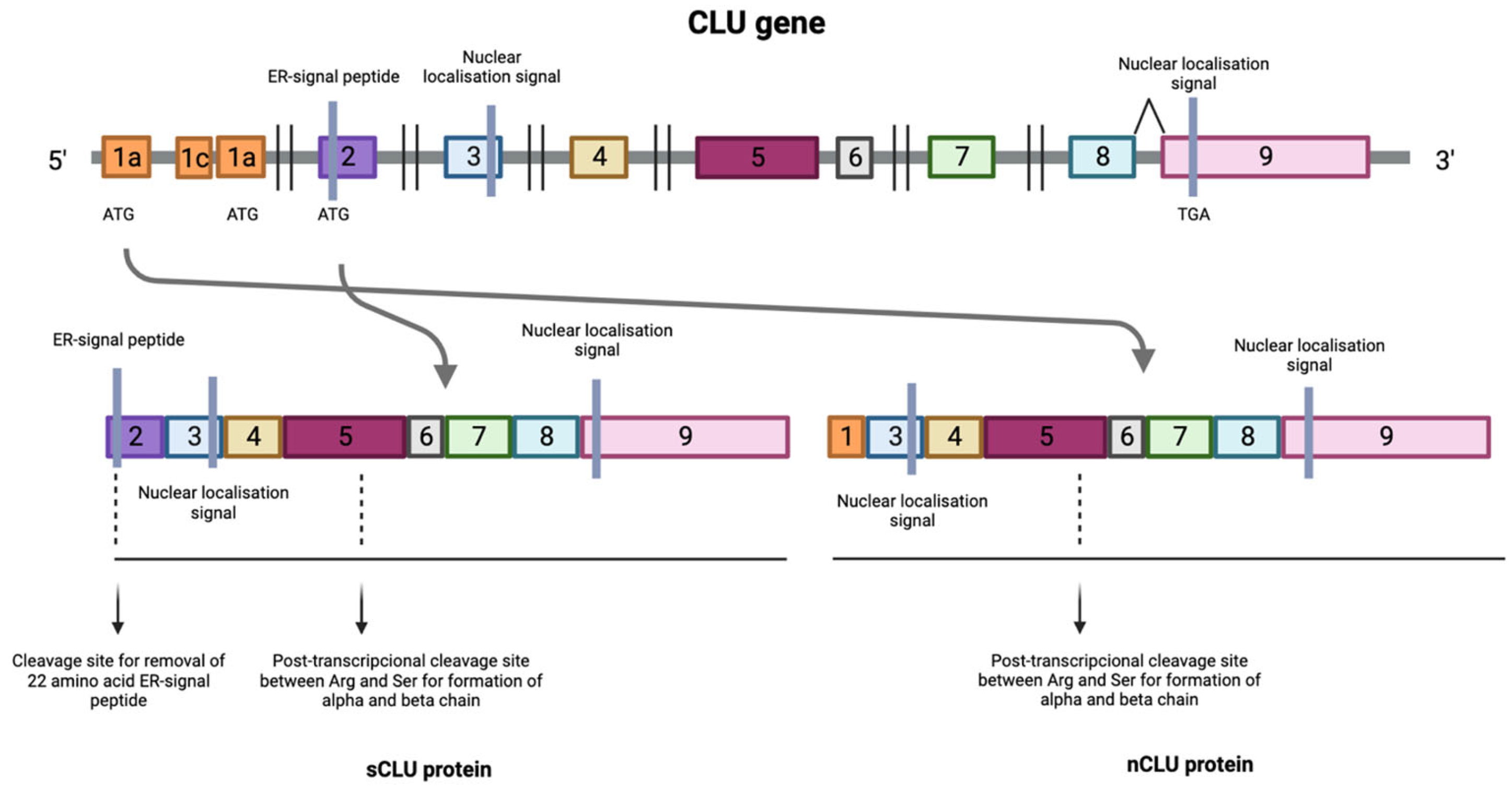

| Cancer Type | Results |
|---|---|
| Lung | More than 80% of non-small-cell lung cancers are immunoreactive for clusterin [34]. |
| Clustering silencing using antisense oligonucleotides (ASOs), or short interfering double-stranded RNAs (siRNAs) significantly improve sensitivity to paclitaxel in vivo [34]. | |
| Gastric | sCLU overexpression is involved in the progression of human gastric carcinomas and it seems that its oncogenic function could be associated with p53 disfunction [35]. |
| Ovary | Clusterin overexpression is more frequently detected in metastatic lesions more frequently than in primary tumors [36]. |
| Cytoplasmatic clusterin overexpression in ovary carcinomas is inversely correlated with tumor apoptotic index [36]. | |
| Breast | Unlike benign lesions, atypical hyperplasias, intraductal carcinomas and invasive carcinomas are characterize by clusterin overexpression [37].Clusterin expression tended to be inversely correlated with the apoptotic rate, indicating that gene expression is not required for apoptotic cell death. This suggests that CLU may play a role in breast carcinogenesis [37,38]. |
| Colon | sCLU is overexpressed while nCLU is downregulated [39]. |
| Clusterin expression may help identify patients with more aggressive tumors who may benefit from targeted therapies [40]. | |
| Bladder | The recurrence-free survival rate of patients with strong clusterin expression is significantly lower than in patients with weak expression [41]. |
| Hepatic | CLU expression in peripheral blood mononuclear cells (PBMC) has been proposed as a prospective screening marker for hepatocellular carcinoma along with other genes [42]. |
| Pancreas | Clusterin is highly expressed in stages I and II (well-differentiated and moderately differentiated cancers) [43] and it expression is not significantly associated with apoptosis [43]. |
| Clusterin-positive patients with pancreatic cancer survived significantly longer [43]. | |
| Melanoma | Clusterin overexpression is associated with increased drug resistance and increased survival of tumor cells [44], and its silencing reduces drug resistance and survival of melanoma cells both in vitro and in vivo [44]. Mice pretreatment with antisense oligonucleotide targeting CLU is associated with a significantly improved tumor response to dacarbazine compared to control [44]. |
| Esophageal squamous cells | High stromal CLU expression is associated with poor locoregional, overall and distant progression-free survival [45]. |
| Esophageal Squamous Cancer Cell patients with high CLU expression in both epithelium and stroma have the shortest survival time [45]. | |
| Anaplastic large cell lymphomas | Clusterin expression is not related to anaplastic lymphoma kinase-1 expression and in reactive lymphoid tissues, only fibroblastic reticular and follicular dendritic cells show positive expression by inmunohistochemistry [46]. |
| Although CLU role in anaplastic large cell lymphoma (ALCL) is unknown, the unique expression of clusterin within this category of lymphoma provides an additional marker for the diagnosis of ALCL [46]. | |
| Osteosarcomas | sCLU is expressed in osteosarcoma and its overexpression is associated with metastasis and chemoresistance [47]. |
| Silencing sCLU inhibits metastasis and improves chemosensitivity in osteosarcoma cells [47]. | |
| Prostate | A significant decrease in clusterin expression compared to corresponding benign tissues has been observed [23] and reduction in hormone-resistant prostate carcinomas [48]. |
| Type | Molecular Target | Immunotherapeutics | Type of Patient |
|---|---|---|---|
| Monoclonal antibodies | VEGF-A | Bevacizumab | CRCm |
| VEGF-A | Ziv-aflibercept | ||
| Extracellular domain of VEGF | Ramucirumab | ||
| Extracellular domain of EGFR | Cetuximab | CRCm with RAS WT tumors | |
| Panitumumad | |||
| BRAF | Encorafenib | CRCm with RAS WT tumors | |
| Immune checkpoint inhibitors | PD-1 | Pembrolizumab | MSI-H/dMMR |
| PD-1 | Nivolumab | CRCm | |
| CTLA4 | Ipilimubab | MSI-H/dMMR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Téllez, T.; Martin-García, D.; Redondo, M.; García-Aranda, M. Clusterin Expression in Colorectal Carcinomas. Int. J. Mol. Sci. 2023, 24, 14641. https://doi.org/10.3390/ijms241914641
Téllez T, Martin-García D, Redondo M, García-Aranda M. Clusterin Expression in Colorectal Carcinomas. International Journal of Molecular Sciences. 2023; 24(19):14641. https://doi.org/10.3390/ijms241914641
Chicago/Turabian StyleTéllez, Teresa, Desirée Martin-García, Maximino Redondo, and Marilina García-Aranda. 2023. "Clusterin Expression in Colorectal Carcinomas" International Journal of Molecular Sciences 24, no. 19: 14641. https://doi.org/10.3390/ijms241914641






